Simple Summary
Infections of potato tubers and roots by Spongospora subterranea result in powdery scab and root diseases. Losses due to infections with S. subterranea are substantial in most potato-growing regions of the world with no fully effective treatments available. Understanding the gene regulation of pathogens in their host is dependent on multidimensional datasets. In this study, we profiled the transcriptome and proteome of S. subterranea within the susceptible and resistant host. Enzyme activity and nucleic acid metabolism appear to be important to the virulence of S. subterranea. Our results provide a good resource for future functional studies of powdery scab and might be useful in S. subterranea inoculum management.
Abstract
Spongospora subterranea is an obligate biotrophic pathogen, causing substantial economic loss to potato industries globally. Currently, there are no fully effective management strategies for the control of potato diseases caused by S. subterranea. To further our understanding of S. subterranea biology during infection, we characterized the transcriptome and proteome of the pathogen during the invasion of roots of a susceptible and a resistant potato cultivar. A total of 7650 transcripts from S. subterranea were identified in the transcriptome analysis in which 1377 transcripts were differentially expressed between two cultivars. In proteome analysis, we identified 117 proteins with 42 proteins significantly changed in comparisons between resistant and susceptible cultivars. The functional annotation of transcriptome data indicated that the gene ontology terms related to the transportation and actin processes were induced in the resistant cultivar. The downregulation of enzyme activity and nucleic acid metabolism in the resistant cultivar suggests a probable influence of these processes in the virulence of S. subterranea. The protein analysis results indicated that the majority of differentially expressed proteins were related to the metabolic processes and transporter activity. The present study provides a comprehensive molecular insight into the multiple layers of gene regulation that contribute to S. subterranea infection and development in planta and illuminates the role of host immunity in affecting pathogen responses.
1. Introduction
The obligate biotrophic Spongospora subterranea is an important pathogen of potato, the world’s third most valuable food crop for human consumption. The pathogen induces root and tuber diseases in potato, the latter known as powdery scab, that impact both tuber yield and quality [1,2]. Currently, there are no effective strategies for the control of these root and tuber diseases. Thus, an understanding of the regulatory principles underlying Spongospora-potato interactions is important, particularly during colonization of potato roots [3]. Spongospora subterranea is a soil-dwelling obligate biotroph. Similar to other plasmodiophorids, S. subterranea has a complex lifestyle that allows the pathogen to persist in soil between host plants with the production of long-life resting spores [4]. In most genera of plasmodiophorids, resting spores are found as aggregates known as sporosori [5]. The resting spores germinate releasing short-lived zoospores in the presence of potato plants. Following attachment and encystment to a host root, the zoospore transfers its cellular contents to a host cell. The pathogen forms a plasmodium within infected cells which develops into a zoosporangium producing and releasing secondary zoospores that will induce further root and tuber infections. Zoospores are the main infective form of S. subterranea that can swim in the soil and search for the potato roots to infect with a specialized extrusome and develop a zoosporangium [6]. This zoospore-mediated root infection can form root galls approximately 1–3 months following the infection, with galls producing sporosori that can be released into the soil as roots degrade following senescence [7].
RNA-sequencing has been widely used to investigate the molecular aspects of plant–pathogen interaction with more focus on the host responses during infection with pathogens [8,9,10]. A deep sequencing approach allows the simultaneous capture of both plant and pathogen transcripts during infection [11]. The transcriptome analysis of the necrotrophic fungus Bipolaris sorghicola inside the sorghum leaves revealed the key genes in the plant–pathogen interaction that included transcriptional factors and plant cell-wall degrading enzymes [12]. Teixeira et al. [13] profiled the transcriptome of Moniliophthora perniciosa in the infected shoots of the Theobroma cacao and identified putative virulence effectors by characterizing the fungal infection strategies. The in planta transcriptome analysis of the bacterial pathogen Pseudomonas syringae in Arabidopsis revealed specific “immune-responsive” bacterial genes and processes [14]. A time-course analysis of the rusty root rot pathogen Ilyonectria robusta transcriptome in planta revealed pathogenicity-related genes, in particular candidate effector genes [15]. Although transcriptome analysis is a useful approach for the study of plant–pathogen interaction, it has been well established that transcript levels do not always reflect protein levels and protein activities. Mass spectrometry (MS) based proteomics has previously been applied to pathogens in vitro and in planta. Previous in planta proteomics analyses of the biotrophic barley fungal pathogen Blumeria graminis identified several proteins related to pathogenicity inside the infected tissues [16]. The proteome analyses of proteases in the biotroph fungal pathogen Cladosporium fulvum confirmed the presence of six proteases at proteome level during host infection [17]. However, the capacity of proteomics to describe pathogens in planta remains limited as the pathogen proteins are diluted by the plant proteome. Therefore, the integration of transcriptomics and proteomics is essential to acquire a precise picture of pathogens during the colonization of their host.
To understand the biology and virulence of obligate biotrophic plant pathogens, a comprehensive analysis of the transcriptome and proteome of these pathogens is a priority. We have researched the transcriptome [18] and proteome [19] dynamics of S. subterranea during the germination of resting spores using an in vitro model. However, to better understand the complex life cycle of the obligate biotrophic pathogens inside their host, a comparable effort to analyse the in planta transcriptome and proteome of S. subterranea during root infection is required. To unravel key components of the infection and interaction with the host during the biotrophic interaction, we simultaneously profiled the transcriptome and proteome of S. subterranea inside the potato roots of a susceptible and a resistant potato cultivar. We also compared the in planta data to our previous in vitro analysis of S. subterranea transcriptome and proteome during the germination of spores.
2. Materials and Methods
2.1. Pathogen Source and Potato Cultivars Used in This Study
Two potato cultivars that differ in resistance to S. subterranea were used in this study. Cultivar ‘Gladiator’ has a strong resistance to both tuber and root disease, while cultivar ‘Iwa’ is known as being highly susceptible to root and tuber infection by S. subterranea [6]. The S. subterranea inoculum was obtained by dry scraping lesions from powdery scab-infected tubers and purified using Ludox® (HS-40 colloidal silica, Sigma, NSW, Australia) as described before [20].
