Contrasting Host-Parasite Population Structure: Morphology and Mitogenomics of a Parasitic Flatworm on Pelagic Deepwater Cichlid Fishes from Lake Tanganyika
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling
2.2. Morphology
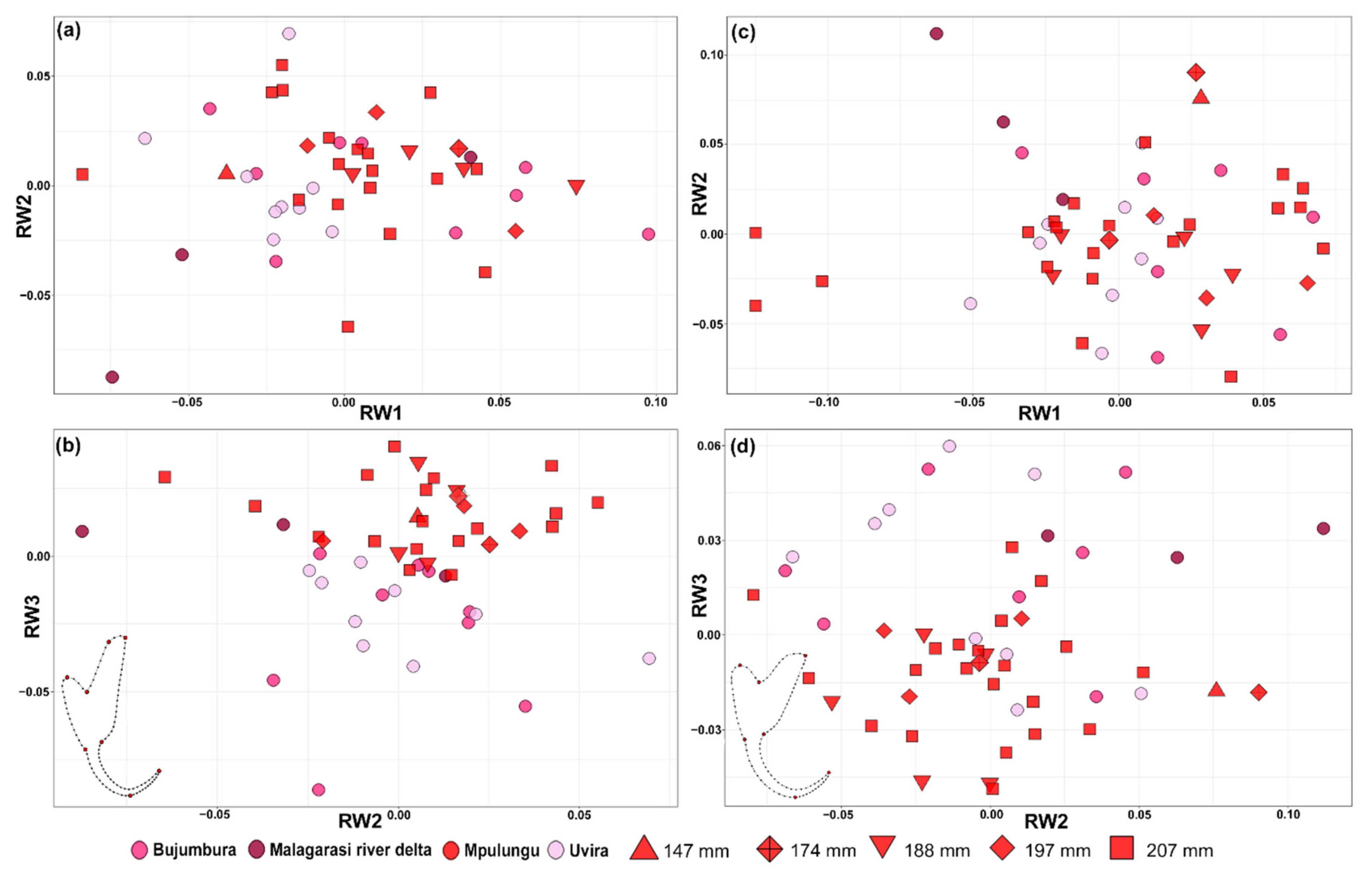
2.2.1. Morphometrics
2.2.2. Geomorphometrics
2.3. Genetics
2.3.1. Data Acquisition
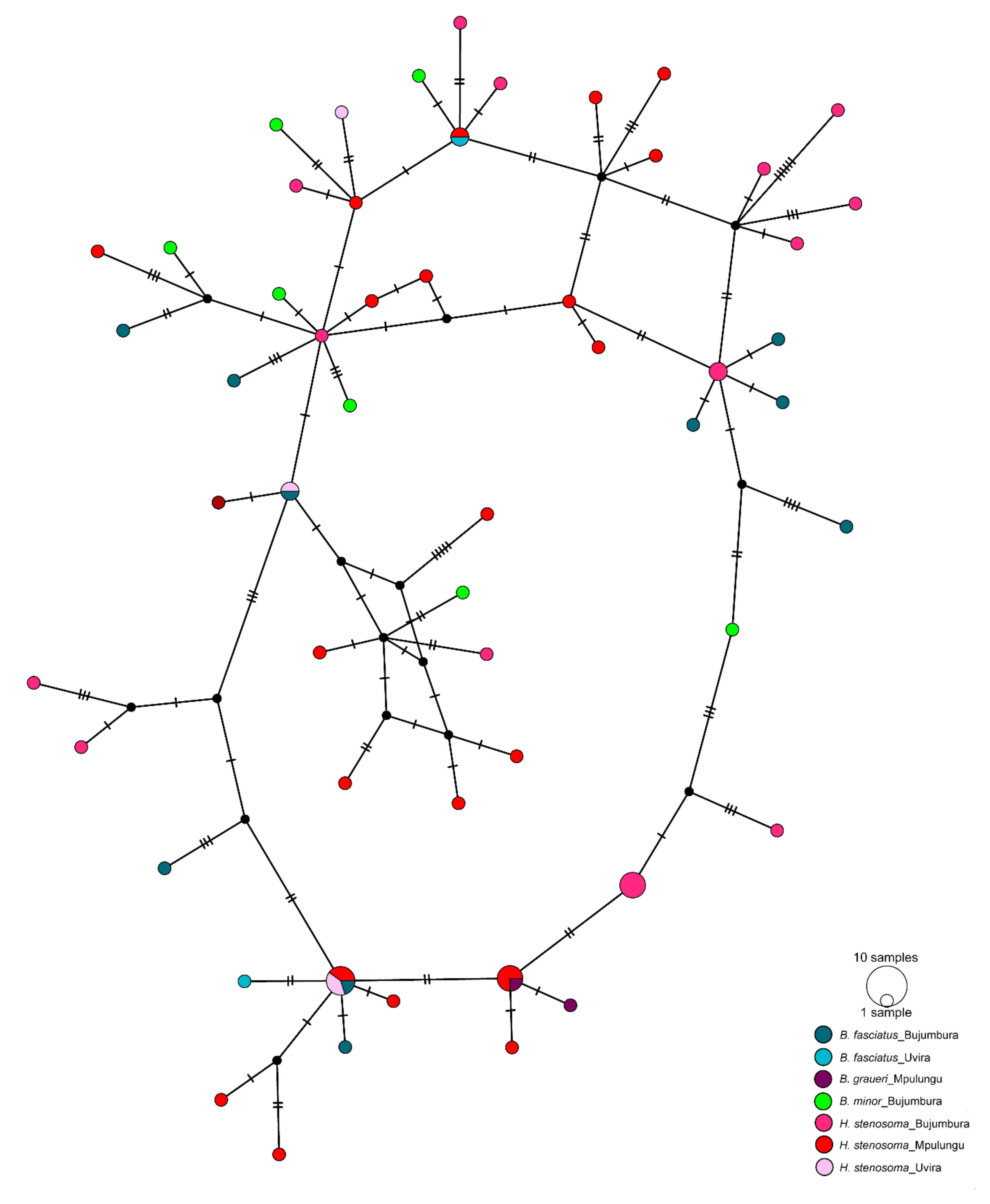
2.3.2. Population Genetic Analyses Based on cox1 Data
2.4. Genomics
2.4.1. Mitogenome Assembly and Annotation
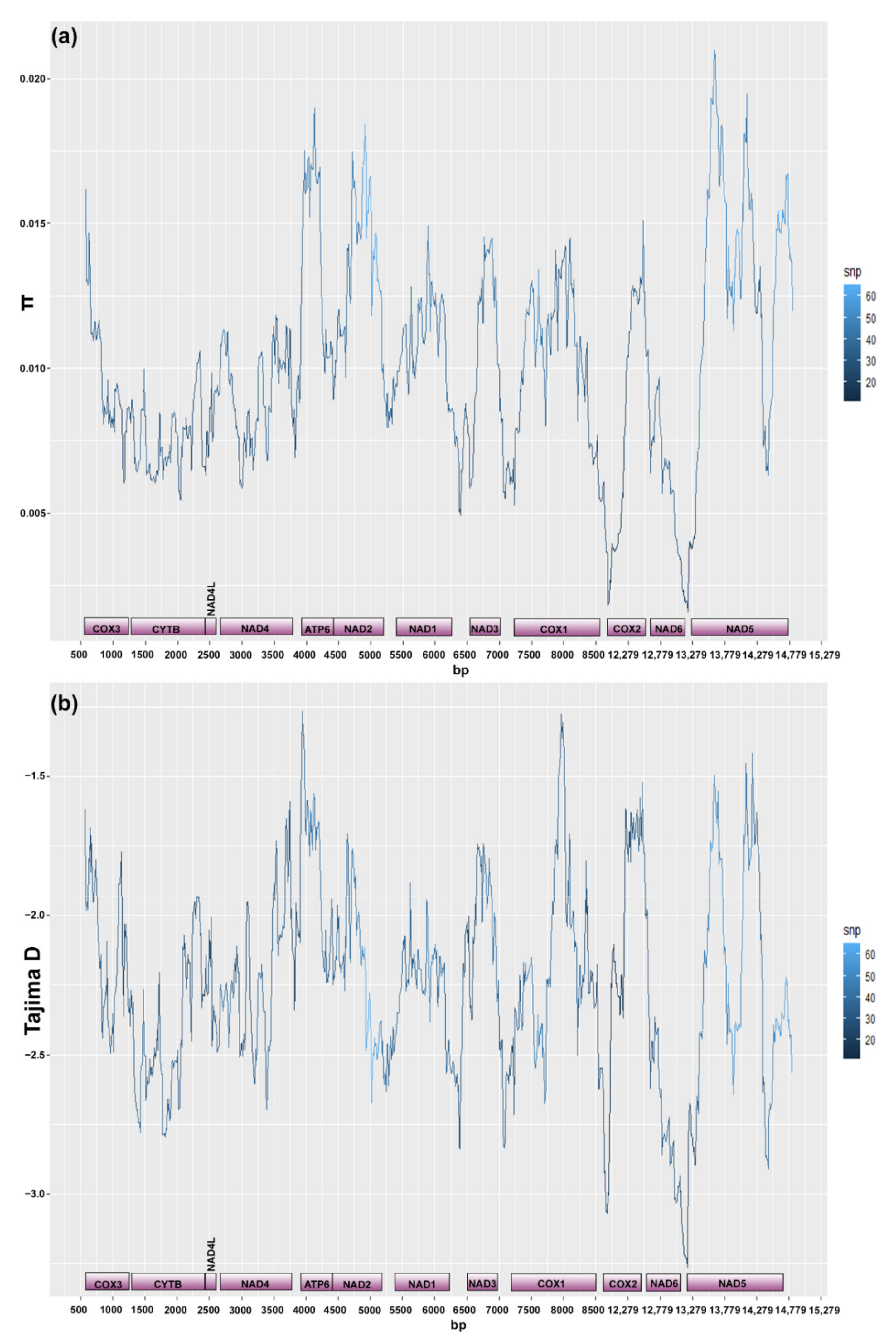
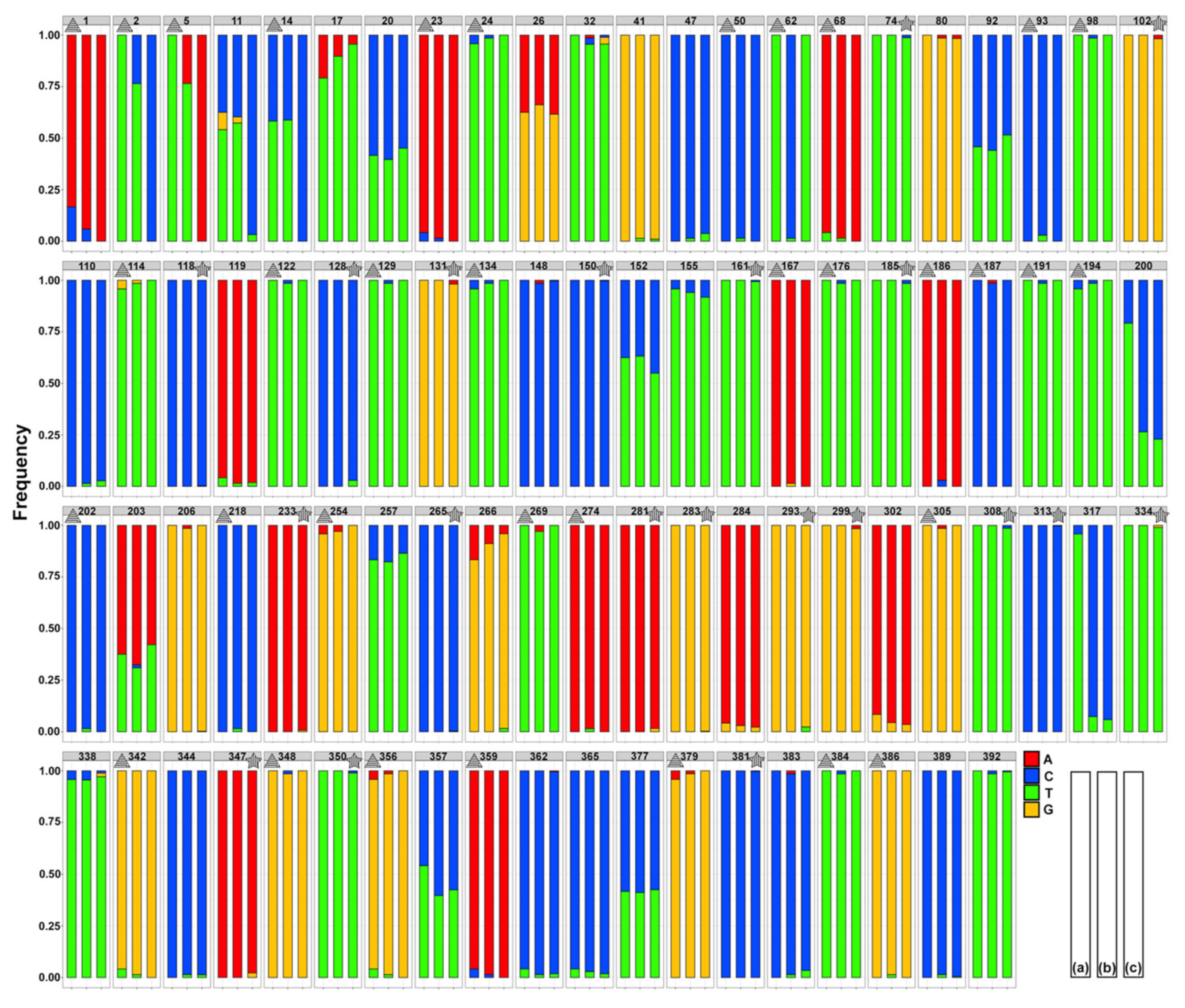
2.4.2. Mitogenome Diversity
2.4.3. Ribosomal Operon
3. Results
3.1. Morphological Variation
3.2. Genetic Diversity and Differentiation
3.3. Mitogenome and Nuclear Ribosomal Operon
4. Discussion
4.1. Morphological and Mitochondrial Diversity and Host Use in Cichlidogyrus casuarinus
4.2. Ribosomal Operon and Its Utility for Future Studies/Research
4.3. Mitochondrial Genome
4.3.1. Lake Tanganyika and the Rest of the Monogenean World
4.3.2. Intraspecific Variation at the Mitogenome Level
4.3.3. Methodological Implications for Future Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Angel, M.V. Biodiversity of the pelagic ocean. Conserv. Biol. 1993, 7, 760–772. [Google Scholar] [CrossRef]
- Lowe-McConnell, R. Fish communities in the African Great Lakes. Environ. Biol. Fishes 1996, 45, 219–235. [Google Scholar] [CrossRef]
- Vadeboncoeur, Y.; Mcintyre, P.B.; Jake Vander Zanden, M. Borders of biodiversity: Life at the edge of the World’s large lakes. Bioscience 2011, 61, 526–537. [Google Scholar] [CrossRef]
- Hellberg, M.E.; Burton, R.S.; Neigel, J.E.; Palumbi, S.R. Genetic assessment of connectivity among marine populations. Bull. Mar. Sci. 2002, 70, 273–290. [Google Scholar]
- Savoca, S.; Grifó, G.; Panarello, G.; Albano, M.; Giacobbe, S.; Capillo, G.; Spanó, N.; Consolo, G. Modelling prey-predator interactions in Messina beachrock pools. Ecol. Modell. 2020, 434, 109206. [Google Scholar] [CrossRef]
- Norris, R.D. Pelagic species diversity, biogeography, and evolution. Paleobiology 2000, 26, 236–258. [Google Scholar] [CrossRef]
- Præbel, K.; Knudsen, R.; Siwertsson, A.; Karhunen, M.; Kahilainen, K.K.; Ovaskainen, O.; Østbye, K.; Peruzzi, S.; Fevolden, S.-E.; Amundsen, P.-A. Ecological speciation in postglacial European whitefish: Rapid adaptive radiations into the littoral, pelagic, and profundal lake habitats. Ecol. Evol. 2013, 3, 4970–4986. [Google Scholar] [CrossRef] [PubMed]
- Boxshall, G.A. Host specificity in copepod parasites of deep-sea fishes. J. Mar. Syst. 1998, 15, 215–223. [Google Scholar] [CrossRef]
- Bray, R.A.; Littlewood, D.T.J.; Herniou, E.A.; Williams, B.; Henderson, R.E. Digenean parasites of deep-sea teleosts: A review and case studies of intrageneric phylogenies. Parasitology 1999, 119, S125–S144. [Google Scholar] [CrossRef]
- Campbell, R.A.; Headrich, R.L.; Munroe, T.A. Parasitism and ecological relationships among deep-see benthic fishes. Mar. Biol. 1980, 57, 301–313. [Google Scholar] [CrossRef]
- Klimpel, S.; Busch, M.W.; Sutton, T.; Palm, H.W. Meso- and bathy-pelagic fish parasites at the Mid-Atlantic Ridge (MAR): Low host specificity and restricted parasite diversity. Deep. Res. Part. I Oceanogr. Res. Pap. 2010, 57, 596–603. [Google Scholar] [CrossRef]
- Klimpel, S.; Busch, M.W.; Kellermans, E.; Kleinertz, S.; Palm, H.W. Metazoan Deep-Sea Fish Parasites; Verlag Natur & Wissenschaft: Solingen, Germany, 2009; Volume 11, ISBN 978-3-936616-61-3. [Google Scholar]
- Klimpel, S.; Palm, H.W.; Busch, M.W.; Kellermanns, E.; Rückert, S. Fish parasites in the Arctic deep-sea: Poor diversity in pelagic fish species vs. heavy parasite load in a demersal fish. Deep Sea Res. Part I Oceanogr. Res. Pap. 2006, 53, 1167–1181. [Google Scholar] [CrossRef]
