Gymnema sylvestre Extract Restores the Autophagic Pathway in Human Glioblastoma Cells U87Mg
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
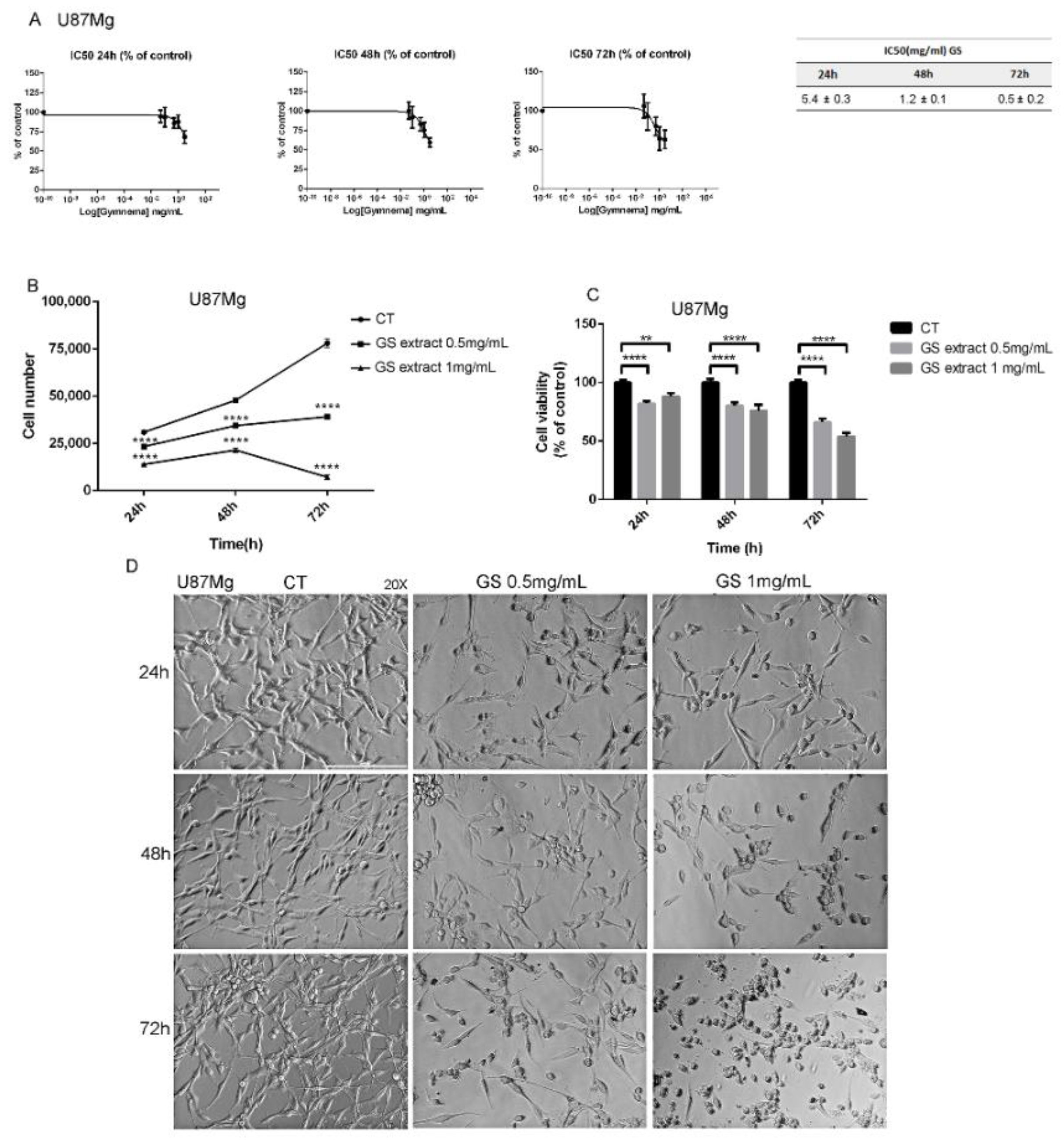
2.2. Evaluation of Half-Maximal Inhibitory Concentration (IC50) of GS Extract
2.3. Treatment of Human GBM Cell Line U87Mg with GS Extract
2.4. Cell Viability Assay
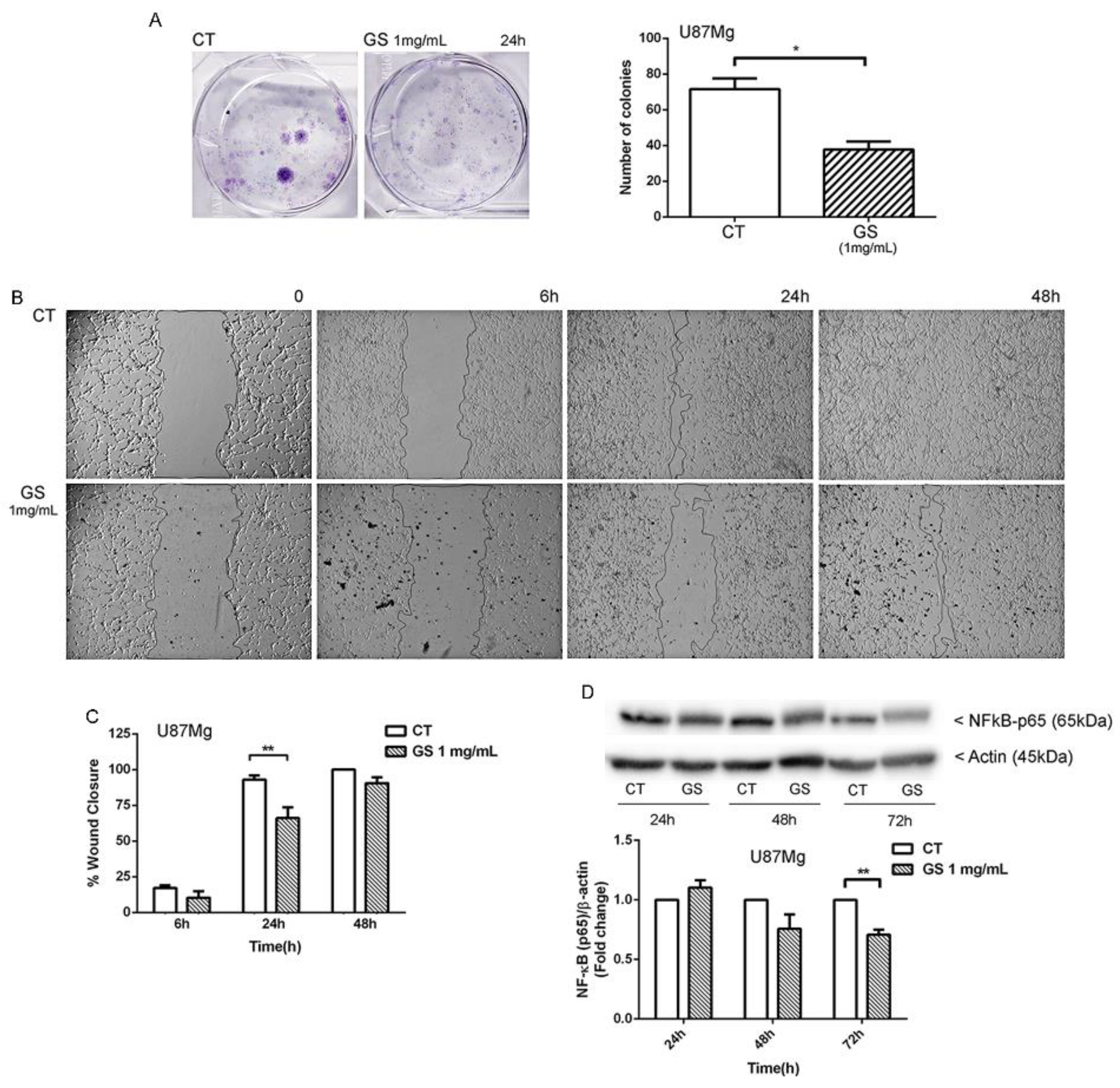
2.5. Clonogenic Assay
2.6. Wound-Healing Assay
2.7. Western Blot Analysis of U87Mg Cells Treated with GS Extract
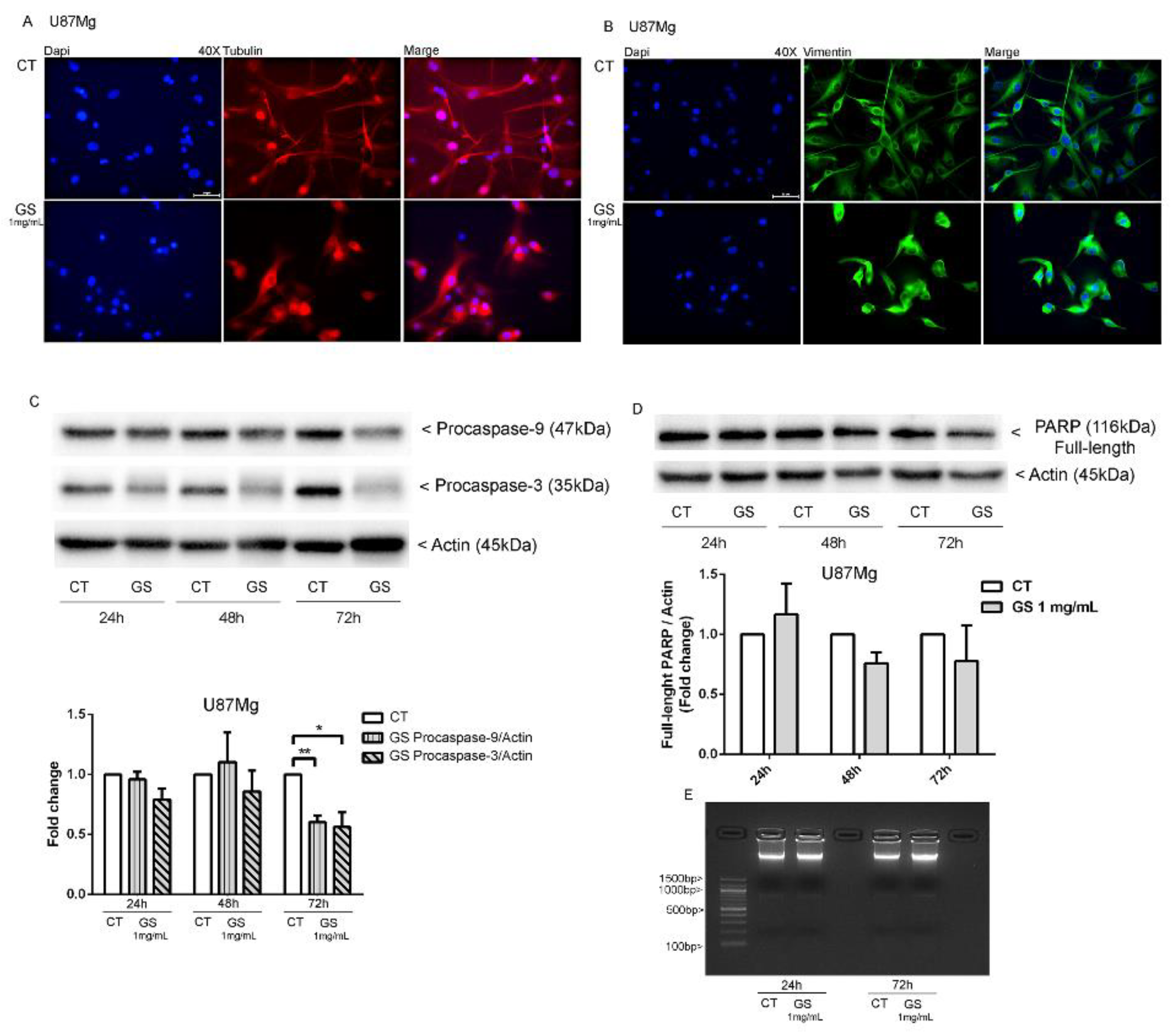
2.8. Western Blot Analysis of Apoptotic Pathway in U87Mg Cells Treated with GS Extract
2.9. DNA Ladder
2.10. Immunofluorescence Detection of Cytoskeletal Proteins in GS-Treated U87Mg Cells
2.11. Immunofluorescence Detection of LC3-Puncta under Bafilomycin Treatment
2.12. Statistical Analysis
3. Results
3.1. Effects of GS Extract on Glioblastoma Cells U87Mg
3.2. Clonogenic Survival and Motility Capacity of GBM Cells after GS Extract Treatment through Potentially Inhibition of NF-κB
3.3. Dynamic Rearrangement of Cytoskeletal Proteins in GS-Treated U87Mg Cells
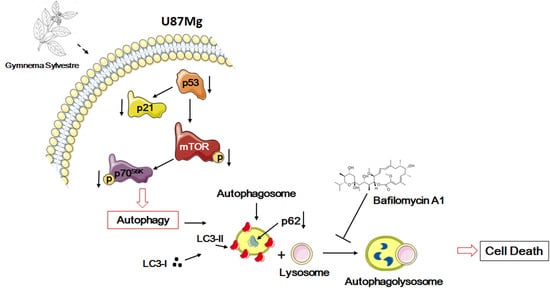
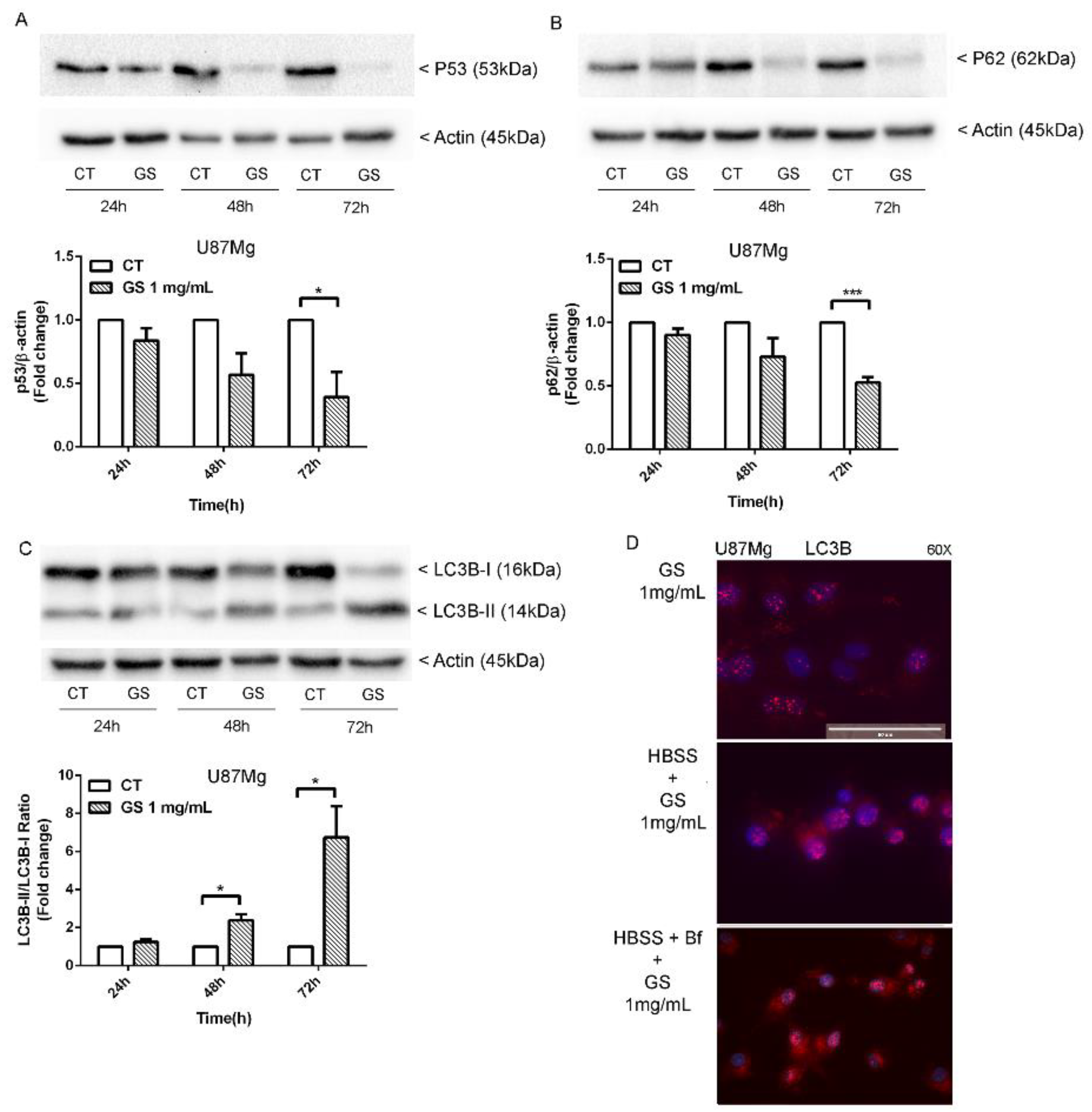
