Insights into Alexandrium minutum Nutrient Acquisition, Metabolism and Saxitoxin Biosynthesis through Comprehensive Transcriptome Survey
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culturing and Harvesting
2.2. RNA Extraction
2.3. cDNA Library Construction and Illumina Sequencing
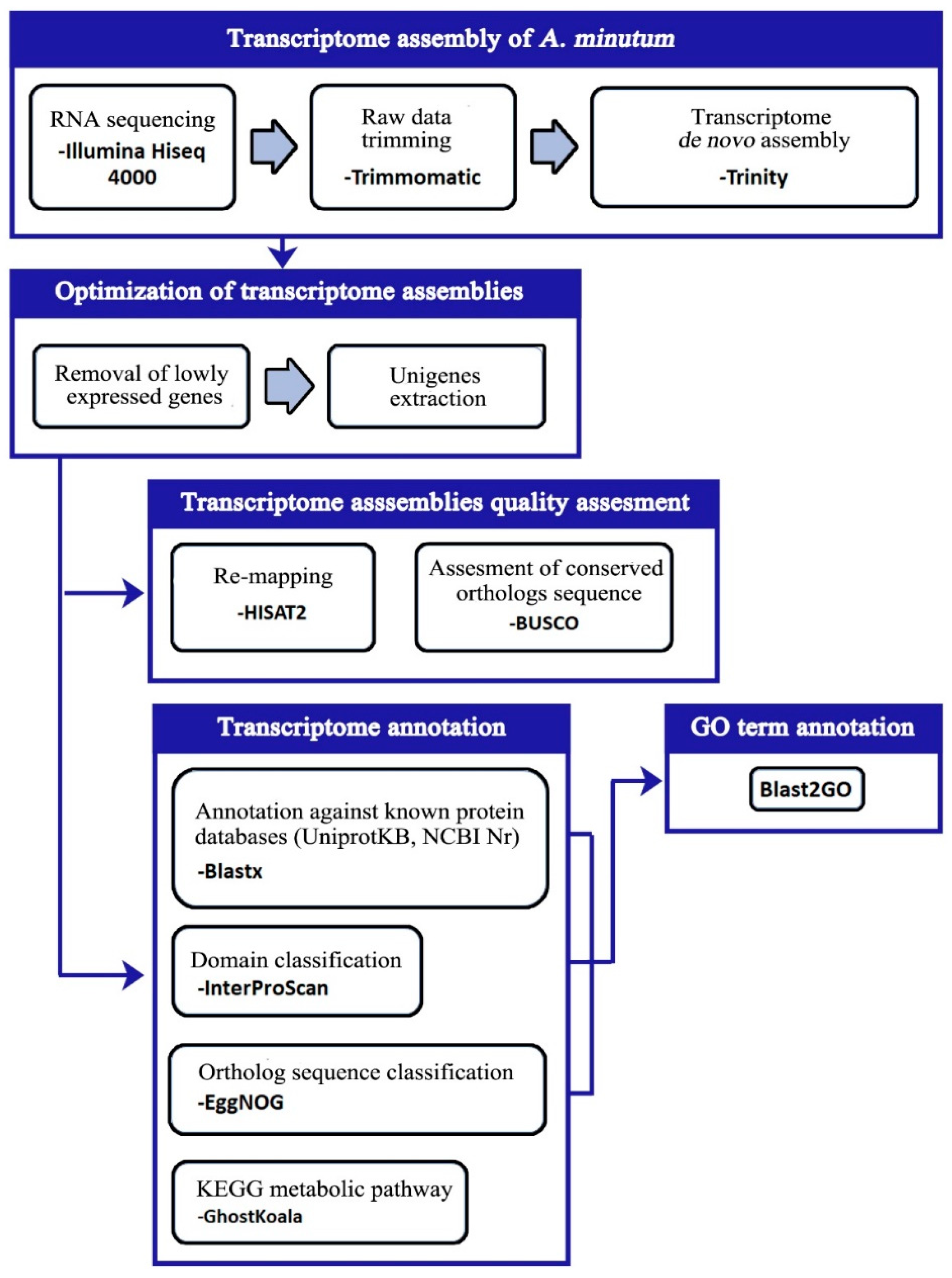
2.4. Transcriptome Assembly
2.5. Quality Evaluation of Transcriptome Assemblies
2.6. Prediction of the Protein-Coding Sequence
2.7. Transcriptome Annotation
2.8. Identification of Putative Saxitoxin Genes
3. Results and Discussions
3.1. Transcriptome Sequencing and De Novo Assembly
3.2. Coding Region Prediction
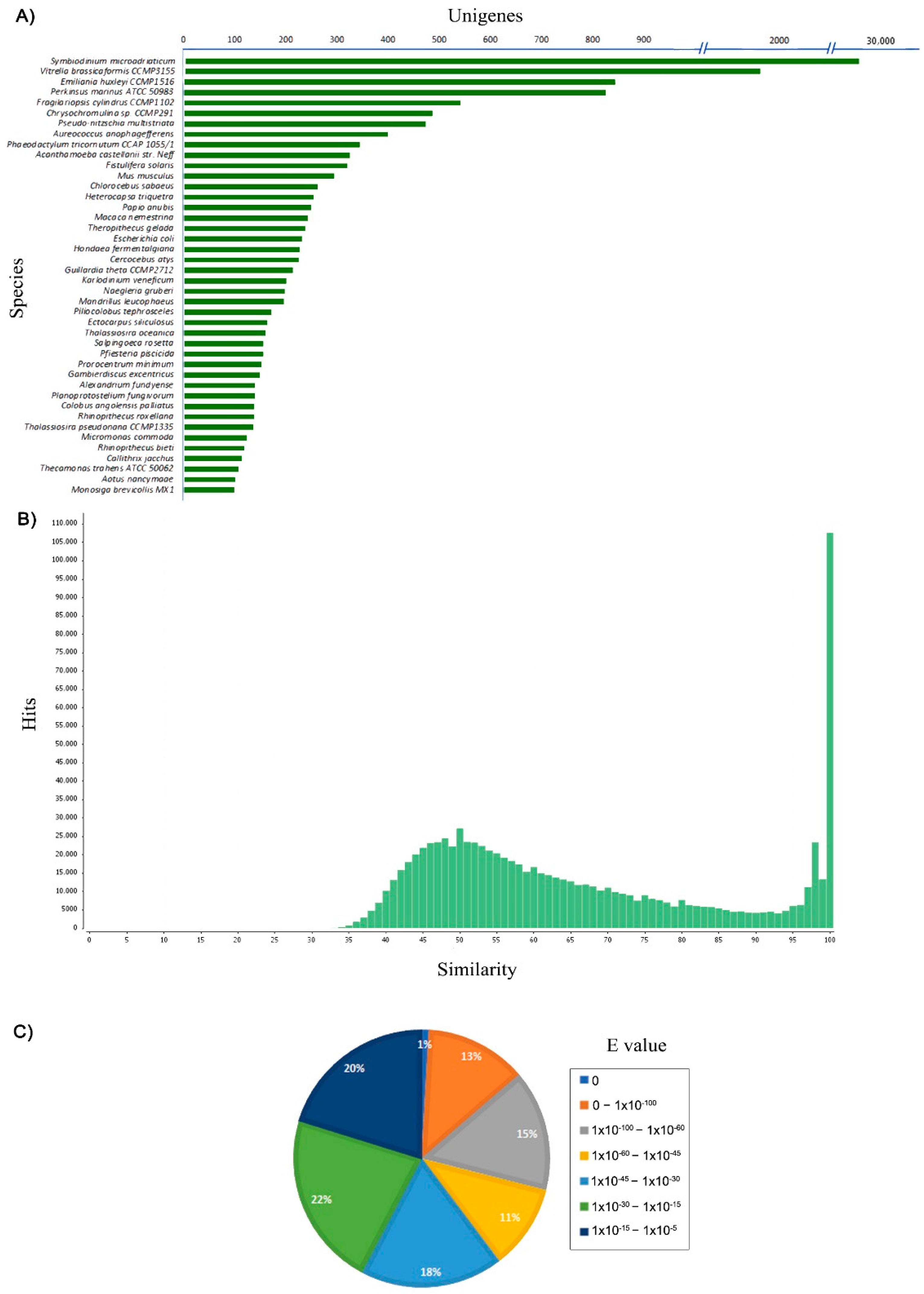
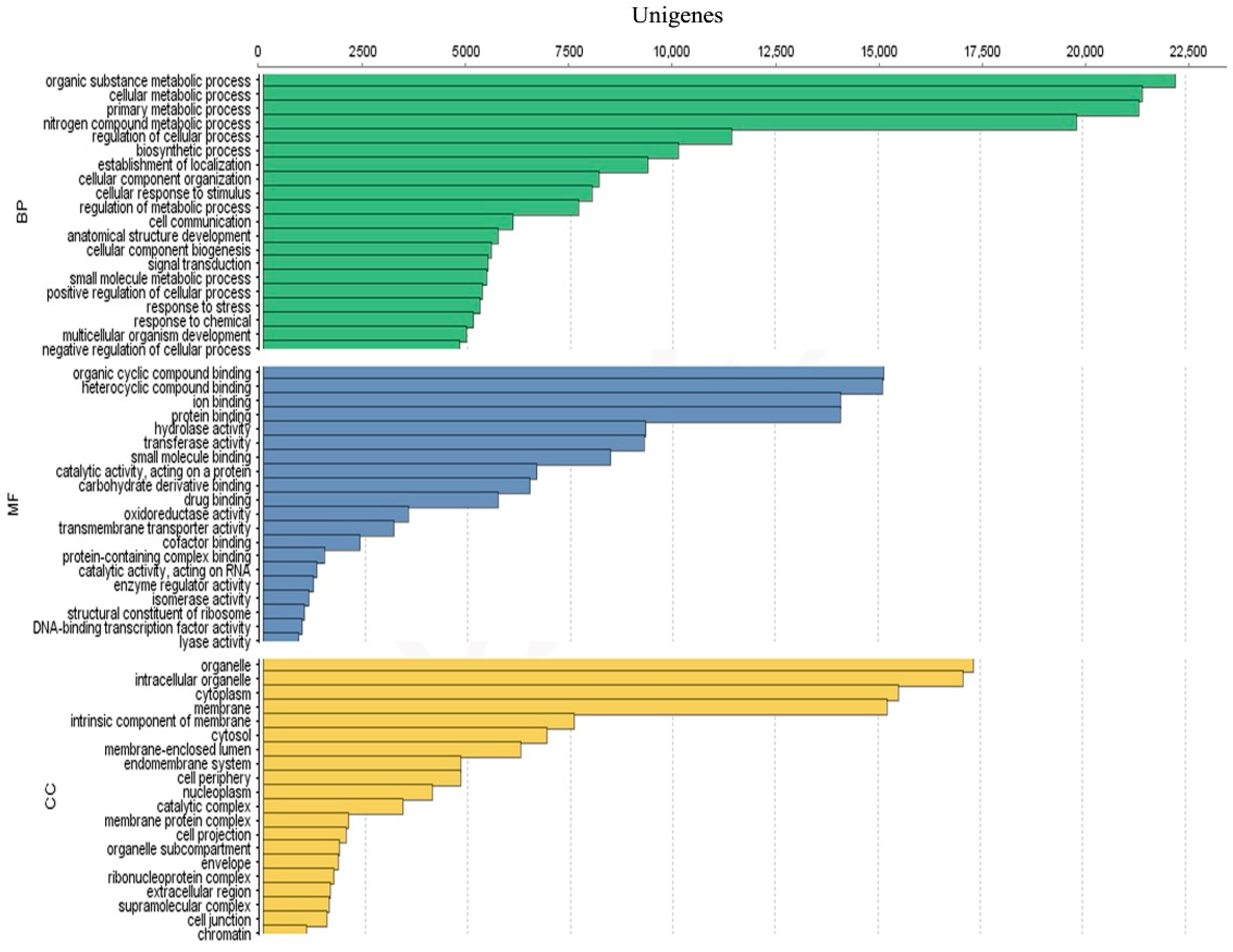
3.3. Unigenes Annotation
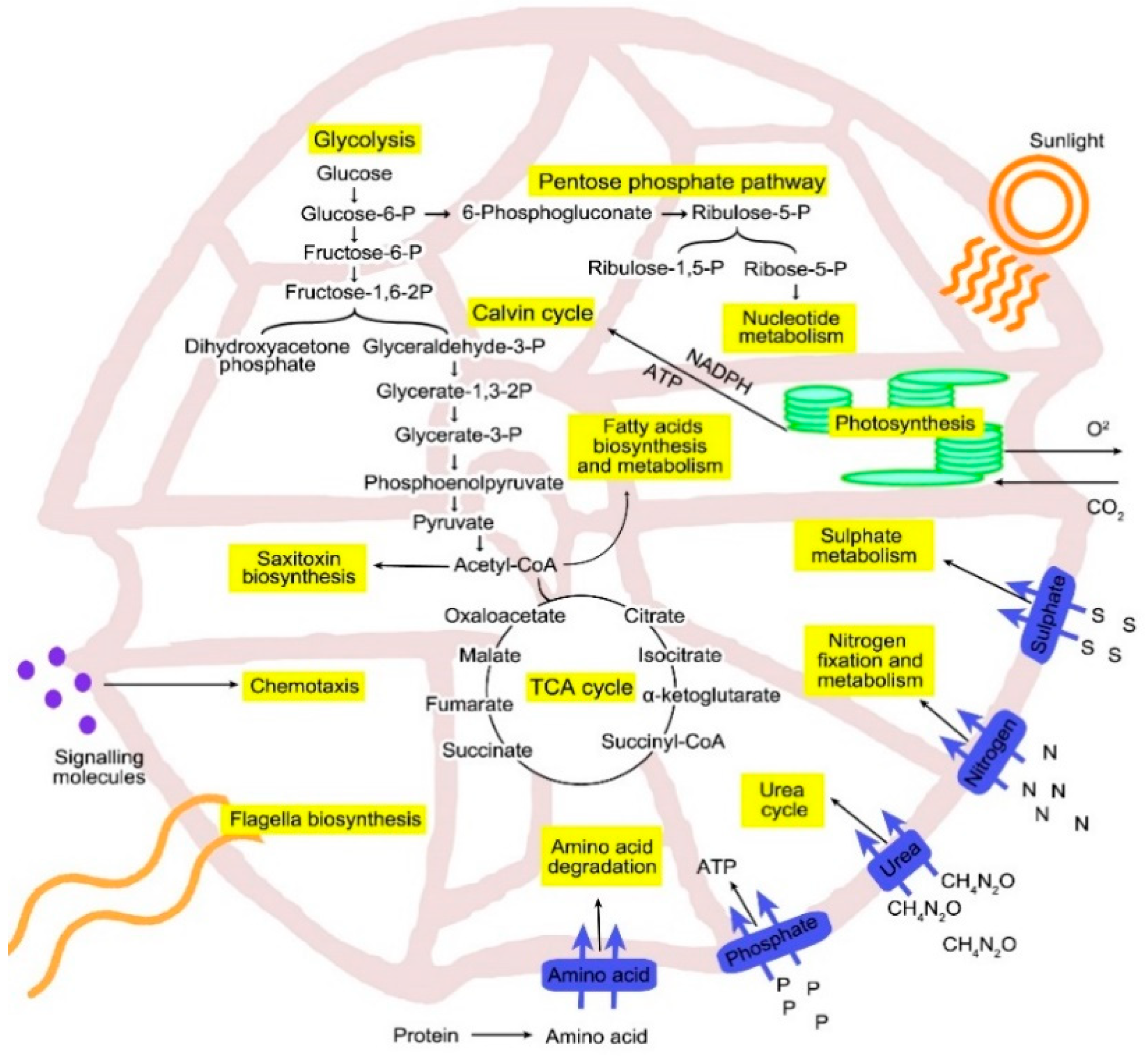
3.4. Carbon Metabolism
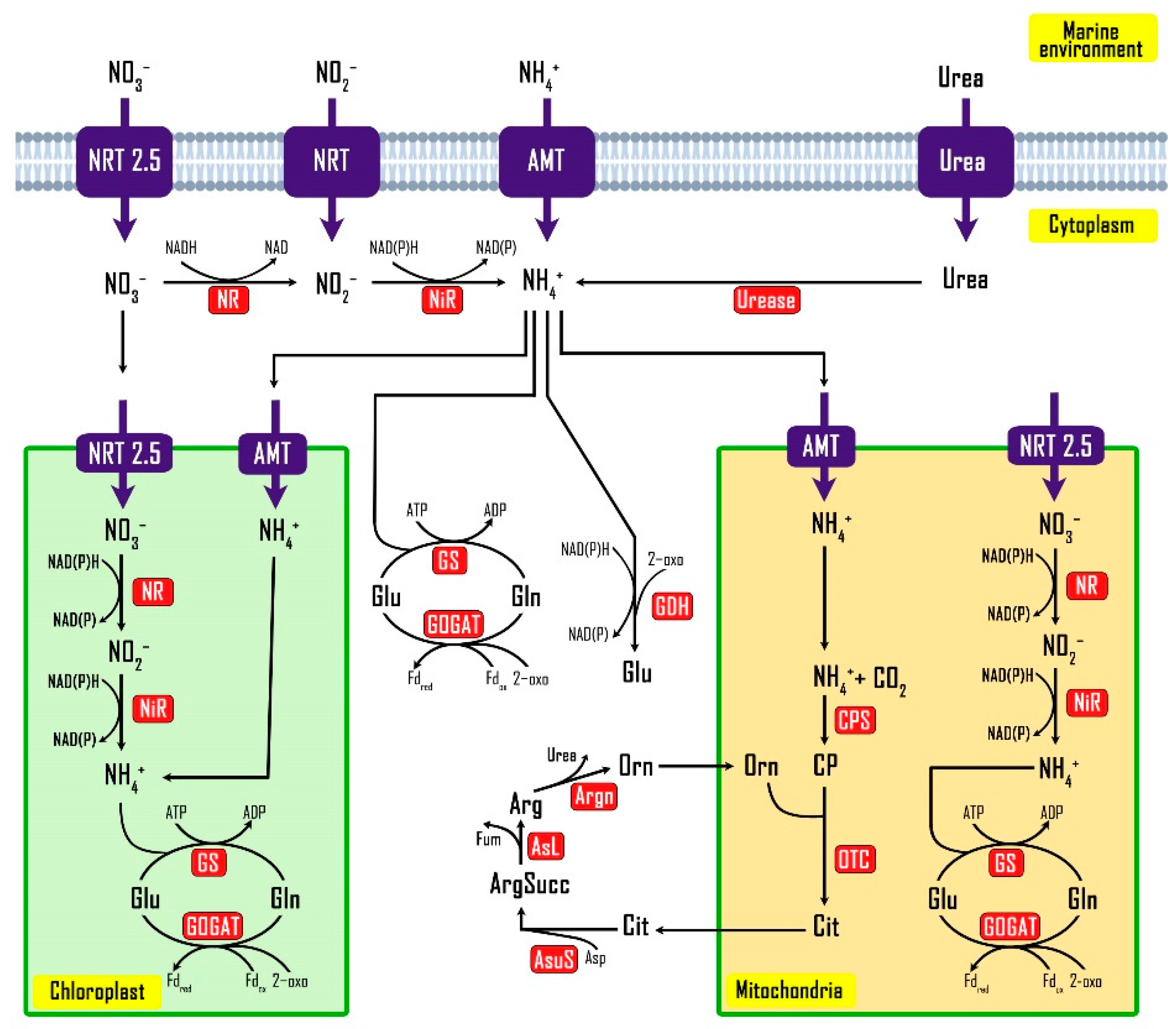
3.5. Nitrogen Metabolism
3.6. Phosphorus Metabolism
3.7. Saxitoxin Biosynthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 2011, 162, 551–569. [Google Scholar] [CrossRef]
- Hou, Y.; Lin, S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: Gene content estimation for dinoflagellate genomes. PLoS ONE 2009, 4, e6978. [Google Scholar] [CrossRef] [Green Version]
- Akbar, M.A.; Ahmad, A.; Usup, G.; Bunawan, H. Current knowledge and recent advances in marine dinoflagellate transcriptomic research. J. Mar. Sci. Eng. 2018, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Akbar, M.A.; Ahmad, A.; Usup, G.; Bunawan, H. RNA-seq as an emerging tool for marine dinoflagellate transcriptome analysis: Process and challenges. Processes 2018, 6, 5. [Google Scholar] [CrossRef] [Green Version]
- LaJeunesse, T.C.; Lambert, G.; Andersen, R.A.; Coffroth, M.A.; Galbraith, D.W. Symbiodinium (Pyrrhophyta) genome sizes (DNA Content) are smallest among dinoflagellates. J. Phycol. 2005, 41, 880–886. [Google Scholar] [CrossRef]
- Yu, L.; Li, T.; Li, L.; Lin, X.; Li, H.; Liu, C.; Guo, C.; Lin, S. SAGER: A database of Symbiodiniaceae and Algal Genomic Resource. Database 2020, 2020, baaa051. [Google Scholar] [CrossRef]
- Stephens, T.G.; González-Pech, R.A.; Cheng, Y.; Mohamed, A.; Burt, D.W.; Bhattacharya, D.; Ragan, M.A.; Chan, C.X. Genomes of the dinoflagellate Polarella glacialis encode tandemly repeated single-exon genes with adaptive functions. BMC Biol. 2020, 18, 1–21. [Google Scholar] [CrossRef]
- Beedessee, G.; Kubota, T.; Arimoto, A.; Nishitsuji, K.; Waller, R.F.; Hisata, K.; Yamasaki, S.; Satoh, N.; Kobayashi, J.; Shoguchi, E. Integrated omics unveil the secondary metabolic landscape of a basal dinoflagellate. BMC Biol. 2020, 18, 1–16. [Google Scholar] [CrossRef]