- Mauchline, J.; Gordon, J.D.M. Incidence of parasitic worms in stomachs of pelagic and demersal fish of the Rockall Trough, northeastern Atlantic Ocean. J. Fish. Biol. 1984, 24, 281–285. [Google Scholar] [CrossRef]
- Nieberding, C.M.; Olivieri, I. Parasites: Proxies for host genealogy and ecology? Trends Ecol. Evol. 2007, 22, 156–165. [Google Scholar] [CrossRef]
- Barson, M.; Přikrylová, I.; Vanhove, M.P.M.; Huyse, T. Parasite hybridization in African Macrogyrodactylus spp. (Monogenea, Platyhelminthes) signals historical host distribution. Parasitology 2010, 137, 1585–1595. [Google Scholar] [CrossRef]
- Criscione, C.D.; Cooper, B.; Blouin, M.S. Parasite genotypes identify source populations of migratory fish more accurately than fish genotypes. Ecology 2006, 87, 823–828. [Google Scholar] [CrossRef]
- Criscione, C.D.; Blouin, M.S. Parasite phylogeographical congruence with salmon host evolutionarily significant units: Implications for salmon conservation. Mol. Ecol. 2006, 16, 993–1005. [Google Scholar] [CrossRef]
- Hoberg, E.P. Phylogeny and historical reconstruction: Host-parasite systems as keystones in biogeography and ecology. In Biodiversity II: Understanding and Protecting Our Biological Resources; Reaka-Kudla, M.L., Wilson, D.E., Wilson, E.O., Henry, A.J., Eds.; Joseph Henry Press: Washington, DC, USA, 1997; pp. 243–261. ISBN 0-309-52075-4. [Google Scholar]
- Kearn, G.C. Parasitism and the Platyhelminths; Chapman & Hall Ltd.: London, UK, 1998. [Google Scholar]
- Kearn, G.C. Evolutionary expansion of the Monogenea. Int. J. Parasitol. 1994, 24, 1227–1271. [Google Scholar] [CrossRef]
- Hecky, R.E. The pelagic ecosystem. In Lake Tanganyika and Its Life; Coulter, G.W., Ed.; Natural History Musem & Oxford University Press: London, UK; Oxford, UK; New York, NY, USA, 1991; pp. 91–110. [Google Scholar]
- Mannini, P.; Aro, E.; Katonda, K.I.; Kassaka, B.; Mambona, C.; Milindi, G.; Paffen, P.; Verburg, P. Pelagic Fish Stocks of Lake Tanganyika: Biology and Exploitation. Research for the Management of the Fisheries of Lake Tanganyika; FAO: Rome, Italy; Finnish International Development Agency: Helsinki, Finland, 1996. [Google Scholar]
- Ronco, F.; Matschiner, M.; Böhne, A.; Boila, A.; Büscher, H.H.; El Taher, A.; Indermaur, A.; Malinsky, M.; Ricci, V.; Kahmen, A.; et al. Drivers and dynamics of a massive adaptive radiation in cichlid fishes. Nature 2021, 589, 76–81. [Google Scholar] [CrossRef]
- Salzburger, W. Understanding explosive diversification through cichlid fish genomics. Nat. Rev. Genet. 2018, 19, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, M.P.M.; Pariselle, A.; Van Steenberge, M.; Raeymaekers, J.A.M.; Hablützel, P.I.; Gillardin, C.; Hellemans, B.; Breman, F.C.; Koblmüller, S.; Sturmbauer, C.; et al. Hidden biodiversity in an ancient lake: Phylogenetic congruence between Lake Tanganyika tropheine cichlids and their monogenean flatworm parasites. Sci. Rep. 2015, 5, 13669. [Google Scholar] [CrossRef]
- Cruz-Laufer, A.J.; Artois, T.; Smeets, K.; Pariselle, A.; Vanhove, M.P.M. The cichlid–Cichlidogyrus network: A blueprint for a model system of parasite evolution. Hydrobiologia 2020, 848, 3847–3863. [Google Scholar] [CrossRef]
- Coulter, G.W. Pelagic Fish. In Lake Tanganyika and Its Life; Coulter, G.W., Ed.; Natural History Musem & Oxford University Press: London, UK; Oxford, UK; New York, NY, USA, 1991; pp. 111–150. [Google Scholar]
- Konings, A. Tanganyika Cichlids in their Natural Habitat, 4th ed.; Hollywood Import & Export Inc.: Gainesville, FL, USA, 2019; ISBN 9789328922607. [Google Scholar]
- Poulin, R. Parasite biodiversity revisited: Frontiers and constraints. Int. J. Parasitol. 2014, 44, 581–589. [Google Scholar] [CrossRef]
- Kmentová, N.; Gelnar, M.; Mendlová, M.; Van Steenberge, M.; Koblmüller, S.; Vanhove, M.P.M. Reduced host-specificity in a parasite infecting non-littoral Lake Tanganyika cichlids evidenced by intraspecific morphological and genetic diversity. Sci. Rep. 2016, 6, 39605. [Google Scholar] [CrossRef]