3.4. Analysis of Autophagy Markers in GS-Induced Cytotoxicity of Glioblastoma Cells U87Mg
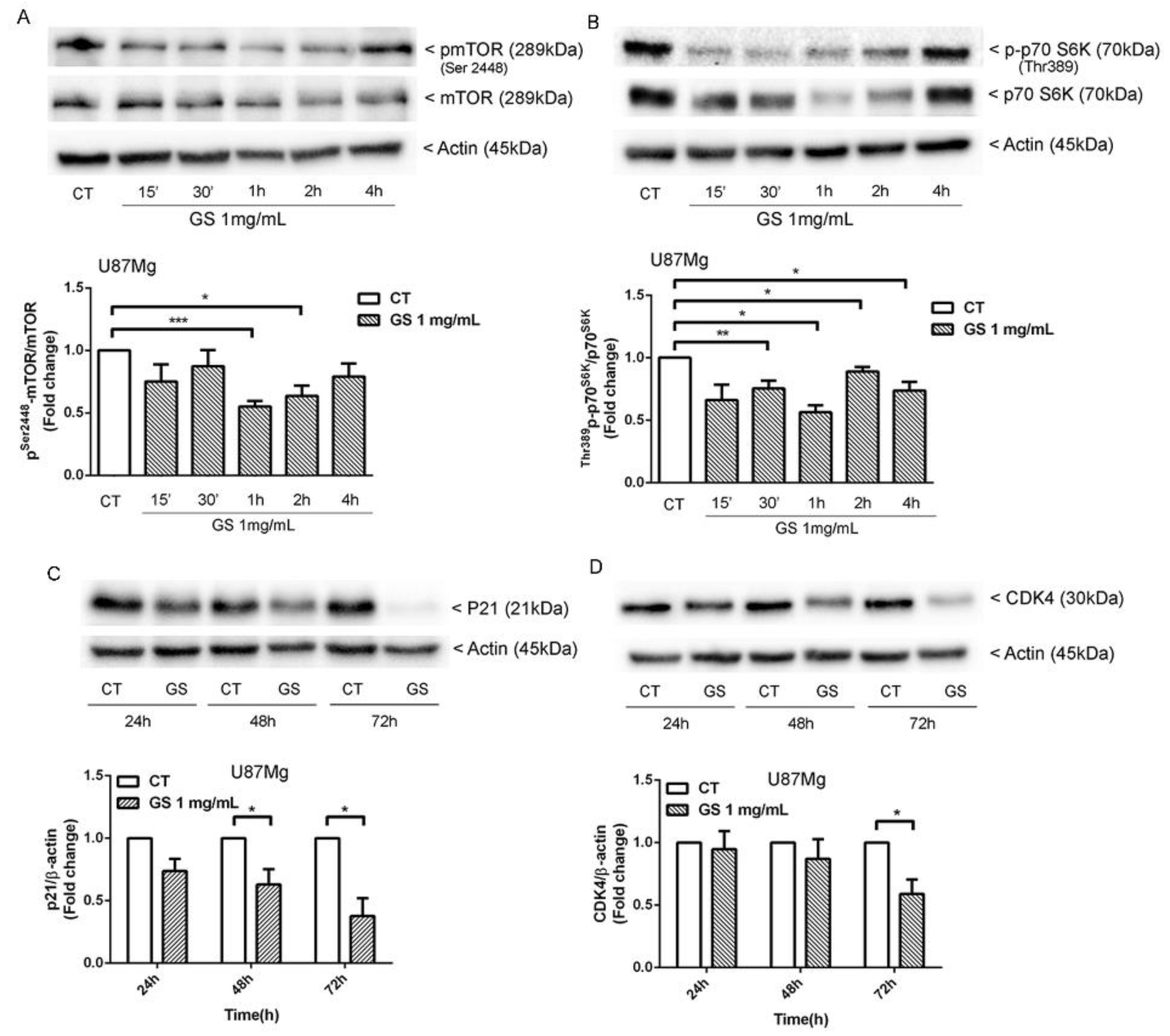
3.5. Inhbition of p21WAF1/CIP1 Expression and CDK4-Induced Macroautophagy in GS-Treated Glioblastoma Cells U87Mg
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Surawicz, T.S.; Davis, F.; Freels, S.; Laws, E.R., Jr.; Menck, H.R. Brain tumor survival: Results from the National Cancer Data Base. J. Neurooncol. 1998, 40, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Friedman, H.S.; Kerby, T.; Calvert, H. Temozolomide and treatment of malignant glioma. Clin. Cancer Res. 2000, 6, 2585–2597. [Google Scholar] [PubMed]
- Xia, Q.; Xu, M.; Zhang, P.; Liu, L.; Meng, X.; Dong, L. Therapeutic Potential of Autophagy in Glioblastoma Treatment With Phosphoinositide 3-Kinase/Protein Kinase B/Mammalian Target of Rapamycin Signaling Pathway Inhibitors. Front. Oncol. 2000, 10, 572904. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, W.; Qin, Z.; Liang, Z. The role of autophagy in sensitizing malignant glioma cells to radiation therapy. Acta Biochim. Biophys. Sin. (Shanghai) 2009, 41, 341–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefranc, F.; Facchini, V.; Kiss, R. Proautophagic