2.2. Plant Growth and Pathogen Infection
Potato plants obtained from single-node cuttings were grown in sterile tissue culture in standard Murashige and Skoog (MS) medium with 30 g/L, 500 mg/L of casein hydrolysate and 40 mg/L of ascorbic acid [21]. During the initial growth stage, plants were maintained under a 16 h light/8 h dark photoperiod at 22 °C. After three weeks of culture, seedlings were carefully uprooted from their medium and inoculated by dipping their roots in an inoculum suspension (two milligrams of dried S. subterranea resting spore in 2 mL of Hoagland solution, incubated at 25 °C for 3 days) for one hour. The seedlings of similar size were then transplanted into plastic pots filled with wetted sterilized potting mix and transferred to a greenhouse with a relative humidity of 80 ± 5% and a temperature of 25 ± 3 °C. To ensure strong disease pressure, an additional 20 mL of the inoculum suspension (one gram of dried S. subterranea resting spore in 1 L of water) was added directly to the potting mix fourteen days after planting. After 6 weeks in the greenhouse, when Spongospora root galls were visible on the roots, potato roots were collected for RNA-seq (n = 3) and proteome (n = 4) analysis. The collected roots were washed thoroughly under running water, frozen in liquid nitrogen and stored at −80 °C until use.
2.3. RNA Extraction and Sequencing
Total RNA was extracted from 50 mg of each sample using the RNeasy plus mini kit (Qiagen, Hilden, Germany) including the gDNA eliminator spin columns to remove genomic DNA. RNA concentration was checked by a Qubit fluorometer using the Qubit RNA Broad-Range kit (Invitrogen, Waltham, MA, USA). The integrity of the RNA samples was analyzed using an Agilent 2100 Bioanalyzer system (Agilent, Palo Alto, Santa Clara, CA, USA). A total of 4 μg of RNA per sample was used as the input material for further analysis. The Illumina TruSeq Stranded Total RNA Library Prep Kit with RiboZero (Illumina, San Diego, CA, USA) was used to produce the sequencing libraries that were then sequenced on the NovaSeq6000 (Illumina, San Diego, CA, USA) platform (50 M, 2 × 100 bp) at the Australian Genome Research Facility (AGRF, Melbourne, Victoria, Australia).
2.4. Transcriptome Assembly and Differential Expression Analysis
The transcriptome analysis was performed using RNA-seq analysis tools available on the Galaxy Australia platform (https://usegalaxy.org.au/ (accessed on 5 February 2021)). Paired-end reads were initially quality checked using FastQC v.0.11.9 tools. Reads of low quality (<20 nucleotides) and of adapter contaminations were trimmed by Trimmomatic v.0.36.6. The quality of trimmed reads was assessed again using FastQC. The processed reads were mapped to the host first (the potato reference genome, downloaded from http://solanaceae.plantbiology.msu.edu/pgsc_download.shtml (accessed on 5 February 2021)) with the remaining reads mapped to the S. subterranea draft genome [22] using the HISAT2 v.2.1.0 tool. Stringtie 2.1.1 was used for de novo transcriptome reconstruction of the aligned reads. The Stringtie-Merge tool was then used to combine the Stringtie outputs and create a transcriptome database. To determine the uniquely mapped reads, FeatureCounts v.2.0.1 was used to generate a count matrix by calculating the transcript’s abundance using the fragments per kilobase of per million mapped reads (FPKM). The mapped reads were then served as input to DESeq2 v.2.11.40.6 tool for the quantification of differential gene expression. The list of differentially expressed genes (DEGs) was filtered with a cut-off for a false discovery rate (FDR) of 0.05. The RNA-seq data were further validated using qRT-PCR as described before [18]. The primer sequences were listed in Supplementary Table S1.
2.5. Protein Extraction and Digestion
Proteins were extracted from 50 mg of frozen roots per sample by adding 150 µL of extraction buffer including 7 M urea and 2 M thiourea, 1% dithiothreitol (DTT), 100 mM NaCl, 40 mM Tris, pH 8.0 and protease inhibitor cocktail (cOmplete Mini EDTA-free; Roche Diagnostics, NSW, Australia). The mixture was homogenized two times for 60 s each using a Fast Prep-24 bead beater (Mp Biomedicals, Seven Hills, NSW, Australia) at 5500× g in room temperature. Samples were then centrifuged at 16,000× g for 10 min at 4 °C and the supernatant was transferred to a new tube. Six volumes of cold acetone were added to the resulting clarified extracts and tubes incubated at −20 °C overnight. The tubes were centrifuged at 10,000× g for 8 min, the protein pellets were left to air dry for 5 min at room temperature and resuspended in denaturing buffer (7 M urea and 2 M thiourea, 40 mM Tris, pH 8.0 and protease inhibitor). Protein concentration was estimated by a Qubit fluorometer using the Qubit protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, DTT was added to a final concentration of 10 mM and proteins were reduced overnight at 4 °C. Samples were then alkylated in dark using 50 mM iodoacetamide for 2 h at room temperature and then digested with trypsin/LysC (Promega, Madison, WI, USA) according to the SP3 method [23]. Peptides were desalted using ZipTips (Merck, Darmstadt, Germany) and dried down in a SpeedVac concentrator.
2.6. Data-Independent Acquisition Mass Spectrometry (DIA-MS)
Peptide samples were dissolved in 12 µL loading buffer (0.05% (w/w) trifluoroacetic acid in water/acetonitrile (98:2)). Approximately 1 μg of each sample was analyzed using an Ultimate 3000 nano RSLC system (Thermo Fisher Scientific, Waltham, MA, USA) coupled to a Q-Exactive HF mass spectrometer fitted with nanospray Flex ion source (Thermo Fisher Scientific, Waltham, MA, USA) and controlled using Xcalibur version 4.3. Peptides were preconcentrated onto a 2 cm PepMap 100 C18 trap column at a flow rate of 5 µL/min and then separated at 300 nL/min on a 25 cm PepMap 100 C18 analytical column held at 45 °C using a 120-min segmented gradient. Mobile phase A comprised 0.1% (w/w) formic acid in water and mobile phase B comprised 0.08% (w/w) formic acid in acetonitrile/water (80:20). Data acquisition parameters were: 2.0 kV spray voltage, S-lens RF level of 60 and heated capillary set to 250 °C. MS1 spectra (390–1240 m/z) were acquired at a scan resolution of 120,000 in profile mode with an AGC target of 3e6 and followed by sequential MS2 scans across 26 DIA × 25 amu windows over the range of 397.5–1027.5 m/z, with 1 amu overlap between sequential windows. MS2 spectra were acquired in centroid mode at a resolution of 30,000 using an AGC target of 1 × 106, maximum IT of 55 ms and normalized collision energy of 27.