- John, U.; Lu, Y.; Wohlrab, S.; Groth, M.; Janouškovec, J.; Kohli, G.S.; Glöckner, G. An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Sci. Adv. 2019, 5, eaav1110. [Google Scholar] [CrossRef] [Green Version]
- Shoguchi, E.; Shinzato, C.; Kawashima, T.; Gyoja, F.; Mungpakdee, S.; Koyanagi, R.; Hamada, M. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013, 23, 1399–1408. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Stephens, T.G.; González-Pech, R.A.; Beltran, V.H.; Lapeyre, B.; Bongaerts, P.; Chan, C.X. Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 2018, 1, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Akbar, M.A.; Yusuf, N.Y.M.; Tahir, N.I.; Ahmad, A.; Usup, G.; Sahrani, F.K.; Bunawan, H. Biosynthesis of saxitoxin in marine dinoflagellates: An omics perspective. Mar. Drugs 2020, 18, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Gac, M.; Metegnier, G.; Chomérat, N.; Malestroit, P.; Quéré, J.; Bouchez, O.; Chapelle, A. Evolutionary processes and cellular functions underlying divergence in Alexandrium minutum. Mol. Ecol. 2016, 25, 5129–5143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, F.Q.; Song, J.T.; Zhou, J.; Cai, Z.H. Transcriptomic profile and sexual reproduction-relevant genes of Alexandrium minutum in response to nutritional deficiency. Front. Microbiol. 2019, 10, 2629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geffroy, S.; Lechat, M.M.; Le Gac, M.; Rovillon, G.A.; Marie, D.; Bigeard, E.; Caruana, A. From the sxtA4 gene to saxitoxin production: What controls the variability among Alexandrium minutum and Alexandrium pacificum Strains? Front. Microbiol. 2021, 12, 341. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kim, H.; Ki, J.S. Transcriptome survey, molecular identification, and expression analysis of stress-responsive genes in the toxic dinoflagellate Alexandrium pacificum under algicidal agents and metal stresses. J. Appl. Phycol. 2021, 33, 1–13. [Google Scholar]
- Vingiani, G.M.; Štālberga, D.; De Luca, P.; Ianora, A.; De Luca, D.; Lauritano, C. De novo transcriptome of the non-saxitoxin producing Alexandrium tamutum reveals new insights on harmful dinoflagellates. Mar. Drugs 2020, 18, 386. [Google Scholar] [CrossRef]