drugs: A novel means to combat apoptosis-resistant cancers, with a special emphasis on glioblastomas. Oncologist 2007, 12, 1395–1403. [Google Scholar] [CrossRef]
- Galan-Moya, E.M.; Le Guelte, A.; Lima-Fernandes, E.; Thirant, C.; Dwyer, J.; Bidere, N.; Couraud, P.-O.; Scott, M.G.H.; Junier, M.-P.; Chneiweiss, H.; et al. Secreted factors from brain endothelial cells maintain glioblastoma stem-like cell expansion through the mTOR pathway. EMBO Rep. 2011, 12, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Arcella, A.; Biagioni, F.; Antonietta Oliva, M.; Bucci, D.; Frati, A.; Esposito, V.; Cantore, G.; Giangaspero, F.; Fornai, F. Rapamycin inhibits the growth of glioblastoma. Brain Res. 2013, 1495, 37–51. [Google Scholar] [CrossRef]
- Miracco, C.; Cosci, E.; Oliveri, G.; Luzi, P.; Pacenti, L.; Monciatti, I.; Mannucci, S.; De Nisi, M.C.; Toscano, M.; Malagnino, V.; et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. Int. J. Oncol. 2007, 30, 429–436. [Google Scholar]
- Huang, X.; Bai, H.M.; Chen, L.; Li, B.; Lu, Y.C. Reduced expression of LC3B-II and Beclin 1 in glioblastoma multiforme indicates a down-regulated autophagic capacity that relates to the progression of astrocytic tumors. J. Clin. Neurosci. 2010, 17, 1515–1519. [Google Scholar] [CrossRef]
