An Alignment-Independent Approach for the Study of Viral Sequence Diversity at Any Given Rank of Taxonomy Lineage
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
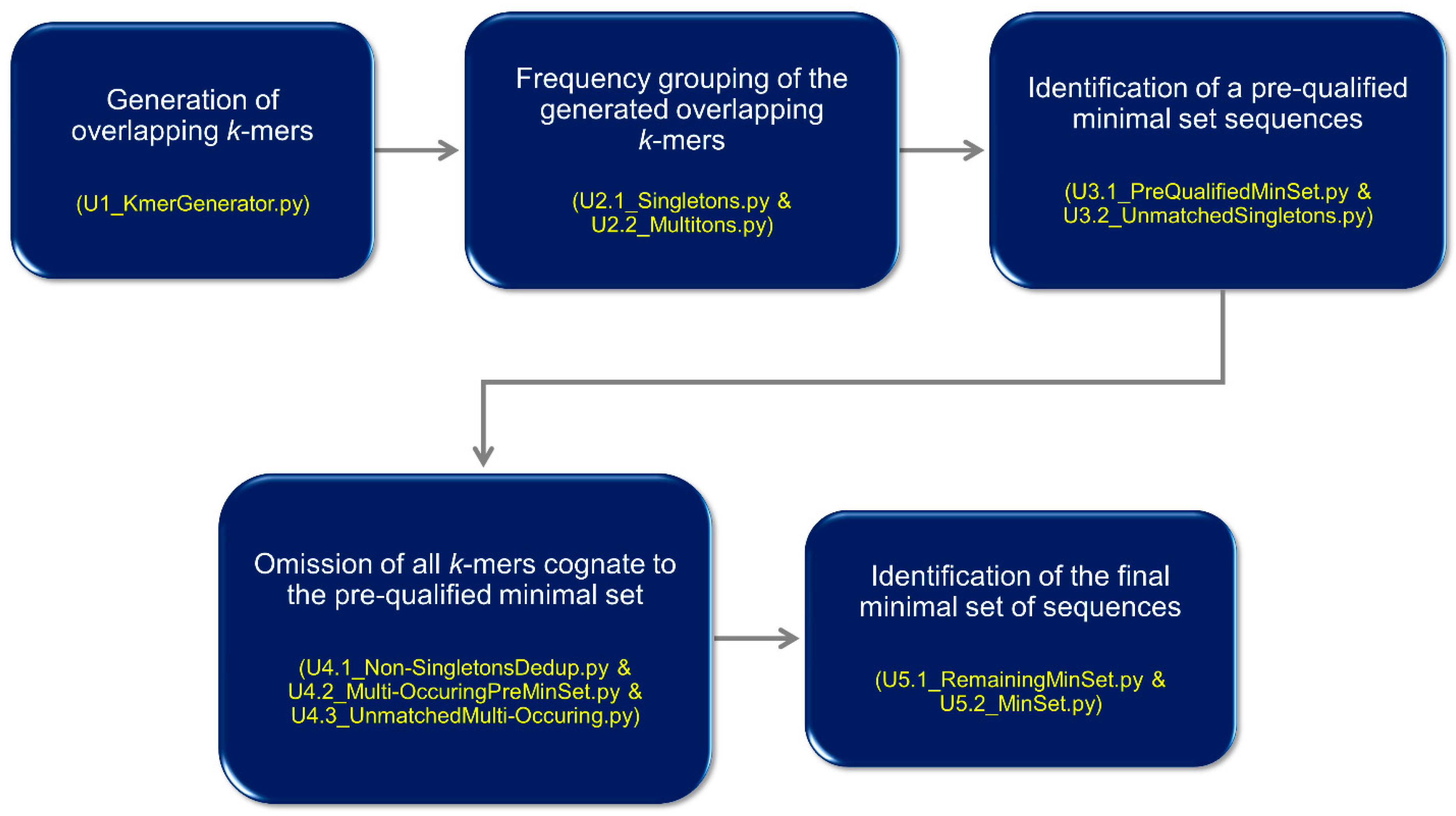
2.1. UNIQmin—Algorithm
2.2. Deployment of UNIQmin
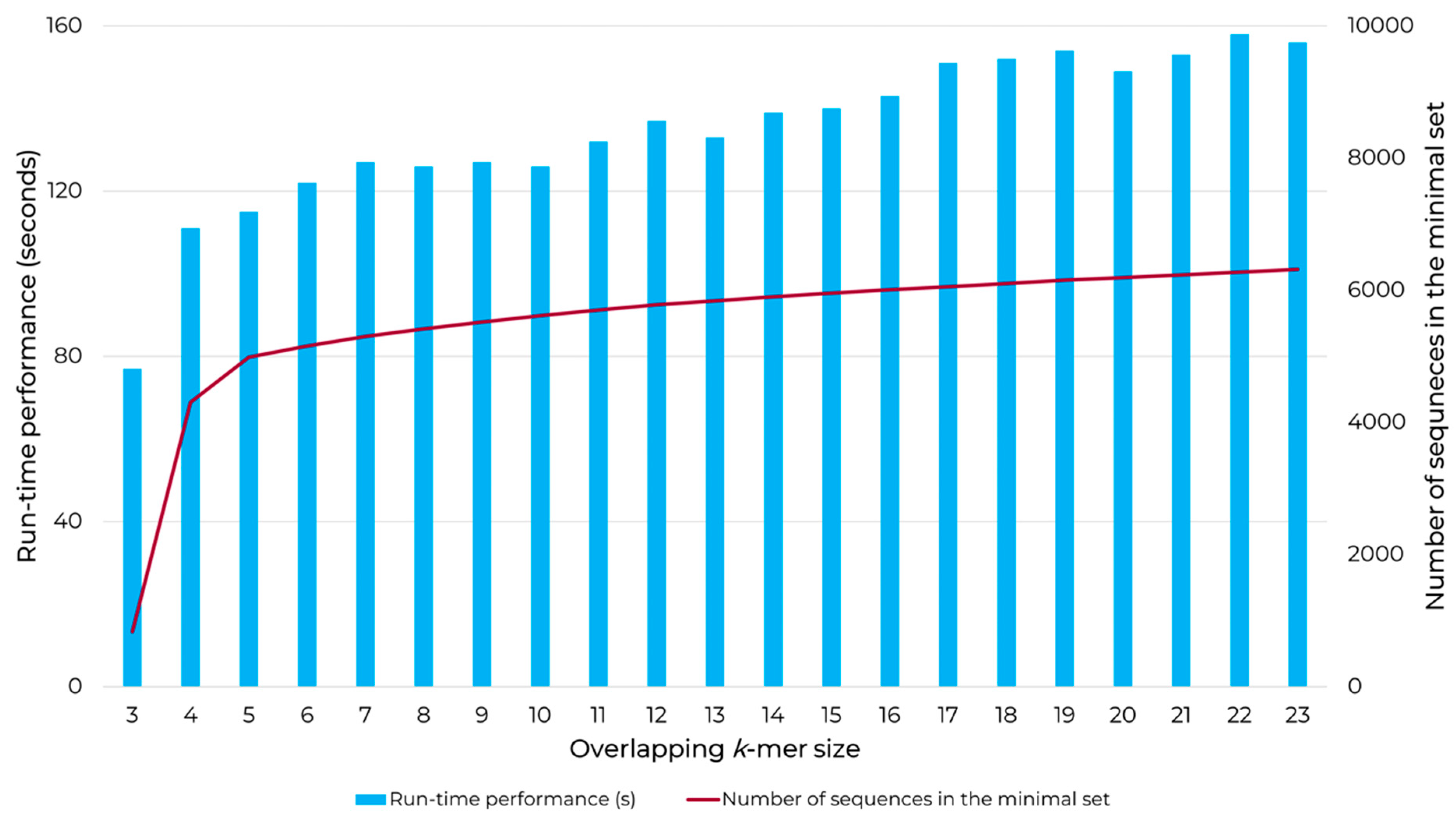
2.3. Determining k-mer Size of Choice for UNIQmin
2.4. Application of UNIQmin—Data Retrieval, Processing, and Data Analysis across Viral Taxonomic Lineages (Species, Genus, and Family Ranks)
2.5. Performance Comparison with Other Existing Alignment Independent Data Compression Methods
3. Results
3.1. Application of UNIQmin—Dengue Virus (DENV) Lineage as a Usage Scenario
3.2. Comparison to Existing Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Keni, R.; Alexander, A.; Nayak, P.G.; Mudgal, J.; Nandakumar, K. COVID-19: Emergence, Spread, Possible Treatments, and Global Burden. Front. Public Health 2020, 8, 216. [Google Scholar] [CrossRef]
- GBD 2019 Diseases and Injuries Collaborator. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1204–1222. [Google Scholar] [CrossRef]
- Steinhauer, D.A. Pathways to human adaptation. Nature 2013, 499, 412–413. [Google Scholar] [CrossRef] [PubMed]
- Wendel, I.; Matrosovich, M.; Klenk, H.D. SnapShot: Evolution of Human Influenza A Viruses. Cell Host Microbe 2015, 17, 416–416.e1. [Google Scholar] [CrossRef][Green Version]
- Thakur, A.; Mikkelsen, H.; Jungersen, G. Intracellular Pathogens: Host Immunity and Microbial Persistence Strategies. J. Immunol. Res. 2019, 2019, 1356540. [Google Scholar] [CrossRef]
- Volkov, I.; Pepin, K.M.; Lloyd-Smith, J.O.; Banavar, J.R.; Grenfell, B.T. Synthesizing within-host and population-level selective pressures on viral populations: The impact of adaptive immunity on viral immune escape. J. R. Soc. Interface 2010, 7, 1311–1318. [Google Scholar] [CrossRef]
- Heiny, A.T.; Miotto, O.; Srinivasan, K.N.; Khan, A.M.; Zhang, G.L.; Brusic, V.; Tan, T.W.; August, J.T. Evolutionarily Conserved Protein Sequences of Influenza A Viruses, Avian and Human, as Vaccine Targets. PLoS ONE 2007, 2, e1190. [Google Scholar] [CrossRef]
- Khan, A.M.; Miotto, O.; Nascimento, E.J.M.; Srinivasan, K.N.; Heiny, A.T.; Zhang, G.L.; Marques, E.; Tan, T.W.; Brusic, V.; Salmon, J.; et al. Conservation and Variability of Dengue Virus Proteins: Implications for Vaccine Design. PLOS Negl. Trop. Dis. 2008, 2, e272. [Google Scholar] [CrossRef]