- Wang, H.; Kim, H.; Ki, J.S. Transcriptome survey and toxin measurements reveal evolutionary modification and loss of saxitoxin biosynthesis genes in the dinoflagellates Amphidinium carterae and Prorocentrum micans. Ecotoxicol. Environ. Saf. 2020, 195, 110474. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.F.; Lin, L.; Wang, D.Z. Comparative transcriptome analysis of a toxin-producing dinoflagellate Alexandrium catenella and its non-toxic mutant. Mar. Drugs 2014, 12, 5698–5718. [Google Scholar] [CrossRef] [Green Version]
- Lim, P.T.; Leaw, C.P.; Usup, G. Harmful algal blooms in Malaysian waters. Sains Malays. 2012, 41, 1509–1515. [Google Scholar]
- Kokinos, J.P.; Anderson, D.M. Morphological development of resting cysts in cultures of the marine dinoflagellate Lingulodinium polyedrum (= L. machaerophorum). Palynology 1995, 19, 143–166. [Google Scholar] [CrossRef]
- Mohd-Din, M.; Abdul-Wahab, M.F.; Mohamad, S.E.; Jamaluddin, H.; Shahir, S.; Ibrahim, Z.; Lim, P.T. Prolonged high biomass diatom blooms induced formation of hypoxic-anoxic zones in the inner part of Johor Strait. Environ. Sci. Pollut. Res. Int. 2020, 27, 42948–42959. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; MacManes, M.D. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Tong, G.X.; Xu, W.; Zhang, Y.Q.; Zhang, Q.Y.; Yin, J.S.; Kuang, Y.Y. De novo assembly and characterization of the Hucho taimen transcriptome. Ecol. Evol. 2018, 8, 1271–1285. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. Busco: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinform. 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.; McAnulla, C.; Pesseat, S. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Von Mering, C. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Kellmann, R.; Mihali, T.K.; Jeon, Y.J.; Pickford, R.; Pomati, F.; Neilan, B.A. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 2008, 74, 4044–4053. [Google Scholar] [CrossRef] [Green Version]
- Galachyants, Y.P.; Zakharova, Y.R.; Volokitina, N.A.; Morozov, A.A.; Likhoshway, Y.V.; Grachev, M.A. De novo transcriptome assembly and analysis of the freshwater araphid diatom Fragilaria radians, Lake Baikal. Sci. Data 2019, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B.; et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Cheng, S.; Song, B.; Zhong, X.; Lin, X.; Li, W.; Cai, M. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 2015, 350, 691–694. [Google Scholar] [CrossRef] [Green Version]
- Shoguchi, E.; Beedessee, G.; Tada, I.; Hisata, K.; Kawashima, T.; Takeuchi, T.; Arakaki, N.; Fujie, M.; Koyanagi, R.; Roy, M.C. Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genom. 2018, 19, 458. [Google Scholar] [CrossRef] [Green Version]