- Escamilla-Ramarez, A.; Castillo-Rodraguez, R.A.; Zavala-Vega, S.; Jimenez-Farfan, D.; Anaya-Rubio, I.; Briseño, E.; Palencia, G.; Guevara, P.; Cruz-Salgado, A.; Sotelo, J.; et al. Autophagy as a Potential Therapy for Malignant Glioma. Pharmaceuticals 2020, 13, 156. [Google Scholar] [CrossRef]
- Law, B.Y.; Chan, W.K.; Xu, S.W.; Wang, J.R.; Bai, L.P.; Liu, L.; Wong, V.K. Natural small-molecule enhancers of autophagy induce autophagic cell death in apoptosis-defective cells. Sci. Rep. 2014, 4, 5510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, D.; Ghosh, S.; Bishayee, K.; Mukherjee, A.; Sikdar, S.; Khuda-Bukhsh, A.R. Antihyperglycemic drug Gymnema sylvestre also shows anticancer potentials in human melanoma A375 cells via reactive oxygen species generation and mitochondria-dependent caspase pathway. Integr. Cancer Ther. 2013, 12, 433–441. [Google Scholar] [CrossRef]
- Arunachalam, K.D.; Arun, L.B.; Annamalai, S.K.; Arunachalam, A.M. Potential anticancer properties of bioactive compounds of Gymnema sylvestre and its biofunctionalized silver nanoparticles. Int. J. Nanomed. 2015, 10, 31–41. [Google Scholar] [CrossRef] [Green Version]