- Bingham, R.J.; Dykeman, E.C.; Twarock, R. RNA Virus Evolution via a Quasispecies-Based Model Reveals a Drug Target with a High Barrier to Resistance. Viruses 2017, 9, 347. [Google Scholar] [CrossRef]
- Chong, L.C.; Khan, A.M. Identification of highly conserved, serotype-specific dengue virus sequences: Implications for vaccine design. BMC Genom. 2019, 20, 921. [Google Scholar] [CrossRef] [PubMed]
- Regional Planning. Influenza Pandemic Plan. The Role of WHO and Guidelines for National and Regional Planning; World Health Organization: Geneva, Switzerland, 1999; pp. 1–66. [Google Scholar]
- Raman, H.S.A.; Tan, S.; August, J.T.; Khan, M.A. Dynamics of Influenza A (H5N1) virus protein sequence diversity. PeerJ 2020, 7, e7954. [Google Scholar] [CrossRef]
- Hackbart, M.; Deng, X.; Baker, S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. USA 2020, 117, 8094–8103. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Kazlauskas, D.; Iranzo, J.; Lucía-Sanz, A.; Kuhn, J.H.; Krupovic, M.; Dolja, V.V.; Koonin, E.V. Origins and Evolution of the Global RNA Virome. mBio 2018, 9, e02329-18. [Google Scholar] [CrossRef]
- Yang, O.O.; Ali, A.; Kasahara, N.; Faure-Kumar, E.; Bae, J.Y.; Picker, L.J.; Park, H. Short Conserved Sequences of HIV-1 Are Highly Immunogenic and Shift Immunodominance. J. Virol. 2015, 89, 1195–1204. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Koo, Q.Y.; Khan, A.M.; Jung, K.-O.; Ramdas, S.; Miotto, O.; Tan, T.W.; Brusic, V.; Salmon, J.; August, J.T. Conservation and Variability of West Nile Virus Proteins. PLoS ONE 2009, 4, e5352. [Google Scholar] [CrossRef]
- Yang, O.O. Candidate Vaccine Sequences to Represent Intra- and Inter-Clade HIV-1 Variation. PLoS ONE 2009, 4, e7388. [Google Scholar] [CrossRef]
- Zielezinski, A.; Vinga, S.; Almeida, J.; Karlowski, W.M. Alignment-free sequence comparison: Benefits, applications, and tools. Genome Biol. 2017, 18, 1–17. [Google Scholar] [CrossRef]
- Chong, L.C.; Khan, A.M. Vaccine Target Discovery. In Encyclopedia of Bioinformatics and Computational Biology; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 241–251. [Google Scholar] [CrossRef]
- Khan, A.M. Mapping Targets of Immune Responses in Complete Dengue Viral Genomes. Master’s Thesis, National University of Singapore, Singapore, 2005; pp. 1–135. [Google Scholar]