2.7. Raw Data Processing and Protein Label-Free Quantitation (LFQ)
DIA-MS raw files were processed using Spectronaut software (ver 14.8) (Biognosys AB, Wagistrasse, Switzerland). A project-specific library was generated using the Pulsar search engine to search the DIA MS2 spectra against the Solanum tuberosum UniProt reference proteome (UP000011115) comprising 53,106 entries concatenated with the UniProtKB protein sequences for S. subterranea (11,129 entries). For library generation, N-terminal acetylation and methionine oxidation were included as variable modifications, and cysteine carbamidomethylation was specified as a fixed modification and up to two missed cleavages were allowed. Peptide, protein, and PSM thresholds were set to 0.01. Mass tolerances were based on first pass calibration and extensive calibration for the calibration and main searches, respectively, with correction factors set to 1 at the MS1 and MS2 levels. Targeted searching of the library based on XIC extraction deployed dynamic retention time alignment with a correction factor of 1. Protein identification deployed 1% q-value thresholds at the precursor and protein levels, and automatic generation of mutated peptide decoys based on 10% of the library and dynamic decoy limitation for protein identification. MS2-level data were used for relative peptide quantitation between experimental samples, using the intensity values for the Top3 peptides (stripped sequences) and cross-run normalization based on median peptide intensity. Proteins identified on the basis of a single peptide (single hit proteins) were excluded while only proteins identified by matching to entries in the S. subterranea database were included in further analysis.
2.8. Gene Ontology Enrichment Analysis
The identified transcripts were subject to BLAST analysis against the UniProt (www.uniprot.org (accessed on 25 April 2021)) and NCBI (non-redundant protein and non-redundant transcript) databases with an e-value cut-off of 10−5. For the differentially expressed transcripts and proteins, gene ontology enrichment and pathway analysis were derived from UniProt, DAVID and the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (www.genome.jp/kegg (accessed on 10 May 2021)). The PCA plot and heatmap were drawn using Perseus software (v. 1.6.14.0). The volcano plot was obtained from the VolcaNoseR app [24].
3. Results and Discussion
3.1. In Planta Transcriptome and Proteome Profiling of S. subterranea
In this study, we have employed RNA-seq and label-free proteomics approaches to profile the transcriptome and proteome of S. subterranea inside potato roots of disease resistant (cv. Gladiator) and susceptible (cv. Iwa) cultivars. The experimental workflow for the RNA and protein preparation and omics data analysis is summarized in Figure 1.
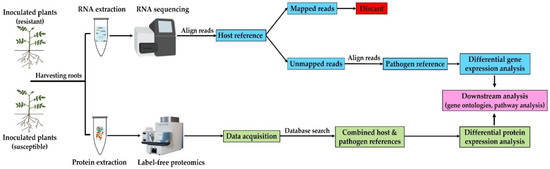
Figure 1.
Workflow of analysis. Transcriptome and proteome of S. subterranea were analyzed simultaneously by using different approaches. The mixed transcriptome obtained from S. subterranea-infected potato roots was sequenced by using Illumina mRNA-Seq technology. The reads were aligned to the potato reference genome and unaligned reads were used to analyse the transcriptome of S. subterranea. For the proteome analysis, proteins were extracted from S. subterranea-infected potato roots, analyzed using a label-free approach and searched in a combined host and pathogen database. Further analysis using multiple bioinformatics approaches identified the functional annotation of the differentially expressed genes and proteins.
Applying these approaches for the in planta transcriptome and proteome analysis of S. subterranea enabled the identification of 7650 transcripts and 117 proteins, respectively (Table 1). The lower number of identified proteins can be explained by the fact that the in planta protein analysis of pathogens is technically challenging as the pathogen proteins are embedded in and diluted by the plant proteome. There are published reports that analyzed the in planta proteome of pathogens, but only a very limited number of pathogen proteins have been identified [16,25].

Table 1.
The number of identified and significantly changing (FDR < 0.05) transcripts and proteins from S. subterranea inside the resistant and susceptible hosts.
3.2. Transcriptome Profile of S. subterranea inside the Resistant and Susceptible Hosts
To analyse the transcriptome of the obligate biotrophic pathogen S. subterranea in potato roots, a deep sequencing approach with an rRNA removal (to enrich mRNA) strategy was employed to overcome the RNA quantity difference between the host plant and pathogen. This method successfully enriched pathogen sequences allowing us to profile high-quality S. subterranea transcriptomes in planta with more than 4 million mean reads using the Illumina HiSeq platform (Supplementary Table S2). The transcriptome results were validated by qPCR measurements of the selected DEGs with a correlation coefficient above 88% (Supplementary Figure S1), indicating the accuracy of our RNA-seq data. Of 7650 identified unigenes, 6458 unigenes (~85%) matched the non-redundant (NR) protein and UniProt databases, suggesting that the de novo assembly approach was reliable. The complete list of identified and annotated transcripts is provided as Supplementary Table S3. The number of identified genes from S. subterranea in this study exceeded what has been reported in other in planta transcriptome analysis of plant pathogens [12,26,27]. According to the hierarchical clustering (sample-to-sample distances) of all six samples (Figure 2A) and principal component analysis (PCA) (Figure 2B), our RNA-seq approach was sensitive enough to capture S. subterranea transcriptome differences between the biological replicates.
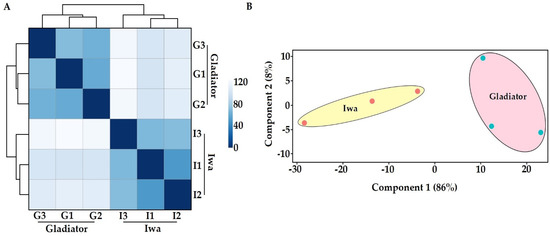
Figure 2.
Hierarchical clustering of the six samples used in RNA-seq study showing two distinct clades (A) and the principal component analysis of RNA-seq data (B). G; Gladiator, I; Iwa.