- Westhoff, M.A.; Zhou, S.; Nonnenmacher, L.; Karpel-Massler, G.; Jennewein, C.; Schneider, M.; Halatsch, M.E.; Carragher, N.O.; Baumann, B.; Krause, A.; et al. Inhibition of NF-kappaB signaling ablates the invasive phenotype of glioblastoma. Mol. Cancer Res. 2013, 11, 1611–1623. [Google Scholar] [CrossRef] [Green Version]
- Povea-Cabello, S.; Oropesa-Avila, M.; de la Cruz-Ojeda, P.; Villanueva-Paz, M.; de la Mata, M.; Suarez-Rivero, J.M.; Alvarez-Cordoba, M.; Villalon-Garcia, I.; Cotan, D.; Ybot-Gonzalez, P.; et al. Dynamic Reorganization of the Cytoskeleton during Apoptosis: The Two Coffins Hypothesis. Int. J. Mol. Sci. 2017, 18, 2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tasdemir, E.; Chiara Maiuri, M.; Morselli, E.; Criollo, A.; D’Amelio, M.; Djavaheri-Mergny, M.; Cecconi, F.; Tavernarakis, N.; Kroemer, G. A dual role of p53 in the control of autophagy. Autophagy 2008, 4, 810–814. [Google Scholar] [CrossRef] [Green Version]