- Cooper, J.T.; Sinclair, G.A.; Wawrik, B. Transcriptome analysis of Scrippsiella trochoidea CCMP 3099 reveals physiological changes related to nitrate depletion. Front. Microbiol. 2016, 7, 639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, S.H.; Jeong, H.J.; Chon, J.K. De novo transcriptome of the newly described phototrophic dinoflagellate Yihiella yeosuensis: Comparison between vegetative cells and cysts. Mar. Biol. 2019, 166, 104. [Google Scholar] [CrossRef]
- Lauritano, C.; De Luca, D.; Ferrarini, A.; Avanzato, C.; Minio, A.; Esposito, F.; Ianora, A. De novo transcriptome of the cosmopolitan dinoflagellate Amphidinium carterae to identify enzymes with biotechnological potential. Sci. Rep. 2017, 7, 11701. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.; Lin, X.; Li, L.; Li, M.; Palenik, B.; Lin, S. Transcriptomic and microRNAomic profiling reveals multi-faceted mechanisms to cope with phosphate stress in a dinoflagellate. ISME J. 2017, 11, 2209–2218. [Google Scholar] [CrossRef] [Green Version]
- Van Dolah, F.M.; Morey, J.S.; Milne, S.; Ung, A.; Anderson, P.E.; Chinain, M. Transcriptomic analysis of polyketide synthases in a highly ciguatoxic dinoflagellate, Gambierdiscus polynesiensis and low toxicity Gambierdiscus pacificus, from French Polynesia. PLoS ONE 2020, 15, e0231400. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Schreiber, K.; Appel, J.; Makowka, A.; Fähnrich, B.; Roettger, M.; Gutekunst, K. The Entner–Doudoroff pathway is an overlooked glycolytic route in cyanobacteria and plants. Proc. Natl. Acad. Sci. USA 2016, 113, 5441–5446. [Google Scholar] [CrossRef] [Green Version]
- Hildebrand, M.; Abbriano, R.M.; Polle, J.E.; Traller, J.C.; Trentacoste, E.M.; Smith, S.R.; Davis, A.K. Metabolic and cellular organization in evolutionarily diverse microalgae as related to biofuels production. Curr. Opin. Chem. Biol. 2013, 17, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.R.; Abbriano, R.M.; Hildebrand, M. Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon partitioning pathways. Algal Res. 2012, 1, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Obata, T.; Fernie, A.R.; Nunes-Nesi, A. The central carbon and energy metabolism of marine diatoms. Metabolites 2013, 3, 325–346. [Google Scholar] [CrossRef] [PubMed]