3.3. Functional Annotation of Differently Expressed Genes
A total of 1151 genes changed significantly in abundance (FDR < 0.05) in the transcriptome of S. subterranea (Gladiator vs. Iwa), including 837 upregulated and 314 downregulated genes (Supplementary Figure S2). Our gene ontology (GO) analysis revealed five major sub-clusters (three up and two downregulated classes) among the DEGs. The Z-scored expression levels of these DEGs were represented in the heatmap (Figure 3). The upregulated DEGs fell into three main clusters including transport (17 genes, 6 GO terms), actin (5 genes, 4 GO terms) and channel/transporter (18 genes, 11 GO terms) (Figure 3A). Upregulation of processes related to transportation and actin emphasized the role of ion exchange and structural rearrangement of the actin cytoskeleton in the pathogen during communication with hosts [28,29]. A transporter is a type of transmembrane protein that absorbs essential nutrients using the energy from ATP hydrolysis and can be divided into importers and exporters [30]. Transporter activity has been associated with spore germination and pathogenicity of Schizosaccharomyces pombe [31], Botrytis cinerea [32], Colletotrichum gloeosporioides [33], and Fusarium graminearum [34]. However, our RNA-seq data provide the first evidence that transporter and channel activities are also involved in Spongospora–potato interaction and potentially during the host infection.
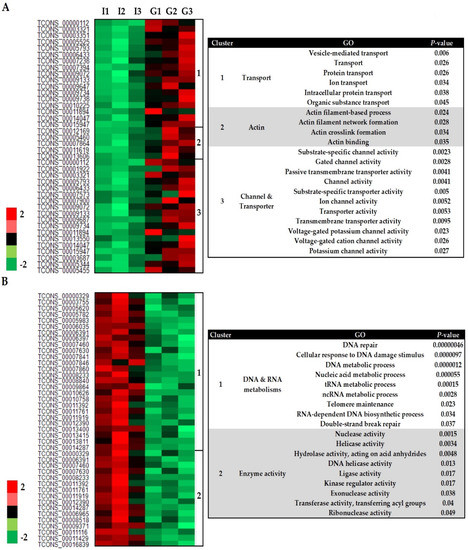
Figure 3.
Heatmap (based on Z-scores of the normalized expression of transcripts) of differently expressed genes from S. subterranea (Gladiator vs. Iwa). The upregulated (A) and downregulated (B) genes and their related GO terms. G; Gladiator, I; Iwa.
The downregulated DEGs formed two main clusters: DNA/RNA metabolism (26 genes, 9 GO terms) and enzyme activity (16 genes, 9 GO terms) (Figure 3B). One of the major significantly over-represented groups in these DEGs encoded for proteins involved in DNA and RNA metabolism (Figure 3B). The successful germination of dormant spores needs the progressive re-starting of transcription and translation [35]. To accelerate the transcription processes during the early stages of germination, DNA relaxes rapidly from the supercoiled state. This requires the activity of helicase enzymes and DNA repair genes [36,37]. The spores deficient in DNA repair in Bacillus subtilis were delayed in the first division of the spore′s nucleoid and severely affected by a ROS-inducer during the germination [38]. Thus, the resistant cultivar, Gladiator, might suppress the germination of S. subterranea resting spores by downregulation of the biological process related to nucleic acid metabolism such as transcription and DNA repair. The transfer RNA (tRNA) metabolic process is another GO term associated with the downregulated DEGs (Figure 3B). The tRNA pathway is an alternative route for cytokinin biosynthesis in many organisms [39]. Plant pathogens have developed several strategies to prevail over their hosts including the production of phytohormones such as cytokinin which interfere with host immune responses [40]. The tRNA-mediated cytokinin production by the rice blast fungus Magnaporthe oryzae reduces the defence responses from the host and alters the amino acid and sugar distribution to the favour of a pathogen [41]. The hemibiotroph Colletotrichum graminicola altered cytokinin responses in maize leaves, either through fungal production of cytokinin or using an indirect route [42]. Cytokinin production contributes to pathogen virulence of the plant pathogen Claviceps purpurea by the induction of susceptibility [43]. Therefore, the resistant cultivar can limit the access of S. subterranea to the plant amino acid and sugar sources by downregulation of the tRNA metabolic process. Hence, our results could lay a foundation for further investigations into the influences of nucleic acid metabolism in potato susceptibility to powdery scab.
Several enzymes including members of the hydrolase, kinase, transferase, ligase, and helicase families were downregulated in the S. subterranea transcriptome in the resistant cultivar (Figure 3B). Hydrolase (EC3) is a class of enzymes that catalyse the cleavage of a covalent bond using water and are known to participate in the germination of spores in fungal and bacterial pathogens by facilitating the reconstruction of the cell wall [44,45]. These enzymes were shown to be involved in the pathogen invasion and pathogenicity of Colletotrichum lagenarium [46] and Cladosporium fulvum [47]. Thus, the downregulation of hydrolase enzymes in the transcriptome of S. subterranea inside the resistant cultivar might be related to their role in the germination of resting spores as well as S. subterranea pathogenicity [48]. The protein kinases (EC2) that catalyse the modification of proteins by phosphorylation were another downregulated enzyme group (Figure 3B). The role of protein kinases in the resting spore germination have been confirmed in Verticillium dahlia [49], Bacillus subtilis [50], Schizosaccharomyces pombe [51], and many other pathogens [52]. Consistent with the above results, our RNA-seq analysis confirmed the downregulation of several enzyme families in the transcriptome of S. subterranea inside the resistant cultivar compared to the susceptible one. Although not enough is known about the Spongospora-potato interaction during the root infection, our RNA-seq data suggest that suppression of enzyme activity, as well as nucleic acid metabolism, can contribute to resistance against S. subterranea.
3.4. In Planta Protein Analysis of S. subterranea
A label-free proteomics approach was used to profile the proteome of S. subterranea in the infected roots of potato cultivars. A total of 117 proteins from S. subterranea were identified in which 42 proteins were differentially expressed between Gladiator and Iwa (Figure 4A). A complete list of all identified and DEPs (Gladiator vs. Iwa) is provided as Supplementary Table S4. The GO analysis (biological process and molecular function) of the total identified and DEPs is shown in Figure 4B. This analysis revealed that DEPs were mainly concentrated on the catabolic and metabolic processes, binding, nitrogen compound metabolic process, and biosynthetic process. The proteins related to the metabolic and catabolic processes are very abundant soluble proteins and are generally well represented in several pathogens such as Phytophthora capsici [53], Colletotrichum acutatum [54], and Botrytis cinerea [55]. Similar to the RNA-seq results, the existence of few molecular functions related to “transporter activity” such as ion transmembrane transporter activity, inorganic transmembrane transporter activity, active transmembrane transporter activity and proton-transporting ATP synthase activity was confirmed in the proteomics study (Figure 4B). Plants have developed a high capacity to recognize pathogens through various strategies and pathogens manipulate the defence response through secretion of virulence effectors [56]. Successful pathogens can suppress the plant responses and thereby cause disease. This suppression is achieved through the deployment of the effector proteins. However, plants are able to recognize the pathogen effectors proteins through an accessory protein or by direct physical association [57]. We identified 34 upregulated and eight downregulated proteins in the resistant cultivar compared to the susceptible cultivar (Figure 4A). The activation of several metabolic and catabolic processes as well as transporter activity was also confirmed among the DEPs (Figure 4B). Thus, we concluded that S. subterranea requires an intensive effort for the suppression of the host immune system in Gladiator which results in a more active plant–pathogen interaction. Unfortunately, 20 proteins out of 42 DEPs were not annotated in databases and presented as “uncharacterized protein” (Supplementary Table S4). The genome annotation is not complete in plasmodiophorids and many of the proteins associated with spore germination and disease development have yet to be characterized in S. subterranea. We believe that the full implication of our present protein analysis will be realised when functional annotation of these DEPs is available.
