Cigarette Smoking Alters the Expression of Circulating microRNAs and Its Potential Diagnostic Value in Female Lung Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Design
2.2. Cigarette Smoking Assessment
2.3. Serum Samples
2.4. RNA Isolation
2.5. MicroRNA Expression Profiling
2.6. Validation by qPCR
2.7. Statistical Analysis
2.8. MiRNA Target Genes and Pathways Enrichment Analysis
3. Results
3.1. Baseline Characteristics of the Study Population
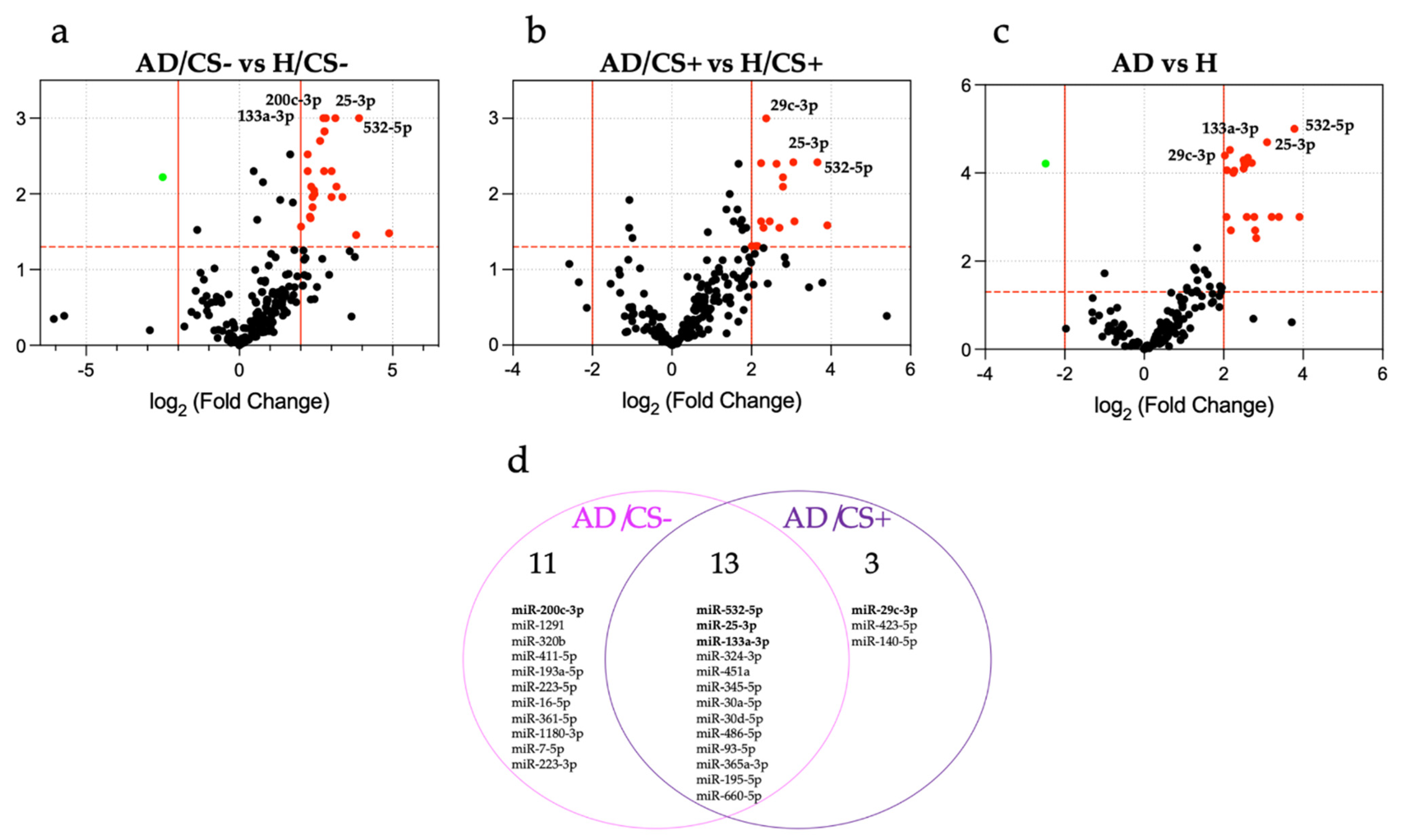
3.2. Expression Profiling of miRNAs in the Sera of AD Patients According to Their Cigarette Smoking Status
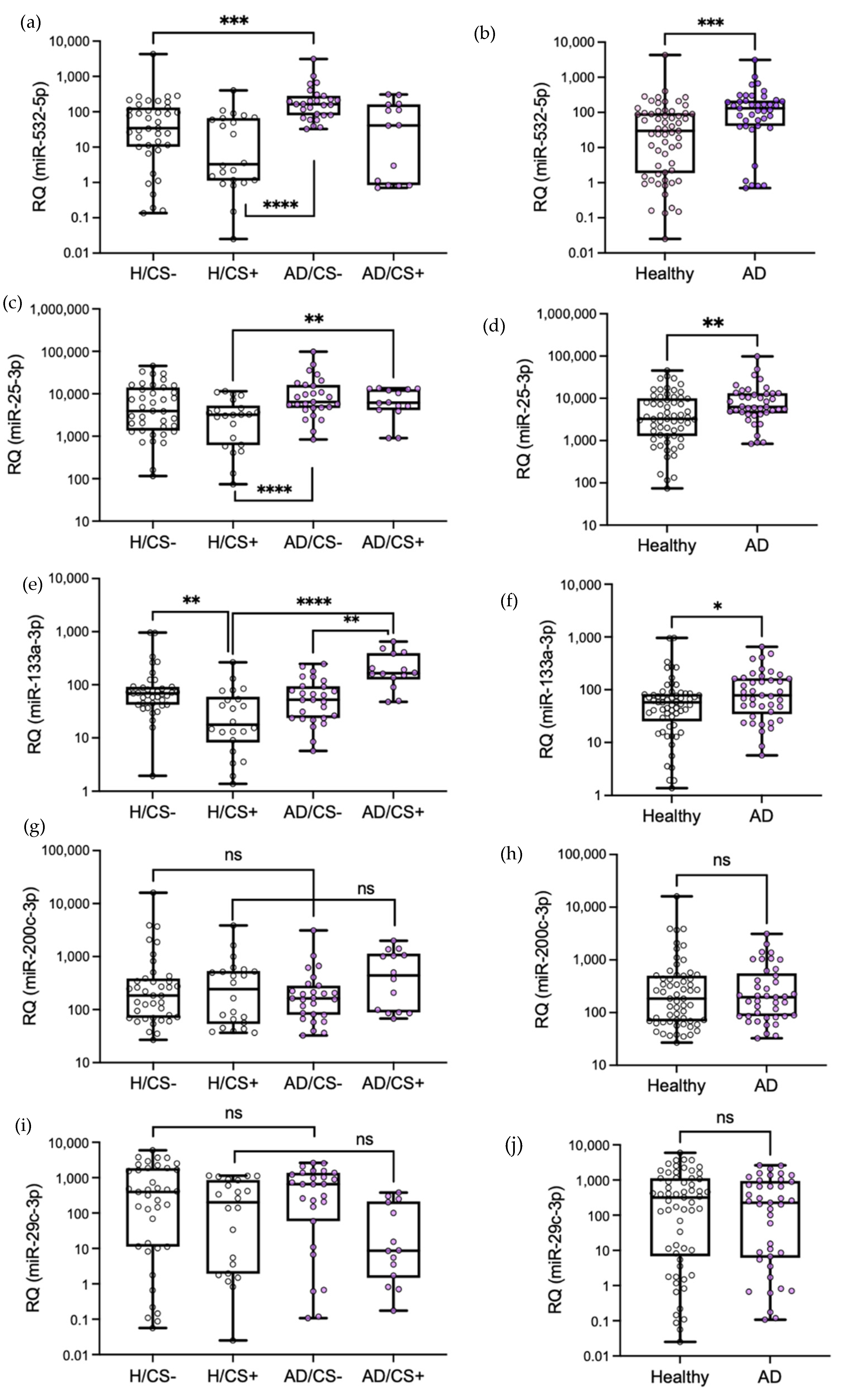
3.3. Validation of Differentially Expressed miRNAs by qPCR
3.4. The Potential Diagnostic Value of the Differentially Expressed miRNAs in