- Andersson, I.; Backlund, A. Structure and function of Rubisco. Plant. Physiol. Biochem. 2008, 46, 275–291. [Google Scholar] [CrossRef]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Brzezinski, M.A. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derelle, E.; Ferraz, C.; Rombauts, S.; Rouzé, P.; Worden, A.Z.; Robbens, S.; Saeys, Y. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 2006, 103, 11647–11652. [Google Scholar] [CrossRef] [Green Version]
- Kroth, P.G.; Chiovitti, A.; Gruber, A.; Martin-Jezequel, V.; Mock, T.; Parker, M.S.; Maheswari, U. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS ONE 2008, 3, e1426. [Google Scholar] [CrossRef] [Green Version]
- Worden, A.Z.; Lee, J.H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Foulon, E. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef] [Green Version]
- Gong, W.; Browne, J.; Hall, N.; Schruth, D.; Paerl, H.; Marchetti, A. Molecular insights into a dinoflagellate bloom. ISME J. 2017, 11, 439–452. [Google Scholar] [CrossRef]
- Sánchez, B.; Zúñiga, M.; González-Candelas, F.; Clara, G.; Margolles, A. Bacterial and eukaryotic phosphoketolases: Phylogeny, distribution and evolution. J. Mol. Microbiol. Biotechnol. 2010, 18, 37–51. [Google Scholar] [CrossRef]
- Fabris, M.; Matthijs, M.; Rombauts, S.; Vyverman, W.; Goossens, A.; Baart, G.J. The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner–Doudoroff glycolytic pathway. Plant. J. 2012, 70, 1004–1014. [Google Scholar] [CrossRef]
- Flamholz, A.; Noor, E.; Bar-Even, A.; Liebermeister, W.; Milo, R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl. Acad. Sci. USA 2013, 110, 10039–10044. [Google Scholar] [CrossRef] [Green Version]
- Peyraud, R.; Cottret, L.; Marmiesse, L.; Genin, S. Control of primary metabolism by a virulence regulatory network promotes robustness in a plant pathogen. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jyoti, P.; Shree, M.; Joshi, C.; Prakash, T.; Ray, S.K.; Satapathy, S.S.; Masakapalli, S.K. The Entner-Doudoroff and nonoxidative pentose phosphate pathways bypass glycolysis and the oxidative pentose phosphate pathway in Ralstonia solanacearum. MSystems 2020, 5, e00091-20. [Google Scholar] [CrossRef] [Green Version]
- Mulholland, M.R.; Morse, R.; Egerton, T.; Bernhardt, P.W.; Filippino, K.C. Blooms of dinoflagellate mixotrophs in a lower Chesapeake Bay tributary: Carbon and nitrogen uptake over diurnal, seasonal, and interannual timescales. Estuaries Coasts 2018, 41, 1744–1765. [Google Scholar] [CrossRef] [Green Version]