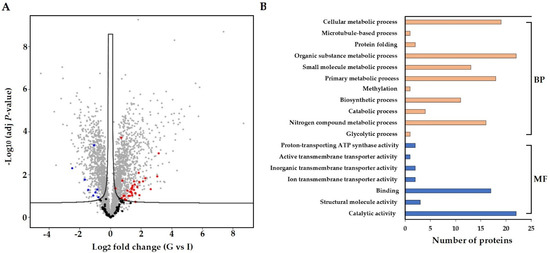
Figure 4.
Comparison of differently abundant proteins (FDR < 0.05) in resistant and susceptible potato cultivars. Proteins from S. subterranea are bolded (black; no significant, red; upregulated and blue downregulated) (A). The GO term for the S. subterranea DEPs (B). BP; biological process, MF; molecular function, G; Gladiator, I; Iwa.
Comparison of the in planta transcriptome and proteome of S. subterranea identified 17 proteins in common between both datasets (Supplementary Table S5). We then plotted the log-transformed fold changes of proteins that were presented in RNA-seq and proteomics data (Figure 5). A high degree of concordance in changes in protein abundance between the two datasets was observed, with only four proteins having opposing changes in abundance between the RNA-seq and proteomics data (red dots in Figure 5). The MS/MS raw data were also searched against the proteome dataset generated from our RNA-seq data based on six-frame translation. A total of 44 proteins were found in this search (Supplementary Table S6) which were categorized into three groups according to functional annotation and pathway analysis (Table 2). The presence of GO terms related to peptide metabolism and translation showed that these processes are vital for the successful germination in planta as well as disease invasion of S. subterranea inside the host. The dependence of spore germination on transcription and translation has been demonstrated in many pathogens [58,59,60]. This finding aligned with the observed upregulation of transcription and protein synthesis in our previous in vitro analysis of transcriptome and proteome of S. subterranea during the germination of resting spores [18,19]. Therefore, transcription and translation may be induced upon the activation of resting spores both in planta and in vitro.
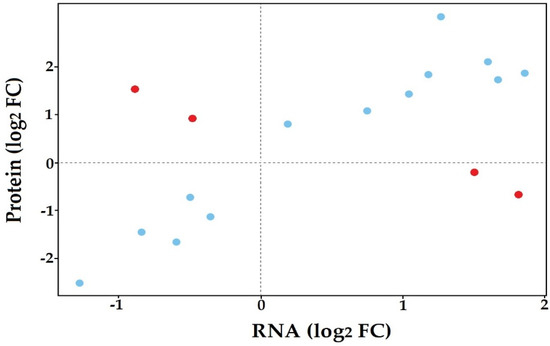
Figure 5.
Distribution of proteins common between transcriptomics and proteomics experiments.

Table 2.
GO term for the proteins from S. subterranea identified in the database created from translated transcripts from RNA-seq analysis.
3.5. Co-Expression of In Planta and In Vitro Analysis of S. subterranea Transcriptome and Proteome
The in planta transcriptomes and proteomes of S. subterranea obtained in this study were compared to our previous in vitro transcriptome [18] and proteomic [19] studies of S. subterranea. The number of identified genes was significantly higher in the in planta analysis of S. subterranea transcriptome (Figure 6A, left). In contrast, the number of identified proteins in the in vitro analysis of S. subterranea proteome was higher compared to in planta (Figure 6A, right). The pathway analysis of identified proteins common between or independent in the in vitro and in planta analysis of S. subterranea transcriptome is presented in Figure 6B. The main pathways identified during this research include nucleotide excision repair, pyrimidine metabolism, transcription, translation, transport, protein degradation, pyrimidine biosynthesis and purine metabolism. The mRNA surveillance pathway, proteasome, fatty acid degradation, spliceosome and basal transcription factors were the prominent pathways in the common proteins between the in vitro and in planta transcriptome of S. subterranea. These pathways are related to transcription, translation and amino acid degradation processes [58,59]. The purine metabolism and ubiquitin-mediated proteolysis were produced in vitro only, and these pathways were not recovered in planta (Figure 6B). On the other hand, some pathways such as RNA transport, inositol phosphate metabolism and phosphatidylinositol signaling system were only recovered in planta [61]. In total, the co-expression of in planta and in vitro transcriptome analysis of S. subterranea revealed that the spore activation in both in planta and in vitro needed the activity of the mRNA synthetase, DNA repair, translation and carbohydrate metabolism [62,63,64].
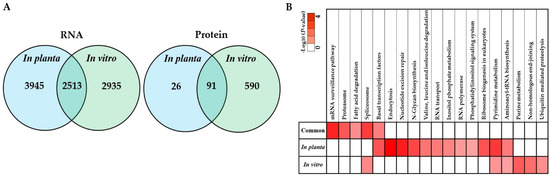
Figure 6.
Venn diagrams depicting the overlap of identified proteins common between or independent in the in vitro and in planta analysis of S. subterranea transcriptome and proteome (A). Pathway analysis of identified proteins common between or independent in the in vitro and in planta analysis of S. subterranea transcriptome (B).