- Rahmouni, C.; Vanhove, M.P.M.; Šimková, A. Seven new species of Cichlidogyrus Paperna, 1960 (Monogenea: Dactylogyridae) parasitizing the gills of Congolese cichlids from northern Lake Tanganyika. PeerJ 2018, 6, e5604. [Google Scholar] [CrossRef] [PubMed]
- Justine, J.-L.; Beveridge, I.; Boxshall, G.A.; Bray, R.A.; Miller, T.L.; Moravec, F.; Trilles, J.-P.; Whittington, I.D. An annotated list of fish parasites (Isopoda, Copepoda, Monogenea, Digenea, Cestoda, Nematoda) collected from Snappers and Bream (Lutjanidae, Nemipteridae, Caesionidae) in New Caledonia confirms high parasite biodiversity on coral reef fish. Aquat. Biosyst. 2012, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Rohde, K. Host specificity indices of parasites and their application. Experientia 1980, 36, 1369–1371. [Google Scholar] [CrossRef]
- Schoelinck, C.; Cruaud, C.; Justine, J.-L. Are all species of Pseudorhabdosynochus strictly host specific? A molecular study. Parasitol. Int. 2012, 61, 356–359. [Google Scholar] [CrossRef] [PubMed]
- Kmentová, N.; Koblmüller, S.; Van Steenberge, M.; Raeymaekers, J.A.M.; Artois, T.; De Keyzer, E.L.R.; Milec, L.; Muterezi Bukinga, F.; N’sibula, T.M.; Mulungula, P.M.; et al. Weak population structure and recent demographic expansion of the monogenean parasite Kapentagyrus spp. infecting clupeid fishes of Lake Tanganyika, East Africa. Int. J. Parasitol. 2020, 50, 471–486. [Google Scholar] [CrossRef]
- Rahmouni, C.; Van Steenberge, M.; Vanhove, M.P.M.; Šimková, A. Intraspecific morphological variation in Cichlidogyrus (Monogenea) parasitizing two cichlid hosts from Lake Tanganyika exhibiting different dispersal capacities. Hydrobiologia 2021, 848, 3833–3845. [Google Scholar] [CrossRef]
- Mendlová, M.; Šimková, A. Evolution of host specificity in monogeneans parasitizing African cichlid fish. Parasit. Vectors 2014, 7, 69. [Google Scholar] [CrossRef]
- Pariselle, A.; Muterezi Bukinga, F.; Van Steenberge, M.; Vanhove, M.P.M. Ancyrocephalidae (Monogenea) of Lake Tanganyika: IV: Cichlidogyrus parasitizing species of Bathybatini (Teleostei, Cichlidae): Reduced host-specificity in the deepwater realm? Hydrobiologia 2015, 748, 99–119. [Google Scholar] [CrossRef]
- Fannes, W.; Vanhove, M.P.M.; Huyse, T.; Paladini, G. A scanning electron microscope technique for studying the sclerites of Cichlidogyrus. Parasitol. Res. 2015, 114, 2031–2034. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Bercial, L.; Bucklin, A. New view of population genetics of zooplankton: RAD-seq analysis reveals population structure of the North Atlantic planktonic copepod Centropages typicus. Mol. Ecol. 2016, 25, 1566–1580. [Google Scholar] [CrossRef]
- Kumar, G.; Kocour, M. Applications of next-generation sequencing in fisheries research: A review. Fish. Res. 2017, 186, 11–22. [Google Scholar] [CrossRef]
- Carlton, J.; Silva, J.; Hall, N. The genome of model malaria parasites, and comparative genomics. Curr. Issues Mol. Biol. 2005, 7, 23–37. [Google Scholar] [CrossRef]
- Young, N.D.; Jex, A.R.; Li, B.; Liu, S.; Yang, L.; Xiong, Z.; Li, Y.; Cantacessi, C.; Hall, R.S.; Xu, X.; et al. Whole-genome sequence of Schistosoma haematobium. Nat. Genet. 2012, 44, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Koblmüller, S.; Zangl, L.; Börger, C.; Daill, D.; Vanhove, M.P.M.; Sturmbauer, C.; Sefc, K.M. Only true pelagics mix: Comparative phylogeography of deepwater bathybatine cichlids from Lake Tanganyika. Hydrobiologia 2019, 832, 93–103. [Google Scholar] [CrossRef]
- Ergens, R.; Lom, J. Causative Agents of Fish Diseases; Academia: Prague, Czech Republic, 1970. [Google Scholar]
- Řehulková, E.; Mendlová, M.; Šimková, A. Two new species of Cichlidogyrus (Monogenea: Dactylogyridae) parasitizing the gills of African cichlid fishes (Perciformes) from Senegal: Morphometric and molecular characterization. Parasitol. Res. 2013, 112, 1399–1410. [Google Scholar] [CrossRef]