- Griffin, J.E.; Park, G.; Dam, H.G. Relative importance of nitrogen sources, algal alarm cues and grazer exposure to toxin production of the marine dinoflagellate Alexandrium catenella. Harmful Algae 2019, 84, 181–187. [Google Scholar] [CrossRef]
- Chakraborty, S.; Pančić, M.; Andersen, K.H.; Kiørboe, T. The cost of toxin production in phytoplankton: The case of PST producing dinoflagellates. ISME J. 2019, 13, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Collos, Y.; Vaquer, A.; Laabir, M.; Abadie, E.; Laugier, T.; Pastoureaud, A.; Souchu, P. Contribution of several nitrogen sources to growth of Alexandrium catenella during blooms in Thau lagoon, southern France. Harmful Algae 2007, 6, 781–789. [Google Scholar] [CrossRef] [Green Version]
- Li, T.S.; Yu, R.C.; Zhou, M.J. Short-term effects of different nitrogen substrates on growth and toxin production of dinoflagellate Alexandrium catenella Balech (strain ACDH). Harmful Algae 2011, 12, 46–54. [Google Scholar] [CrossRef]
- Jing, X.; Lin, S.; Zhang, H.; Koerting, C.; Yu, Z. Utilization of urea and expression profiles of related genes in the dinoflagellate Prorocentrum donghaiense. PLoS ONE 2017, 12, e0187837. [Google Scholar] [CrossRef] [Green Version]
- Pechkovskaya, S.A.; Knyazev, N.A.; Matantseva, O.V.; Emelyanov, A.K.; Telesh, I.V.; Skarlato, S.O.; Filatova, N.A. Dur3 and nrt2 genes in the bloom-forming dinoflagellate Prorocentrum minimum: Transcriptional responses to available nitrogen sources. Chemosphere 2020, 241, 125083. [Google Scholar] [CrossRef]
- Bellefeuille, S.D.; Morse, D. The main nitrate transporter of the dinoflagellate Lingulodinium polyedrum is constitutively expressed and not responsible for daily variations in nitrate uptake rates. Harmful Algae 2016, 55, 272–281. [Google Scholar] [CrossRef]
- Cuartero, M.; Crespo, G.; Cherubini, T.; Pankratova, N.; Confalonieri, F.; Massa, F.; Bakker, E. In situ detection of macronutrients and chloride in seawater by submersible electrochemical sensors. Anal. Chem. 2018, 90, 4702–4710. [Google Scholar] [CrossRef] [Green Version]
- Dagenais-Bellefeuille, S.; Morse, D. Putting the N in dinoflagellates. Front. Microbiol. 2013, 4, 369. [Google Scholar] [CrossRef] [Green Version]
- Glibert, P.M.; Wilkerson, F.P.; Dugdale, R.C.; Raven, J.A.; Dupont, C.L.; Leavitt, P.R.; Kana, T.M. Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol. Oceanogr. 2016, 61, 165–197. [Google Scholar] [CrossRef]
- Allen, A.E.; Dupont, C.L.; Oborník, M.; Horák, A.; Nunes-Nesi, A.; McCrow, J.P.; Bowler, C. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 2011, 473, 203–207. [Google Scholar] [CrossRef]