4. Conclusions
In conclusion, our detailed in planta analysis of the S. subterranea transcriptome and proteome revealed broad-scale differences between susceptible and resistant potato cultivars. The upregulation of transporters/channels and actin metabolism in the resistant cultivar was discovered in this study. Downregulation of the enzyme activity and nucleic acid metabolism in S. subterranea transcriptome in the resistant cultivar is one of the most important findings of the present study. The suppression of these processes in S. subterranea within roots of the resistant cultivar provides new insight into Spongospora–potato interaction and may lead to the development of novel approaches for the resistance against powdery scab disease in potato breeding programs. The proteome data reported in this study provides the first in planta analysis of S. subterranea inside the potato roots and validates our transcriptome results. However, many of the identified proteins in the proteome analysis were not annotated according to publicly available databases. The value of this protein analysis will be understood when the genome annotation is completed in plasmodiophorids. In total, our study contributes to the significant progress in our understanding of the interaction between the obligate biotrophic pathogens and their host plants and increased the availability of “omic” data in such a complex interaction.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biology10090840/s1, Figure S1: Correlation between RNA-seq and qPCR, Figure S2: Comparison of differently abundant transcripts from S. subterranea in resistant and susceptible potato cultivars, Table S1: List of primers used in this study, Table S2: RNA-seq information, Table S3: List of identified, annotated and differentially expressed transcripts, Table S4: List of identified and differentially expressed proteins, Table S5: List of common proteins in both RNA-seq and proteomics, Table S6: List of identified proteins in our transcriptome database.
Author Contributions
S.B., C.R.W., R.W., R.S.T. and D.S.N. designed the experiments. S.B. performed the experiments, analysed the data and prepared the paper draft. R.W. acquired the proteomics data. C.R.W., R.W., R.S.T. and D.S.N. reviewed and edited the manuscript. C.R.W. and D.S.N. financially supported the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research and the APC was funded by The Australian Research Council, Discovery Grant program, DP180103337.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study can be found here: the transcriptome data is deposited in the NCBI BioSample Submissions (SRA) repository, accession number PRJNA747755. The proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [65] partner repository with the dataset identifier PXD027266.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Amponsah, J.; Tegg, R.S.; Thangavel, T.; Wilson, C.R. Moments of weaknesses–exploiting vulnerabilities between germination and encystment in the Phytomyxea. Biol. Rev. 2021, 96, 1603–1615. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Fulladolsa, A.C.; Cordova, A.M.; O’Neill, P.; Gray, S.M.; Charkowski, A.O. Evaluation of effects of chemical soil treatments and potato cultivars on Spongospora subterranea soil inoculum and incidence of powdery scab and potato mop-top virus in potato. Plant Dis. 2020, 104, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Balendres, M.; Tegg, R.; Wilson, C. Key events in pathogenesis of Spongospora diseases in potato: A review. Australas. Plant Pathol. 2016, 45, 229–240. [Google Scholar] [CrossRef]
- Neuhauser, S.; Bulman, S.; Kirchmair, M. Plasmodiophorids: The challenge to understand soil-borne, obligate biotrophs with a multiphasic life cycle. In Molecular Identification of Fungi; Springer: Berlin/Heidelberg, Germany, 2010; pp. 51–78. [Google Scholar]
- Balendres, M.; Clark, T.; Tegg, R.; Wilson, C. Germinate to exterminate: Chemical stimulation of Spongospora subterranea resting spore germination and its potential to diminish soil inoculum. Plant Pathol. 2018, 67, 902–908. [Google Scholar] [CrossRef]
- Falloon, R.; Merz, U.; Butler, R.; Curtin, D.; Lister, R.; Thomas, S. Root infection of potato by Spongospora subterranea: Knowledge review and evidence for decreased plant productivity. Plant Pathol. 2016, 65, 422–434. [Google Scholar] [CrossRef]
- Balendres, M.A.; Tegg, R.S.; Amponsah, J.; Wilson, C.R. Zoosporangial root infection of tomato by Spongospora subterranea in hydroponic and glasshouse culture results in diminished plant growth. J. Phytopathol. 2018, 166, 412–419. [Google Scholar] [CrossRef]
- Gao, L.; Tu, Z.J.; Millett, B.P.; Bradeen, J.M. Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions. BMC Genom. 2013, 14, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, Y.; Oono, Y.; Kanamori, H.; Matsumoto, T.; Itoh, T.; Minami, E. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS ONE 2012, 7, e49423. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-J.; Sniezko, R.A.; Zamany, A.; Williams, H.; Omendja, K.; Kegley, A.; Savin, D.P. Comparative transcriptomics and RNA-seq-based bulked segregant analysis reveals genomic basis underlying Cronartium ribicola vcr2 Virulence. Front. Microbiol. 2021, 12, 309. [Google Scholar]
- Naidoo, S.; Visser, E.A.; Zwart, L.; Du Toit, Y.; Bhadauria, V.; Shuey, L.S. Dual RNA-seq to elucidate the plant–pathogen duel. Curr. Issues Mol. Biol. 2018, 27, 127–142. [Google Scholar] [CrossRef]