- Tasdemir, E.; Maiuri, M.C.; Galluzzi, L.; Vitale, I.; Djavaheri-Mergny, M.; D’Amelio, M.; Criollo, A.; Morselli, E.; Zhu, C.; Harper, F.; et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008, 10, 676–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Liu, W.; Hao, H.; Wang, Q.; Xue, L. Rapamycin enhanced the antitumor effects of doxorubicin in myelogenous leukemia K562 cells by downregulating the mTOR/p70S6K pathway. Oncol. Lett. 2019, 18, 2694–2703. [Google Scholar] [CrossRef] [Green Version]
- Beuvink, I.; Boulay, A.; Fumagalli, S.; Zilbermann, F.; Ruetz, S.; O’Reilly, T.; Natt, F.; Hall, J.; Lane, H.A.; Thomas, G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell 2005, 120, 747–759. [Google Scholar] [CrossRef] [Green Version]
- Iriyama, N.; Hino, H.; Moriya, S.; Hiramoto, M.; Hatta, Y.; Takei, M.; Miyazawa, K. The cyclin-dependent kinase 4/6 inhibitor, abemaciclib, exerts dose-dependent cytostatic and cytocidal effects and induces autophagy in multiple myeloma cells. Leuk. Lymphoma 2017, 59, 1439–1450. [Google Scholar] [CrossRef]
- Sumi, N.J.; Kuenzi, B.M.; Knezevic, C.E.; Remsing Rix, L.L.; Rix, U. Chemoproteomics Reveals Novel Protein and Lipid Kinase Targets of Clinical CDK4/6 Inhibitors in Lung Cancer. ACS Chem. Biol. 2015, 10, 