- Khan, A.M.; Heiny, A.T.; Lee, K.X.; Srinivasan, K.N.; Tan, T.W.; August, J.T.; Brusic, V. Large-scale analysis of antigenic diversity of T-cell epitopes in dengue virus. BMC Bioinform. 2006, 7, S4. [Google Scholar] [CrossRef]
- Özer, O.; Lenz, T.L. Unique Pathogen Peptidomes Facilitate Pathogen-Specific Selection and Specialization of MHC Alleles. Mol. Biol. Evolution. 2021, msab176. [Google Scholar] [CrossRef]
- Heiny, A.T. The Antigenic Diversity Analysis of Complete Viral Genome of Influenza A Virus. Bachelor’s Thesis, National University of Singapore, Singapore, 2005; pp. 1–95. [Google Scholar]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Mahram, A.; Herbordt, M.C. Fast and accurate NCBI BLASTP: Acceleration with multiphase FPGA-based prefiltering. In Proceedings of the 24th ACM International Conference on Supercomputing—ICS’10, Tsukuba, Ibaraki, Japan, 2–4 June 2010; ACM Press: New York, NY, USA, 2010; p. 73. [Google Scholar]
- Nicholson, L.B. The immune system. Essays Biochem. 2016, 60, 275–301. [Google Scholar] [CrossRef]
- Trolle, T.; McMurtrey, C.P.; Sidney, J.; Bardet, W.; Osborn, S.C.; Kaever, T.; Sette, A.; Hildebrand, W.H.; Nielsen, M.; Peters, B. The Length Distribution of Class I–Restricted T Cell Epitopes Is Determined by Both Peptide Supply and MHC Allele–Specific Binding Preference. J. Immunol. 2016, 196, 1480–1487. [Google Scholar] [CrossRef]
- Gfeller, D.; Guillaume, P.; Michaux, J.; Pak, H.-S.; Daniel, R.T.; Racle, J.; Coukos, G.; Bassani-Sternberg, M. The Length Distribution and Multiple Specificity of Naturally Presented HLA-I Ligands. J. Immunol. 2018, 201, 3705–3716. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Trincado, J.L.; Gomez-Perosanz, M.; Reche, P.A. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J. Immunol. Res. 2017, 2017, 2680160. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef] [PubMed]
- EL-Manzalawy, Y.; Honavar, V. Major Histocompatibility Complex (MHC), Binder Prediction. In Encyclopedia of Systems Biology; Springer: New York, NY, USA, 2013; pp. 1162–1166. [Google Scholar]
- Lim, W.C.; Khan, A.M. Mapping HLA-A2, -A3 and -B7 supertype-restricted T-cell epitopes in the ebolavirus proteome. BMC Genom. 2018, 19, 42. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Tan, P.T.; Tan, T.W.; August, J.T.; Khan, A.M. Dissecting the Dynamics of HIV-1 Protein Sequence Diversity. PLoS ONE 2013, 8, e59994. [Google Scholar] [CrossRef]