- Yazawa, T.; Kawahigashi, H.; Matsumoto, T.; Mizuno, H. Simultaneous transcriptome analysis of Sorghum and Bipolaris sorghicola by using RNA-seq in combination with de novo transcriptome assembly. PLoS ONE 2013, 8, e62460. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, P.J.P.L.; de Toledo Thomazella, D.P.; Reis, O.; do Prado, P.F.V.; do Rio, M.C.S.; Fiorin, G.L.; José, J.; Costa, G.G.L.; Negri, V.A.; Mondego, J.M.C. High-resolution transcript profiling of the atypical biotrophic interaction between Theobroma cacao and the fungal pathogen Moniliophthora perniciosa. Plant Cell 2014, 26, 4245–4269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nobori, T.; Velásquez, A.C.; Wu, J.; Kvitko, B.H.; Kremer, J.M.; Wang, Y.; He, S.Y.; Tsuda, K. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. USA 2018, 115, E3055–E3064. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Chen, M.; Ma, Y.; Du, Z.; Yuan, N.; Li, Y.; Xiao, J.; Zhang, Y. Whole-genome and time-course dual RNA-Seq analyses reveal chronic pathogenicity-related gene dynamics in the ginseng rusty root rot pathogen Ilyonectria robusta. Sci. Rep. 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindschedler, L.V.; Burgis, T.A.; Mills, D.J.; Ho, J.T.; Cramer, R.; Spanu, P.D. In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei. Mol. Cell. Proteom. 2009, 8, 2368–2381. [Google Scholar] [CrossRef] [Green Version]
- Jashni, M.K.; van der Burgt, A.; Battaglia, E.; Mehrabi, R.; Collemare, J.; de Wit, P.J. Transcriptome and proteome analyses of proteases in biotroph fungal pathogen Cladosporium fulvum. J. Plant Pathol. 2020, 102, 377–386. [Google Scholar] [CrossRef]
- Balotf, S.; Tegg, R.S.; Nichols, D.S.; Wilson, C.R. Spore germination of the obligate biotroph Spongospora subterranea: Transcriptome analysis reveals germination associated genes. Front. Microbiol. 2021, 12, 1557. [Google Scholar] [CrossRef] [PubMed]
- Balotf, S.; Wilson, R.; Tegg, R.S.; Nichols, D.S.; Wilson, C.R. Quantitative proteomics provides an insight into germination-related proteins in the obligate biotrophic plant pathogen Spongospora subterranea. Environ. Microbiol. Rep. 2021, 13, 521–532. [Google Scholar] [CrossRef]
- Balotf, S.; Wilson, R.; Tegg, R.S.; Nichols, D.S.; Wilson, C.R. Optimisation of sporosori purification and protein extraction techniques for the biotrophic protozoan plant pathogen Spongospora subterranea. Molecules 2020, 25, 3109. [Google Scholar] [CrossRef] [PubMed]
- Balendres, M.A.; Nichols, D.S.; Tegg, R.S.; Wilson, C.R. Metabolomes of potato root exudates: Compounds that stimulate resting spore germination of the soil-borne pathogen Spongospora subterranea. J. Agric. Food Chem. 2016, 64, 7466–7474. [Google Scholar] [CrossRef]
- Ciaghi, S.; Neuhauser, S.; Schwelm, A. Draft genome resource for the potato powdery scab pathogen Spongospora subterranea. Mol. Plant-Microbe Interact. 2018, 31, 1227–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, C.S.; Moggridge, S.; Müller, T.; Sorensen, P.H.; Morin, G.B.; Krijgsveld, J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019, 14, 68–85. [Google Scholar] [CrossRef] [PubMed]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 1–5. [Google Scholar] [CrossRef]
- Zhou, W.; Eudes, F.; Laroche, A. Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 2006, 6, 4599–4609. [Google Scholar] [CrossRef]
- Gétaz, M.; Puławska, J.; Smits, T.H.; Pothier, J.F. Host–pathogen interactions between Xanthomonas fragariae and its host Fragaria × ananassa investigated with a dual RNA-Seq analysis. Microorganisms 2020, 8, 1253. [Google Scholar] [CrossRef]
- Rudd, J.J.; Kanyuka, K.; Hassani-Pak, K.; Derbyshire, M.; Andongabo, A.; Devonshire, J.; Lysenko, A.; Saqi, M.; Desai, N.M.; Powers, S.J. Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 2015, 167, 1158–1185. [Google Scholar]
- Gao, S.; Zeng, R.; Xu, L.; Song, Z.; Gao, P.; Dai, F. Genome sequence and spore germination-associated transcriptome analysis of Corynespora cassiicola from cucumber. BMC Microbiol. 2020, 20, 1–20. [Google Scholar] [CrossRef]
- Galhano, R.; Illana, A.; Ryder, L.S.; Rodriguez-Romero, J.; Demuez, M.; Badaruddin, M.; Martinez-Rocha, A.L.; Soanes, D.M.; Studholme, D.J.; Talbot, N.J. Tpc1 is an important Zn (II) 2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus. PLoS Pathog. 2017, 13, e1006516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beis, K. Structural basis for the mechanism of ABC transporters. Biochem. Soc. Trans. 2015, 43, 889–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plante, S.; Labbé, S. Spore germination requires ferrichrome biosynthesis and the siderophore transporter Str1 in Schizosaccharomyces pombe. Genetics 2019, 211, 893–911. [Google Scholar] [CrossRef] [Green Version]
- Doehlemann, G.; Molitor, F.; Hahn, M. Molecular and functional characterization of a fructose specific transporter from the gray mold fungus Botrytis cinerea. Fungal Genet. Biol. 2005, 42, 601–610. [Google Scholar] [CrossRef]
- Chagué, V.; Maor, R.; Sharon, A. CgOpt1, a putative oligopeptide transporter from Colletotrichum gloeosporioides that is involved in responses to auxin and pathogenicity. BMC Microbiol. 2009, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Seong, K.-Y.; Zhao, X.; Xu, J.-R.; Güldener, U.; Kistler, H.C. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 2008, 45, 389–399. [Google Scholar] [CrossRef]
- Sephton-Clark, P.C.; Voelz, K. Spore germination of pathogenic filamentous fungi. Adv. Appl. Microbiol. 2018, 102, 117–157. [Google Scholar]
- Setlow, P. I will survive: Protecting and repairing spore DNA. J. Bacteriol. 1992, 174, 2737–2741. [Google Scholar] [CrossRef] [Green Version]
- Walker, C.A.; Köppe, M.; Grenville-Briggs, L.J.; Avrova, A.O.; Horner, N.R.; McKinnon, A.D.; Whisson, S.C.; Birch, P.R.; Van West, P. A putative DEAD-box RNA-helicase is required for normal zoospore development in the late blight pathogen Phytophthora infestans. Fungal Genet. Biol. 2008, 45, 954–962. [Google Scholar] [CrossRef]
- Valenzuela-García, L.I.; Ayala-García, V.M.; Regalado-García, A.G.; Setlow, P.; Pedraza-Reyes, M. Transcriptional coupling (Mfd) and DNA damage scanning (DisA) coordinate excision repair events for efficient Bacillus subtilis spore outgrowth. MicrobiologyOpen 2018, 7, e00593. [Google Scholar] [CrossRef] [Green Version]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hluska, T.; Hlusková, L.; Emery, R. The Hulks and the Deadpools of the cytokinin universe: A dual strategy for cytokinin production, translocation, and signal transduction. Biomolecules 2021, 11, 209. [Google Scholar] [CrossRef]