- Thioulouse, J.; Dufour, A.B.; Jombart, T.; Dray, S.; Siberchicot, A.; Pavoine, S. Multivariate Analysis of Ecological Data with Ade4; Springer: New York, NY, USA, 2018; ISBN 9781493988501. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; The R Foundation for Statistical Computing: Vienna, Austria, 2019; ISBN 3-900051-07-0. [Google Scholar]
- Rohlf, F. Tpsdig, Digitize Landmarks and Outlines, Version 2.10; State University: Stony Brook, NY, USA, 2006. [Google Scholar]
- Cox, C.F.; Cox, M.A.A. Procrustes analysis. In Multidimentional Scaling, 2nd ed.; Cox, C.F., Cox, M.A.A., Eds.; Chapman & Hall: London, UK, 1989; pp. 123–139. ISBN 9780412406508. [Google Scholar]
- Zelditch, M.; Swiderski, D.L.; Sheets, H.D. Geometric Morphometrics for Biologists: A Primer; Elsevier: London, UK, 2012; ISBN 9780123869043. [Google Scholar]
- Klingenberg, C.P. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef]
- Rohlf, F.J.; Marcus, L.F. A revolution morphometrics. Trends Ecol. Evol. 1993, 8, 129–132. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98140-6. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Littlewood, D.T.J.; Rohde, K.; Clough, K.A. Parasite speciation within or between host species? Phylogenetic evidence from site-specific polystome monogeneans. Int. J. Parasitol. 1997, 27, 1289–1297. [Google Scholar] [CrossRef]
- Lockyer, A.E.; Olson, P.D.; Littlewood, D.T.J. Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): Implications and a review of the cercomer theory. Biol. J. Linn. Soc. 2003, 78, 155–171. [Google Scholar] [CrossRef]
- Hassouna, N.; Michot, B.; Bachellerie, J.-P.; Narbonne, D. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984, 12, 3563–3583. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Tajima, F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.; Chan, P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Kofler, R.; Nolte, V.; Schlötterer, C. The impact of library preparation protocols on the consistency of allele frequency estimates in Pool-Seq data. Mol. Ecol. Resour. 2016, 16, 118–122. [Google Scholar] [CrossRef]
- Faust, G.G.; Hall, I.M. SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 2014, 30, 2503–2505. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kofler, R.; Orozco-terWengel, P.; De Maio, N.; Pandey, R.V.; Nolte, V.; Futschik, A.; Kosiol, C.; Schlötterer, C. PoPoolation: A toolbox for population genetic analysis of Next Generation Sequencing data from pooled individuals. PLoS ONE 2011, 6, e15925. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, M.P.M.; Briscoe, A.G.; Jorissen, M.W.P.; Littlewood, D.T.J.; Huyse, T. The first next-generation sequencing approach to the mitochondrial phylogeny of African monogenean parasites (Platyhelminthes: Gyrodactylidae and Dactylogyridae). BMC Genom. 2018, 19, 520. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Cruz-Laufer, A.J.; Pariselle, A.; Jorissen, M.W.P.; Muterezi, F.; Al Assadi, A.; Van Steenberge, M.; Koblmüller, S.; Smeets, K.; Huyse, T.; Artois, T.; et al. Somewhere I belong: Phylogenetic comparative methods and machine learning to investigate the evolution of a species-rich lineage of parasites. bioRxiv 2021. [Google Scholar] [CrossRef]
- Chevreux, B.; Suhai, S. Genome sequence assembly using trace signals and additional sequence information. In Proceedings of the German Conference on Bioinformatics, Hannover, Germany, 4–6 October 1999; pp. 45–56. [Google Scholar]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Caña-Bozada, V.; Llera-Herrera, R.; Fajer-Ávila, E.J.; Morales-Serna, F.N. Mitochondrial genome of Scutogyrus longicornis (Monogenea: Dactylogyridea), a parasite of Nile tilapia Oreochromis niloticus. Parasitol. Int. 2021, 81, 102281. [Google Scholar] [CrossRef]