- Tan, S.; Sjaugi, M.; Fong, S.; Chong, L.; Raman, H.A.; Mohamed, N.N.; August, J.; Khan, A. Avian Influenza H7N9 Virus Adaptation to Human Hosts. Viruses 2021, 13, 871. [Google Scholar] [CrossRef]
- Pornputtapong, N.; Acheampong, D.A.; Patumcharoenpol, P.; Jenjaroenpun, P.; Wongsurawat, T.; Jun, S.-R.; Yongkiettrakul, S.; Chokesajjawatee, N.; Nookaew, I. KITSUNE: A Tool for Identifying Empirically Optimal K-mer Length for Alignment-Free Phylogenomic Analysis. Front. Bioeng. Biotechnol. 2020, 8, 556413. [Google Scholar] [CrossRef]
- Zhang, Q.; Jun, S.-R.; Leuze, M.; Ussery, D.; Nookaew, I. Viral Phylogenomics Using an Alignment-Free Method: A Three-Step Approach to Determine Optimal Length of k-mer. Sci. Rep. 2017, 7, 40712. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.; McK Bird, D. Optimizing k-mer size using a variant grid search to enhance de novo genome assembly. Bioinformation 2016, 12, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Chikhi, R.; Medvedev, P. Informed and automated k-mer size selection for genome assembly. Bioinformation 2014, 30, 31–37. [Google Scholar] [CrossRef]
- Khan, A.M.; Hu, Y.; Miotto, O.; Thevasagayam, N.M.; Sukumaran, R.; Raman, H.S.A.; Brusic, V.; Tan, T.W.; August, J.T. Analysis of viral diversity for vaccine target discovery. BMC Med. Genom. 2017, 10, 78. [Google Scholar] [CrossRef]
- Oliveira, S.C.; de Magalhães, M.T.Q.; Homan, E.J. Immunoinformatic Analysis of SARS-CoV-2 Nucleocapsid Protein and Identification of COVID-19 Vaccine Targets. Front. Immunol. 2020, 11, 587615. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, M.; Pratas, D.; Pinho, A.J. AC: A Compression Tool for Amino Acid Sequences. Interdiscip. Sci. Comput. Life Sci. 2019, 11, 68–76. [Google Scholar] [CrossRef]
- Kryukov, K.; Ueda, M.T.; Nakagawa, S.; Imanishi, T. Sequence Compression Benchmark (SCB) database—A comprehensive evaluation of reference-free compressors for FASTA-formatted sequences. GigaScience 2020, 9, giaa072. [Google Scholar] [CrossRef]
- Hategan, A.; Tabus, I. Protein is compressible. In Proceedings of the 6th Nordic Signal Processing Symposium—NORSIG 2004, Espoo, Finland, 9–11 June 2004; pp. 192–195. [Google Scholar]
- Adjeroh, D.; Nan, F. On Compressibility of Protein Sequences. In Proceedings of the Data Compression Conference (DCC’06), Snowbird, UT, USA, 28–30 March 2006; pp. 422–434. [Google Scholar]