- Ou, L.J.; Huang, K.X.; Li, J.J.; Jing, W.Y.; Dong, H.P. Transcriptomic responses of harmful dinoflagellate Prorocentrum donghaiense to nitrogen and light. Mar. Pollut. Bull. 2019, 149, 110617. [Google Scholar] [CrossRef]
- Price, D.C.; Farinholt, N.; Gates, C.; Shumaker, A.; Wagner, N.E.; Bienfang, P.; Bhattacharya, D. Analysis of Gambierdiscus transcriptome data supports ancient origins of mixotrophic pathways in dinoflagellates. Environ. Microbiol. 2016, 18, 4501–4510. [Google Scholar] [CrossRef]
- Lin, S.J.; Litaker, R.W.; Sunda, W.G. Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J. Phycol. 2016, 52, 10–36. [Google Scholar] [CrossRef]
- Falhof, J.; Pedersen, J.T.; Fuglsang, A.T.; Palmgren, M. Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant. 2016, 9, 323–337. [Google Scholar] [CrossRef] [Green Version]
- Haley, S.T.; Alexander, H.; Juhl, A.R.; Dyhrman, S.T. Transcriptional response of the harmful raphidophyte Heterosigma akashiwo to nitrate and phosphate stress. Harmful Algae 2017, 68, 258–270. [Google Scholar] [CrossRef]
- Hothorn, M.; Neumann, H.; Lenherr, E.D.; Wehner, M.; Rybin, V.; Hassa, P.O.; Ladurner, A.G. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 2009, 324, 513–516. [Google Scholar] [CrossRef]
- Stüken, A.; Orr, R.J.; Kellmann, R.; Murray, S.A.; Neilan, B.A.; Jakobsen, K.S. Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE 2011, 6, e20096. [Google Scholar] [CrossRef] [Green Version]
- Orr, R.J.S.; Stüken, A.; Murray, S.A.; Jakobsen, K.S. Evolution and distribution of saxitoxin biosynthesis in dinoflagellates. Mar. Drugs 2013, 11, 2814–2828. [Google Scholar] [CrossRef] [Green Version]
- Hackett, J.D.; Wisecaver, J.H.; Brosnahan, M.L.; Kulis, D.M.; Anderson, D.M.; Bhattacharya, D.; Erdner, D.L. Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates. Mol. Biol. Evol. 2013, 30, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, S.F.; Lin, L.; Wang, D.Z. Whole transcriptomic analysis provides insights into molecular mechanisms for toxin biosynthesis in a toxic dinoflagellate Alexandrium catenella (ACHK-T). Toxins 2017, 9, 213. [Google Scholar] [CrossRef] [Green Version]
| Statistic | Unigenes |
|---|---|
| Total unigenes | 124,977 |
| GC% | 6430 |
| N50 (bp) | 1566 |
| Unigenes median lenght (bp) | 468 |
| Unigenes mean lenght (bp) | 89,669 |
| Total base pairs | 112,065,669 |
| BUSCO score | 83.5% |
| Databases | Annotated Unigenes |
|---|---|
| NCBI-Nr | 58,802 (47.05%) |