- Koblmüller, S.; Duftner, N.; Katongo, C.; Phiri, H.; Sturmbauer, C. Ancient divergence in bathypelagic Lake Tanganyika deepwater cichlids: Mitochondrial phylogeny of the tribe Bathybatini. J. Mol. Evol. 2005, 60, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Pereya, R.; Taylor, M.I.; Turner, G.F.; Rico, C. Variation in habitat preference and population structure among three species of the Lake Malawi cichlid genus Protomelas. Mol. Ecol. 2004, 13, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Coulter, G.W. The biology of Lates species (Nile perch) in Lake Tanganyika, and the status of the pelagic fishery for Lates species and Luciolates stappersii (Blgr.). J. Fish. Biol. 1976, 9, 235–259. [Google Scholar] [CrossRef]
- Thompson, A.B.; Allison, E.H.; Ngatunga, B.P. Distribution and breeding biology of offshore pelagic cyprinids and catfish in Lake Malawi/Niassa. Environ. Biol. Fishes 1996, 47, 27–42. [Google Scholar] [CrossRef]
- Hahn, C.; Genner, M.J.; Turner, G.F.; Joyce, D.A. The genomic basis of cichlid fish adaptation within the deepwater “twilight zone” of Lake Malawi. Evol. Lett. 2017, 1, 184–198. [Google Scholar] [CrossRef]
- Kirchberger, P.C.; Sefc, K.M.; Sturmbauer, C.; Koblmüller, S. Evolutionary history of Lake Tanganyika’s predatory deepwater cichlids. Int. J. Evol. Biol. 2012, 2012, 716209. [Google Scholar] [CrossRef]
- Anderson, C.; Werdenig, A.; Koblmüller, S.; Sefc, K.M. Same school, different conduct: Rates of multiple paternity vary within a mixed-species breeding school of semi-pelagic cichlid fish (Cyprichromis spp.). Ecol. Evol. 2016, 6, 37–45. [Google Scholar] [CrossRef]
- Buchmann, K. Interactions between monogenean parasites and their fish hosts. Int. J. Parasitol. 2002, 32, 309–319. [Google Scholar] [CrossRef]
- Poulin, R. The evolution of monogenean diversity. Int. J. Parasitol. 2002, 32, 245–254. [Google Scholar] [CrossRef]
- Brazenor, A.K.; Saunders, R.J.; Miller, T.L.; Hutson, K.S. Morphological variation in the cosmopolitan fish parasite Neobenedenia girellae (Capsalidae: Monogenea). Int. J. Parasitol. 2018, 48, 125–134. [Google Scholar] [CrossRef]
- Ergens, R.; Gelnar, M. Experimental verification of the effect of temperature on the size of the hard parts of haptor of Gyrodactylus katharineri Malberg 1964. Folia Parasitol. 1985, 32, 377–380. [Google Scholar]
- Kmentová, N.; Gelnar, M.; Koblmüller, S.; Vanhove, M.P.M. First insights into the diversity of gill monogeneans of “Gnathochromis” and Limnochromis (Teleostei, Cichlidae) in Burundi: Do the parasites mirror host ecology and phylogenetic history? PeerJ 2016, 4, e1629. [Google Scholar] [CrossRef]
- Kmentová, N.; Van Steenberge, M.; Raeymaekers, J.A.R.; Koblmüller, S.; Hablützel, P.I.; Muterezi Bukinga, F.; Mulimbwa N’sibula, T.; Masilya Mulungula, P.; Nzigidahera, B.; Ntakimazi, G.; et al. Monogenean parasites of sardines in Lake Tanganyika: Diversity, origin and intra-specific variability. Contrib. Zool. 2018, 87, 105–132. [Google Scholar] [CrossRef]
- Kaci-Chaouch, T.; Verneau, O.; Desdevises, Y. Host specificity is linked to intraspecific variability in the genus Lamellodiscus (Monogenea). Parasitology 2008, 135, 607–616. [Google Scholar] [CrossRef]
- Rohde, K. Size differences in hamuli of Kuhnia scombri (Monogenea: Polyopisthocotylea) from different geographical areas not due to differences in host size. Int. J. Parasitol. 1991, 21, 113–114. [Google Scholar] [CrossRef]
- Schedel, F.D.B.; Schliewen, U.K. Hemibates koningsi spec. nov: A new deep-water cichlid (Teleostei: Cichlidae) from Lake Tanganyika. Zootaxa 2017, 4312, 92–112. [Google Scholar] [CrossRef]
- Mendoza-Palmero, C.A.; Blasco-Costa, I.; Scholz, T. Molecular phylogeny of Neotropical monogeneans (Platyhelminthes: Monogenea) from catfishes (Siluriformes). Parasit. Vectors 2015, 8, 164. [Google Scholar] [CrossRef]