AD Patients, miR-532-5p, miR-25-3p, and miR-133a-3p, Is Affected by Cigarette Smoking Status
3.5. Levels of miR-133a-3p Were Independently Associated with Cigarette Smoking in AD Patients
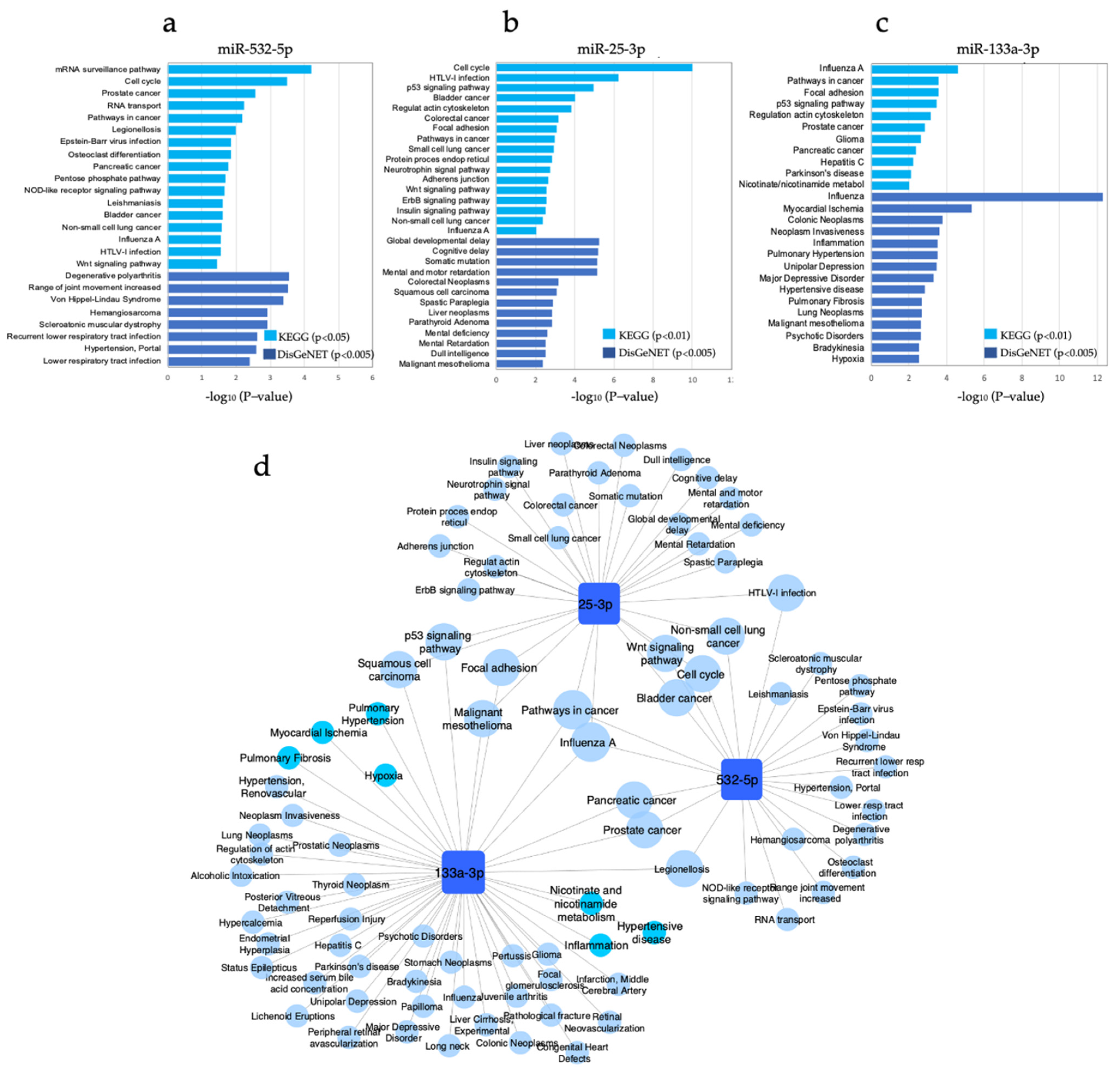
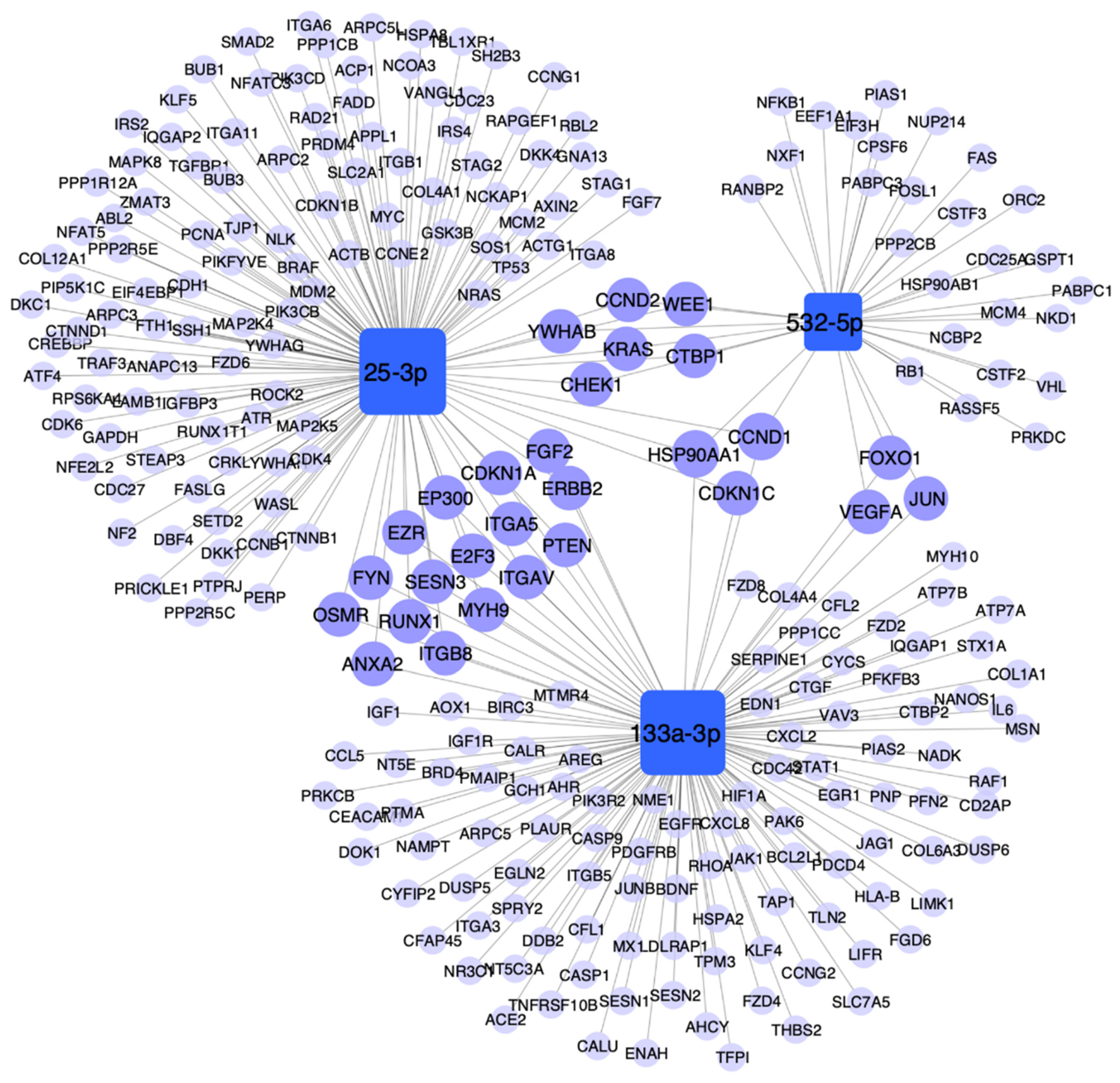
3.6. Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Agency for Research on Cancer. GLOBOCAN 2018. Available online: http://gco.iarc.fr (accessed on 21 October 2020).
- Bade, B.C.; Dela Cruz, C.S. Lung cancer 2020: Epidemiology, etiology, and prevention. Clin. Chest Med. 2020, 41, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Courtney, R. In the Health Consequences of Smoking-50 Years of Progress: A Report of the Surgeon General; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2014; 1081.