2680–2686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, K.L.; Ciacci, J.D.; Ammirati, M.; Selch, M.T.; Becker, D.P. Clinical trial of Serratia marcescens extract and radiation therapy in patients with malignant astrocytoma. J. Clin. Oncol. 1993, 11, 1746–1750. [Google Scholar] [CrossRef]
- Oehler, C.; Frei, K.; Rushing, E.J.; McSheehy, P.M.; Weber, D.; Allegrini, P.R.; Weniger, D.; Lutolf, U.M.; Knuth, A.; Yonekawa, Y.; et al. Patupilone (epothilone B) for recurrent glioblastoma: Clinical outcome and translational analysis of a single-institution phase I/II trial. Oncology 2012, 83, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Krakhmal, N.V.; Zavyalova, M.V.; Denisov, E.V.; Vtorushin, S.V.; Perelmuter, V.M. Cancer Invasion: Patterns and Mechanisms. Acta Nat. 2015, 7, 17–28. [Google Scholar] [CrossRef] [Green Version]
- Ryter, S.W.; Cloonan, S.M.; Choi, A.M. Autophagy: A critical regulator of cellular metabolism and homeostasis. Mol. Cells 2013, 36, 7–16. [Google Scholar] [CrossRef] [Green Version]
- Deng, D.; Luo, K.; Liu, H.; Nie, X.; Xue, L.; Wang, R.; Xu, Y.; Cui, J.; Shao, N.; Zhi, F. p62 acts as an oncogene and is targeted by miR-124-3p in glioma. Cancer Cell Int. 2019, 19, 280. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Fang, L.; Zhang, H.; Zhang, W.S.; Li, X.O.; Du, S.Y. Metformin Induces Autophagy via the AMPK-mTOR Signaling Pathway in Human Hepatocellular Carcinoma Cells. Cancer Manag. Res. 2020, 12, 5803–5811. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rotondo, R.; Castaldo, S.; Oliva, M.A.; Arcella, A. Gymnema sylvestre Extract Restores the Autophagic Pathway in Human Glioblastoma Cells U87Mg. Biology 2021, 10, 870. https://doi.org/10.3390/biology10090870
Rotondo R, Castaldo S, Oliva MA, Arcella A. Gymnema sylvestre Extract Restores the Autophagic Pathway in Human Glioblastoma Cells U87Mg. Biology. 2021; 10(9):870. https://doi.org/10.3390/biology10090870
Chicago/Turabian StyleRotondo, Rossella, Salvatore Castaldo, Maria Antonietta Oliva, and Antonietta Arcella. 2021. "Gymnema sylvestre Extract Restores the Autophagic Pathway in Human Glioblastoma Cells U87Mg" Biology 10, no. 9: 870. https://doi.org/10.3390/biology10090870
APA StyleRotondo, R., Castaldo, S., Oliva, M. A., & Arcella, A. (2021). Gymnema sylvestre Extract Restores the Autophagic Pathway in Human Glioblastoma Cells U87Mg. Biology, 10(9), 870. https://doi.org/10.3390/biology10090870