- Willems, W.R.; Wallberg, A.; Jondelius, U.; Littlewood, D.T.J.; Backeljau, T.; Schockaert, E.R.; Artois, T.J. Filling a gap in the phylogeny of flatworms: Relationships within the Rhabdocoela (Platyhelminthes), inferred from 18S ribosomal DNA sequences. Zool. Scr. 2006, 35, 1–17. [Google Scholar] [CrossRef]
- Nolan, M.J.; Cribb, T.H. The use and implications of ribosomal DNA sequencing for the discrimination of digenean species. Adv. Parasitol. 2005, 60, 101–163. [Google Scholar] [CrossRef]
- Monnens, M.; Thijs, S.; Briscoe, A.G.; Clark, M.; Frost, E.J.; Littlewood, D.T.J.; Sewell, M.; Smeets, K.; Artois, T.; Vanhove, M.P.M. The first mitochondrial genomes of endosymbiotic rhabdocoels illustrate evolutionary relaxation of atp8 and genome plasticity in flatworms. Int. J. Biol. Macromol. 2020, 162, 454–469. [Google Scholar] [CrossRef]
- Solà, E.; Álvarez-Presas, M.; Frías-López, C.; Littlewood, D.T.J.; Rozas, J.; Riutort, M. Evolutionary analysis of mitogenomes from parasitic and free-living flatworms. PLoS ONE 2015, 10, e0120081. [Google Scholar] [CrossRef]
- Littlewood, D.T.J.; Lockyer, A.E.; Webster, B.L.; Johnston, D.A.; Le, T.H. The complete mitochondrial genomes of Schistosoma haematobium and Schistosoma spindale and the evolutionary history of mitochondrial genome changes among parasitic flatworms. Mol. Phylogenet. Evol. 2006, 39, 452–467. [Google Scholar] [CrossRef]
- Ross, E.; Blair, D.; Guerrero-Hernández, C.; Alvarado, A.S. Comparative and transcriptome analyses uncover key aspects of coding- and long noncoding RNAs in flatworm mitochondrial genomes. G3 Genes Genomes Genet. 2016, 6, 1191–1200. [Google Scholar] [CrossRef]
- Caña-Bozada, V.; Llera-Herrera, R.; Fajer-Ávila, E.J.; Morales-Serna, F.N. Mitochondrial genome of Rhabdosynochus viridisi (Monogenea: Diplectanidae), a parasite of Pacific white snook Centropomus viridis. J. Helminthol. 2021, 95, e21. [Google Scholar] [CrossRef]
- Zhang, D.; Li, W.X.; Zou, H.; Wu, S.G.; Li, M.; Jakovlić, I.; Zhang, J.; Chen, R.; Wang, G.T. Mitochondrial genomes and 28S rDNA contradict the proposed obsoletion of the order Tetraonchidea (Platyhelminthes: Monogenea). Int. J. Biol. Macromol. 2020, 143, 891–901. [Google Scholar] [CrossRef]
- Shaughnessy, D.T.; McAllister, K.; Worth, L.; Haugen, A.C.; Meyer, J.N.; Domann, F.E.; Van Houten, B.; Mostoslavsky, R.; Bultman, S.J.; Baccarelli, A.A.; et al. Mitochondria, energetics, epigenetics, and cellular responses to stress. Environ. Health Perspect. 2015, 122, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Le, T.H.; Blair, D.; McManus, D.P. Mitochondrial genomes of parasitic flatworms. Trends Parasitol. 2002, 18, 206–213. [Google Scholar] [CrossRef]
- Wolstenholme, D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992, 141, 173–216. [Google Scholar] [CrossRef]
- Nakao, M.; Sako, Y.; Ito, A. The mitochondrial genome of the tapeworm Taenia solium: A finding of the abbreviated stop codon U. J. Parasitol. 2003, 89, 633–635. [Google Scholar] [CrossRef]
- Egger, B.; Bachmann, L.; Fromm, B. Atp8 is in the ground pattern of flatworm mitochondrial genomes. BMC Genom. 2017, 18, 414. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Trillo, I.; Riutort, M.; Fourcade, H.M.; Baguñà, J.; Boore, J.L. Mitochondrial genome data support the basal position of Acoelomorpha and the polyphyly of the Platyhelminthes. Mol. Phylogenet. Evol. 2004, 33, 321–332. [Google Scholar] [CrossRef]
- Bachmann, L.; Fromm, B.; Patella De Azambuja, L.; Boeger, W.A. The mitochondrial genome of the egg-laying flatworm Aglaiogyrodactylus forficulatus (Platyhelminthes: Monogenoidea). Parasites Vectors 2016, 9, 285. [Google Scholar] [CrossRef]
- Ye, F.; King, S.D.; Cone, D.K.; You, P. The mitochondrial genome of Paragyrodactylus variegatus (Platyhelminthes: Monogenea): Differences in major non-coding region and gene order compared to Gyrodactylus. Parasites Vectors 2014, 7, 377. [Google Scholar] [CrossRef]