- Hecht, S.S. Lung carcinogenesis by tobacco smoke. Int. J. Cancer 2012, 131, 2724–2732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.; Golpon, H.; Zardo, P.; Borlak, J. miRNAs in lung cancer. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl. Res. 2021, 230, 164–196. [Google Scholar] [CrossRef]
- Gayosso-Gomez, L.V.; Ortiz-Quintero, B. Circulating MicroRNAs in blood and other body fluids as biomarkers for diagnosis, prognosis, and therapy response in lung cancer. Diagnostics 2021, 11, 421. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Wang, G.; Wang, R.; Strulovici-Barel, Y.; Salit, J.; Staudt, M.R.; Ahmed, J.; Tilley, A.E.; Yee-Levin, J.; Hollmann, C.; Harvey, B.G.; et al. Persistence of smoking-induced dysregulation of miRNA expression in the small airway epithelium despite smoking cessation. PLoS ONE 2015, 10, e0120824. [Google Scholar] [CrossRef]
- Graff, J.W.; Powers, L.S.; Dickson, A.M.; Kim, J.; Reisetter, A.C.; Hassan, I.H.; Kremens, K.; Gross, T.J.; Wilson, M.E.; Monick, M.M. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS ONE 2012, 7, e44066. [Google Scholar] [CrossRef] [Green Version]
- Schembri, F.; Sridhar, S.; Perdomo, C.; Gustafson, A.M.; Zhang, X.; Ergun, A.; Lu, J.; Liu, G.; Zhang, X.; Bowers, J.; et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. USA 2009, 106, 2319–2324. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.B.; Park, Y.S.; Sung, J.S.; Lee, J.W.; Kim, B.; Kim, Y.H. Tumor suppressor miR-584-5p inhibits migration and invasion in smoking related non-small cell lung cancer cells by targeting YKT6. Cancers 2021, 13, 1159. [Google Scholar] [CrossRef]
- Wang, J.; Yu, X.F.; Ouyang, N.; Zhao, S.; Yao, H.; Guan, X.; Tong, J.; Chen, T.; Li, J.X. MicroRNA and mRNA interaction network regulates the malignant transformation of human bronchial epithelial cells induced by cigarette smoke. Front. Oncol. 2019, 9, 1029. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yokota, S.I.; Tatsumi, N.; Fukami, T.; Yokoi, T.; Nakajima, M. Cigarette smoking substantially alters plasma microRNA profiles in healthy subjects. Toxicol. Appl. Pharmacol. 2013, 272, 154–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, B.A.; Sayardoust, S.; Lofgren, S.; Rutqvist, L.E.; Laytragoon-Lewin, N. Cigarette smoking affects microRNAs and inflammatory biomarkers in healthy individuals and an association to single nucleotide polymorphisms is indicated. Biomarkers 2019, 24, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Badrnya, S.; Baumgartner, R.; Assinger, A. Smoking alters circulating plasma microvesicle pattern and microRNA signatures. Thromb. Haemost. 2014, 112, 128–136. [Google Scholar] [CrossRef] [Green Version]
- Willinger, C.M.; Rong, J.; Tanriverdi, K.; Courchesne, P.L.; Huan, T.; Wasserman, G.A.; Lin, H.; Dupuis, J.; Joehanes, R.; Jones, M.R.; et al. MicroRNA signature of cigarette smoking and evidence for a putative causal role of MicroRNAs in smoking-related inflammation and target organ damage. Circ. Cardiovasc. Genet. 2017, 10, e001678. [Google Scholar] [CrossRef]
- Marabita, F.; de Candia, P.; Torri, A.; Tegner, J.; Abrignani, S.; Rossi, R.L. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief. Bioinform. 2016, 17, 204–212. [Google Scholar] [CrossRef] [Green Version]
- Shah, J.S.; Soon, P.S.; Marsh, D.J. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS ONE 2016, 11, e0153200. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Kryczka, J.; Migdalska-Sek, M.; Kordiak, J.; Kiszalkiewicz, J.M.; Pastuszak-Lewandoska, D.; Antczak, A.; Brzezianska-Lasota, E. Serum extracellular vesicle-derived miRNAs in patients with non-small cell lung cancer-search for non-invasive diagnostic biomarkers. Diagnostics 2021, 11, 425. [Google Scholar] [CrossRef]