| Taxonomic Lineage Rank # | Number of Retrieved Sequences | Number of nr Sequences #| Percentage of Deduplication (Relative to the Retrieved Sequences) | Number of Sequences in the Minimal Set | Compression Using UNIQmin (Relative to the Retrieved Sequences|nr Dataset) ^ |
|---|---|---|---|---|
| Species: Dengue virus | 26,205 | 9800|~62.6% | 5519 | ~16.3%|~43.7% |
| Genus: Flavivirus | 45,593 | 17,771|~61.0% | 9763 | ~17.6%|~45.1% |
| Family: Flaviviridae | 273,463 | 141,200|~48.4% | 66,707 | ~27.2%|~52.8% |
| Input Dataset (Number of nr Sequences) # | Measure | Algorithm Implementation * | |
|---|---|---|---|
| ITERmin | UNIQmin | ||
| 1000 sequences ** | Run-time performance (minutes) | 273 | <1 (14 s) |
| Number of sequences in the minimal set | 851 | 851 | |
| Compression ^ | ~14.9% | ~14.9% | |
| 9800 sequences | Run-time performance (minutes) | ~194,400 | >2 (127 s) |
| Number of sequences in the minimal set | 5534 | 5519 | |
| Compression ^ | ~43.5% | ~43.7% | |
| Compressors | Human | Viruses | |||
|---|---|---|---|---|---|
| HS2019 | HS2020 | AP2019 | EP2019 | All Viruses ^ | |
| Gzip # | 4.61 | 1.55 | 4.59 | 4.69 | 1.61 |
| bzip2 # | 4.26 | - | 4.27 | 4.49 | - |
| 7zip # | 4.03 | - | 4.14 | 4.59 | - |
| Lzma # | 4.03 | - | 4.14 | 4.43 | - |
| Paq81 # | 3.90 | - | 3.97 | 4.30 | - |
| AC #,$ | 3.79 | 0.94 | 3.99 | 4.52 | 0.63 |
| UNIQmin | - | 1.93 | - | - | 2.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chong, L.C.; Lim, W.L.; Ban, K.H.K.; Khan, A.M. An Alignment-Independent Approach for the Study of Viral Sequence Diversity at Any Given Rank of Taxonomy Lineage. Biology 2021, 10, 853. https://doi.org/10.3390/biology10090853
Chong LC, Lim WL, Ban KHK, Khan AM. An Alignment-Independent Approach for the Study of Viral Sequence Diversity at Any Given Rank of Taxonomy Lineage. Biology. 2021; 10(9):853. https://doi.org/10.3390/biology10090853
Chicago/Turabian StyleChong, Li Chuin, Wei Lun Lim, Kenneth Hon Kim Ban, and Asif M. Khan. 2021. "An Alignment-Independent Approach for the Study of Viral Sequence Diversity at Any Given Rank of Taxonomy Lineage" Biology 10, no. 9: 853. https://doi.org/10.3390/biology10090853
APA StyleChong, L. C., Lim, W. L., Ban, K. H. K., & Khan, A. M. (2021). An Alignment-Independent Approach for the Study of Viral Sequence Diversity at Any Given Rank of Taxonomy Lineage. Biology, 10(9), 853. https://doi.org/10.3390/biology10090853