- Zhang, D.; Li, W.X.; Zou, H.; Wu, S.G.; Li, M.; Jakovlić, I.; Zhang, J.; Chen, R.; Wang, G.T. Mitochondrial genomes of two diplectanids (Platyhelminthes: Monogenea) expose paraphyly of the order Dactylogyridea and extensive tRNA gene rearrangements. Parasites Vectors 2018, 11, 601. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [CrossRef]
- Castellana, S.; Vicario, S.; Saccone, C. Evolutionary patterns of the mitochondrial genome in Metazoa: Exploring the role of mutation and selection in mitochondrial protein–coding genes. Genome Biol. Evol. 2011, 3, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Castoe, T.A.; Jiang, Z.J.; Gu, W.; Wang, Z.O.; Pollock, D.D. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS ONE 2008, 3, e2201. [Google Scholar] [CrossRef]
- Kmentová, N.; Koblmüller, S.; Van Steenberge, M.; Artois, T.; Muterezi Bukinga, F.; Mulimbwa N’sibula, T.; Muzumani Risasi, D.; Masilya Mulungula, P.; Gelnar, M.; Vanhove, M.P.M. Failure to diverge in African Great Lakes: The case of Dolicirroplectanum lacustre gen. nov. comb. nov. (Monogenea, Diplectanidae) infecting latid hosts. J. Great Lakes Res. 2020, 46, 1113–1130. [Google Scholar] [CrossRef]
- Zhang, D.; Zou, H.; Wu, S.G.; Li, M.; Jakovlić, I.; Zhang, J.; Chen, R.; Li, W.X.; Wang, G.T. Evidence for adaptive selection in the mitogenome of a mesoparasitic monogenean flatworm Enterogyrus malmbergi. Genes 2019, 10, 863. [Google Scholar] [CrossRef] [PubMed]
- Consuegra, S.; John, E.; Verspoor, E.; De Leaniz, C.G. Patterns of natural selection acting on the mitochondrial genome of a locally adapted fish species. Genet. Sel. Evol. 2015, 47, 58. [Google Scholar] [CrossRef]
- Theis, A.; Ronco, F.; Indermaur, A.; Salzburger, W.; Egger, B. Adaptive divergence between lake and stream populations of an East African cichlid fish. Mol. Ecol. 2014, 23, 5304–5322. [Google Scholar] [CrossRef] [PubMed]
- Seguin-Orlando, A.; Schubert, M.; Clary, J.; Stagegaard, J.; Alberdi, M.T.; Prado, J.L.; Prieto, A.; Willerslev, E.; Orlando, L. Ligation bias in illumina next-generation DNA libraries: Implications for sequencing ancient genomes. PLoS ONE 2013, 8, e78575. [Google Scholar] [CrossRef] [PubMed]
- Ekblom, R.; Smeds, L.; Ellegren, H. Patterns of sequencing coverage bias revealed by ultra-deep sequencing of vertebrate mitochondria. BMC Genom. 2014, 15, 467. [Google Scholar] [CrossRef]
- Roberts, D.M.; Boag, B.; Hunter, F.; Tarlton, J.; Mackenzie, K.; Neilson, R. Genetic variability of Arthurdendyus triangulatus (Dendy, 1894), a non-native invasive land planarian. Zootaxa 2020, 4808, 38–50. [Google Scholar] [CrossRef]
- Kowal, K.; Tkaczyk, A.; Pierzchała, M.; Bownik, A.; Ślaska, B. Identification of mitochondrial DNA (NUMTs) in the nuclear genome of Daphnia magna. Int. J. Mol. Sci. 2020, 21, 8725. [Google Scholar] [CrossRef]
- Lopez, J.V.; Yuhki, N.; Masuda, R.; Modi, W.; O’Brien, S.J. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J. Mol. Evol. 1994, 39, 174–190. [Google Scholar] [CrossRef]
- Rellstab, C.; Zoller, S.; Tedder, A.; Gugerli, F.; Fischer, M.C. Validation of SNP allele frequencies determined by pooled next-generation sequencing in natural populations of a non-model plant species. PLoS ONE 2013, 8, e80422. [Google Scholar] [CrossRef]
- Futschik, A.; Schlötterer, C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics 2010, 186, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Tilk, S.; Bergland, A.; Goodman, A.; Schmidt, P.; Petrov, D.; Greenblum, S. Accurate allele frequencies from ultra-low coverage Pool-seq samples in evolve-and-resequence experiments. G3 Genes Genomes Genet. 2019, 9, 4159–4168. [Google Scholar] [CrossRef] [PubMed]
- Neethiraj, R.; Hornett, E.A.; Hill, J.A.; Wheat, C.W. Investigating the genomic basis of discrete phenotypes using a Pool-Seq-only approach: New insights into the genetics underlying colour variation in diverse taxa. Mol. Ecol. 2017, 26, 4990–5002. [Google Scholar] [CrossRef] [PubMed]