| UniProtKB | 58,495 (46.80%) |
| EggNOG | 34,602 (27.69%) |
| KEGG | 18,381 (14.70%) |
| InterPro | 57,605 (46.09%) |
| Gene Ontology (GO) | 57,128 (45.71%) |
| Categories | Genes | Putative Function | Unigenes |
|---|---|---|---|
| Core genes | sxtA | Methylation, loading of ACP, Claisen condensation | 14 |
| sxtB | Cyclization | 1 | |
| sxtC | Regulatory | - | |
| sxtD | Desaturation | 1 | |
| sxtG | Amidinotransfer | 4 | |
| sxtH/T | C-12 hydroxylation | 21 | |
| sxtI | Carbamoylation | 5 | |
| sxtJ | Regulatory | 1 | |
| sxtK | Regulatory | - | |
| sxtS | Ring formation | 8 | |
| sxtU | Short-chain alcohol dehydrogenase | 131 | |
| sxtV | Dioxygenase reductase | 2 | |
| sxtW | Ferredoxin | 10 | |
| Modification genes | sxtL | Decarbamoylation | 1 |
| sxtN | Sulfotransferase | 1 | |
| sxtO | PAPS biosynthesis | 9 | |
| sxtX | N-1 hydroxylation | 3 | |
| Regulatory genes | sxtY | Signal transduction | - |
| sxtZ | Signal transduction | - | |
| Transporter genes | sxtF/M | Export of PSTs | 4 |
| sxtP | Binding of PSTs | 2 | |
| Unknown | sxtE | Unknown | - |
| sxtQ | Unknown | - | |
| sxtR | Unknown | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbar, M.A.; Yusof, N.Y.M.; Sahrani, F.K.; Usup, G.; Ahmad, A.; Baharum, S.N.; Muhammad, N.A.N.; Bunawan, H. Insights into Alexandrium minutum Nutrient Acquisition, Metabolism and Saxitoxin Biosynthesis through Comprehensive Transcriptome Survey. Biology 2021, 10, 826. https://doi.org/10.3390/biology10090826
Akbar MA, Yusof NYM, Sahrani FK, Usup G, Ahmad A, Baharum SN, Muhammad NAN, Bunawan H. Insights into Alexandrium minutum Nutrient Acquisition, Metabolism and Saxitoxin Biosynthesis through Comprehensive Transcriptome Survey. Biology. 2021; 10(9):826. https://doi.org/10.3390/biology10090826
Chicago/Turabian StyleAkbar, Muhamad Afiq, Nurul Yuziana Mohd Yusof, Fathul Karim Sahrani, Gires Usup, Asmat Ahmad, Syarul Nataqain Baharum, Nor Azlan Nor Muhammad, and Hamidun Bunawan. 2021. "Insights into Alexandrium minutum Nutrient Acquisition, Metabolism and Saxitoxin Biosynthesis through Comprehensive Transcriptome Survey" Biology 10, no. 9: 826. https://doi.org/10.3390/biology10090826
APA StyleAkbar, M. A., Yusof, N. Y. M., Sahrani, F. K., Usup, G., Ahmad, A., Baharum, S. N., Muhammad, N. A. N., & Bunawan, H. (2021). Insights into Alexandrium minutum Nutrient Acquisition, Metabolism and Saxitoxin Biosynthesis through Comprehensive Transcriptome Survey. Biology, 10(9), 826. https://doi.org/10.3390/biology10090826