- Chanclud, E.; Kisiala, A.; Emery, N.R.J.; Chalvon, V.; Ducasse, A.; Romiti-Michel, C.; Gravot, A.; Kroj, T.; Morel, J.-B. Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLoS Pathog. 2016, 12, e1005457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Behr, M.; Motyka, V.; Weihmann, F.; Malbeck, J.; Deising, H.B.; Wirsel, S.G. Remodeling of cytokinin metabolism at infection sites of Colletotrichum graminicola on maize leaves. Mol. Plant-Microbe Interact. 2012, 25, 1073–1082. [Google Scholar] [CrossRef]
- Hinsch, J.; Galuszka, P.; Tudzynski, P. Functional characterization of the first filamentous fungal tRNA-isopentenyltransferase and its role in the virulence of Claviceps purpurea. New Phytol. 2016, 211, 980–992. [Google Scholar] [CrossRef]
- Keep, N.; Ward, J.; Robertson, G.; Cohen-Gonsaud, M.; Henderson, B. Bacterial resuscitation factors: Revival of viable but non-culturable bacteria. Cell. Mol. Life Sci. 2006, 63, 2555. [Google Scholar] [CrossRef]
- Wrońska, A.K.; Boguś, M.I.; Włóka, E.; Kazek, M.; Kaczmarek, A.; Zalewska, K. Cuticular fatty acids of Galleria mellonella (Lepidoptera) inhibit fungal enzymatic activities of pathogenic Conidiobolus coronatus. PLoS ONE 2018, 13, e0192715. [Google Scholar] [CrossRef] [Green Version]
- Takano, Y.; Kubo, Y.; Shimizu, K.; Mise, K.; Okuno, T.; Furusawa, I. Structural analysis of PKS1, a polyketide synthase gene involved in melanin biosynthesis in Colletotrichum lagenarium. Mol. Gen. Genet. MGG 1995, 249, 162–167. [Google Scholar] [CrossRef]
- Ökmen, B.; Bachmann, D.; De Wit, P.J. A conserved GH17 glycosyl hydrolase from plant pathogenic Dothideomycetes releases a DAMP causing cell death in tomato. Mol. Plant Pathol. 2019, 20, 1710–1721. [Google Scholar] [CrossRef]
- Talukdar, P.K.; Sarker, M.R. The serine proteases CspA and CspC are essential for germination of spores of Clostridium perfringens SM101 through activating SleC and cortex hydrolysis. Food Microbiol. 2020, 86, 103325. [Google Scholar] [CrossRef]
- Tzima, A.; Paplomatas, E.J.; Rauyaree, P.; Kang, S. Roles of the catalytic subunit of cAMP-dependent protein kinase A in virulence and development of the soilborne plant pathogen Verticillium dahliae. Fungal Genet. Biol. 2010, 47, 406–415. [Google Scholar] [CrossRef]
- Nguyen, K.B.; Sreelatha, A.; Durrant, E.S.; Lopez-Garrido, J.; Muszewska, A.; Dudkiewicz, M.; Grynberg, M.; Yee, S.; Pogliano, K.; Tomchick, D.R. Phosphorylation of spore coat proteins by a family of atypical protein kinases. Proc. Natl. Acad. Sci. USA 2016, 113, E3482–E3491. [Google Scholar] [CrossRef] [Green Version]
- Hatanaka, M.; Shimoda, C. The cyclic AMP/PKA signal pathway is required for initiation of spore germination in Schizosaccharomyces pombe. Yeast 2001, 18, 207–217. [Google Scholar] [CrossRef]
- Rekadwad, B. Applications of serine/threonine protein kinases (STPK): A bus for dormancy exit. In Quorum Sensing and Its Biotechnological Applications; Springer: Berlin/Heidelberg, Germany, 2018; pp. 271–278. [Google Scholar]
- Pang, Z.; Srivastava, V.; Liu, X.; Bulone, V. Quantitative proteomics links metabolic pathways to specific developmental stages of the plant-pathogenic oomycete Phytophthora capsici. Mol. Plant Pathol. 2017, 18, 378–390. [Google Scholar] [CrossRef] [Green Version]
- Brown, S.H.; Yarden, O.; Gollop, N.; Chen, S.; Zveibil, A.; Belausov, E.; Freeman, S. Differential protein expression in Colletotrichum acutatum: Changes associated with reactive oxygen species and nitrogen starvation implicated in pathogenicity on strawberry. Mol. Plant Pathol. 2008, 9, 171–190. [Google Scholar] [CrossRef]
- Fernández-Acero, F.J.; Colby, T.; Harzen, A.; Carbú, M.; Wieneke, U.; Cantoral, J.M.; Schmidt, J. 2-DE proteomic approach to the Botrytis cinerea secretome induced with different carbon sources and plant-based elicitors. Proteomics 2010, 10, 2270–2280. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Duplessis, S.; Lorrain, C.; Petre, B.; Figueroa, M.; Dodds, P.N.; Aime, M.C. Host adaptation and virulence in heteroecious rust fungi. Annu. Rev. Phytopathol. 2021, 59. [Google Scholar] [CrossRef]
- Zhou, B.; Semanjski, M.; Orlovetskie, N.; Bhattacharya, S.; Alon, S.; Argaman, L.; Jarrous, N.; Zhang, Y.; Macek, B.; Sinai, L. Arginine dephosphorylation propels spore germination in bacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 14228–14237. [Google Scholar] [CrossRef] [Green Version]
- Osherov, N.; May, G. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 2000, 155, 647–656. [Google Scholar] [CrossRef]
- Sinai, L.; Rosenberg, A.; Smith, Y.; Segev, E.; Ben-Yehuda, S. The molecular timeline of a reviving bacterial spore. Mol. Cell 2015, 57, 695–707. [Google Scholar] [CrossRef] [Green Version]
- Holtappels, M.; Noben, J.-P.; Van Dijck, P.; Valcke, R. Fire blight host-pathogen interaction: Proteome profiles of Erwinia amylovora infecting apple rootstocks. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Djouiai, B.; Thwaite, J.E.; Laws, T.R.; Commichau, F.M.; Setlow, B.; Setlow, P.; Moeller, R. Role of DNA repair and protective components in Bacillus subtilis spore resistance to inactivation by 400-nm-wavelength blue light. Appl. Environ. Microbiol. 2018, 84, e01604–e01618. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Huber, N.; Moeller, R.; Stülke, J.; Dubovcova, B.; Akepsimaidis, G.; Meneses, N.; Drissner, D.; Mathys, A. Role of DNA repair in Bacillus subtilis spore resistance to high energy and low energy electron beam treatments. Food Microbiol. 2020, 87, 103353. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, S.; Leveau, J.H.; Stergiopoulos, I. Repeated exposure of Aspergillus niger spores to the antifungal bacterium Collimonas fungivorans Ter331 selects for delayed spore germination. Appl. Environ. Microbiol. 2021, 87, e00233-21. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).