- Rode, N.O.; Holtz, Y.; Loridon, K.; Santoni, S.; Ronfort, J.; Gay, L. How to optimize the precision of allele and haplotype frequency estimates using pooled-sequencing data. Mol. Ecol. Resour. 2018, 18, 194–203. [Google Scholar] [CrossRef]




| Host Species | Morphological Characterisation | Genetic Characterisation (cox1 mtDNA) | Genomic Characterisation (PoolSeq) |
|---|---|---|---|
| Hemibates stenosoma | 43 | 24 | 80 |
| Bathybates graueri | 5 | 2 | - |
| Region | Position | Length | Start/Stop Codon | Anticodon | Min–Max Coverage |
|---|---|---|---|---|---|
| trnG | 430–491 | 62 | TTC | 463–562 | |
| cox3 | 536–1181 | 646 | ATG/T | 571–803 | |
| trnH | 1182–1243 | 62 | GTG | 690–763 | |
| cytb | 1244–2320 | 1077 | ATG/TAG | 620–818 | |
| nad4L | 2322–2582 | 261 | ATG/TAG | 494–674 | |
| nad4 | 2621–3754 | 1134 | GTG/TAG | 411–750 | |
| trnQ | 3757–3817 | 60 | TTG | 650–794 | |
| trnF | 3816–3871 | 56 | GAA | 785–841 | |
| trnM | 3872–3935 | 64 | CAT | 683–814 | |
| atp6 | 3938–4447 | 510 | ATG/TAA | 518–876 | |
| nad2 | 4452–5275 | 824 | ATG/TA | 609–916 | |
| trnV | 5276–5337 | 62 | TAC | 780–833 | |
| trnA | 5350–5419 | 70 | TGC | 771–831 | |
| trnD | 5436–5500 | 65 | GTC | 758–799 | |
| nad1 | 5501–6388 | 876 | ATT/TAA | 415–860 | |
| trnN | 6395–6460 | 66 | GTT | 677–815 | |
| trnP | 6466–6528 | 63 | TGG | 800–867 | |
| trnI | 6528–6594 | 67 | GAT | 820–878 | |
| trnK | 6595–6659 | 65 | CTT | 767–822 | |
| nad3 | 6661–7008 | 348 | GTG/TAG | 688–774 | |
| trnS1 | 7014–7070 | 57 | GTC | 722–809 | |
| trnW | 7073–7135 | 63 | TCA | 702–811 | |
| cox1 | 7139–8692 | 1554 | ATG/TAA | 602–1088 | |
| trnT | 8693–8758 | 66 | TGT | 744–843 | |
| AT-rich region | 8763–9858 | 1096 | 181–1701 | ||
| rrnL | 9859–10,809 | 951 | 1253–1800 | ||
| trnC | 10,820–10,881 | 62 | GCA | 1879–2213 | |
| rrnS | 10,882–11,601 | 720 | 1069–2284 | ||
| AT-rich region | 11,602–11,955 | 354 | 590–1214 | ||
| cox2 | 11,956–12,531 | 576 | ATG/TAG | 857–1298 | |
| trnE | 12,531–12,595 | 65 | TTC | 821–965 | |
| nad6 | 12,621–13,082 | 462 | GTG/TAG | 606–800 | |
| trnY | 13,083–13,145 | 63 | GTA | 791–885 | |
| trnL1 | 13,147–13,211 | 65 | TAG | 879–921 | |
| trnS2 | 13,212–13,274 | 63 | TGA | 837–897 | |
| trnL2 | 13,275–13,339 | 65 | TAA | 792–881 | |
| trnR | 13,340–13,407 | 68 | TCG | 657–815 | |
| nad5 | 13,409–14,953 | 1545 | ATG/TAA | 430–862 | |
| Repeat region | 14,965–426 | 1037 | 12–829 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kmentová, N.; Hahn, C.; Koblmüller, S.; Zimmermann, H.; Vorel, J.; Artois, T.; Gelnar, M.; Vanhove, M.P.M. Contrasting Host-Parasite Population Structure: Morphology and Mitogenomics of a Parasitic Flatworm on Pelagic Deepwater Cichlid Fishes from Lake Tanganyika. Biology 2021, 10, 797. https://doi.org/10.3390/biology10080797
Kmentová N, Hahn C, Koblmüller S, Zimmermann H, Vorel J, Artois T, Gelnar M, Vanhove MPM. Contrasting Host-Parasite Population Structure: Morphology and Mitogenomics of a Parasitic Flatworm on Pelagic Deepwater Cichlid Fishes from Lake Tanganyika. Biology. 2021; 10(8):797. https://doi.org/10.3390/biology10080797
Chicago/Turabian StyleKmentová, Nikol, Christoph Hahn, Stephan Koblmüller, Holger Zimmermann, Jiří Vorel, Tom Artois, Milan Gelnar, and Maarten P. M. Vanhove. 2021. "Contrasting Host-Parasite Population Structure: Morphology and Mitogenomics of a Parasitic Flatworm on Pelagic Deepwater Cichlid Fishes from Lake Tanganyika" Biology 10, no. 8: 797. https://doi.org/10.3390/biology10080797
APA StyleKmentová, N., Hahn, C., Koblmüller, S., Zimmermann, H., Vorel, J., Artois, T., Gelnar, M., & Vanhove, M. P. M. (2021). Contrasting Host-Parasite Population Structure: Morphology and Mitogenomics of a Parasitic Flatworm on Pelagic Deepwater Cichlid Fishes from Lake Tanganyika. Biology, 10(8), 797. https://doi.org/10.3390/biology10080797