- Huang, J.; Wu, J.; Li, Y.; Li, X.; Yang, T.; Yang, Q.; Jiang, Y. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed. Res. Int. 2014, 2014, 364316. [Google Scholar] [CrossRef]
- Lebanony, D.; Benjamin, H.; Gilad, S.; Ezagouri, M.; Dov, A.; Ashkenazi, K.; Gefen, N.; Izraeli, S.; Rechavi, G.; Pass, H.; et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J. Clin. Oncol. 2009, 27, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, J.; Soejima, K.; Yoda, S.; Naoki, K.; Nakayama, S.; Satomi, R.; Terai, H.; Ikemura, S.; Sato, T.; Yasuda, H.; et al. Identification of microRNAs differentially expressed between lung squamous cell carcinoma and lung adenocarcinoma. Mol. Med. Rep. 2013, 8, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Hartmann, N.; Baeriswyl, L.; Andreasen, D.; Bernard, N.; Chen, C.; Cheo, D.; D’Andrade, P.; DeMayo, M.; Dennis, L.; et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods 2014, 11, 809–815. [Google Scholar] [CrossRef]
- Kanaan, Z.; Roberts, H.; Eichenberger, M.R.; Billeter, A.; Ocheretner, G.; Pan, J.; Rai, S.N.; Jorden, J.; Williford, A.; Galandiuk, S. A plasma microRNA panel for detection of colorectal adenomas: A step toward more precise screening for colorectal cancer. Ann. Surg. 2013, 258, 400–408. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Cai, J.; Xu, Z.; Zhou, S.; Ye, L.; Yan, Q.; Zhang, Y.; Fang, Y.; Liu, Y.; Tu, C.; et al. MiR-532-5p suppresses colorectal cancer progression by disrupting the ETS1/TGM2 axis-mediated Wnt/beta-catenin signaling. Cell Death Dis. 2019, 10, 739. [Google Scholar] [CrossRef]
- Zhao, T.; Meng, W.; Chin, Y.; Gao, L.; Yang, X.; Sun, S.; Pan, X.; He, L. Identification of miR253p as a tumor biomarker: Regulation of cellular functions via TOB1 in breast cancer. Mol. Med. Rep. 2021, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Li, Y.; Pan, Y.; Lan, X.; Song, F.; Sun, J.; Zhou, K.; Liu, X.; Ren, X.; Wang, F.; et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018, 9, 5395. [Google Scholar] [CrossRef] [Green Version]
- Hua, Y.T.; Xu, W.X.; Li, H.; Xia, M. Emerging roles of MiR-133a in human cancers. J. Cancer 2021, 12, 198–206. [Google Scholar] [CrossRef]
- Chiyomaru, T.; Enokida, H.; Tatarano, S.; Kawahara, K.; Uchida, Y.; Nishiyama, K.; Fujimura, L.; Kikkawa, N.; Seki, N.; Nakagawa, M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br. J. Cancer 2010, 102, 883–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.W.; Choi, C.H.; Choi, J.J.; Park, Y.A.; Kim, S.J.; Hwang, S.Y.; Kim, W.Y.; Kim, T.J.; Lee, J.H.; Kim, B.G.; et al. Altered MicroRNA expression in cervical carcinomas. Clin. Cancer Res. 2008, 14, 2535–2542. [Google Scholar] [CrossRef] [Green Version]
- Moriya, Y.; Nohata, N.; Kinoshita, T.; Mutallip, M.; Okamoto, T.; Yoshida, S.; Suzuki, M.; Yoshino, I.; Seki, N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J. Hum. Genet. 2012, 57, 38–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambs, S.; Prueitt, R.L.; Yi, M.; Hudson, R.S.; Howe, T.M.; Petrocca, F.; Wallace, T.A.; Liu, C.G.; Volinia, S.; Calin, G.A.; et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008, 68, 6162–6170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; He, X.; Xie, Y.; Huang, Y.; Wolff, D.W.; Abel, P.W.; Tu, Y. Up-regulated miR-133a orchestrates epithelial-mesenchymal transition of airway epithelial cells. Sci. Rep. 2018, 8, 15543. [Google Scholar] [CrossRef] [PubMed]
- Escate, R.; Padro, T.; Suades, R.; Camino, S.; Muniz, O.; Diaz-Diaz, J.L.; Sionis, A.; Mata, P.; Badimon, L. High miR-133a levels in the circulation anticipates presentation of clinical events in familial hypercholesterolaemia patients. Cardiovasc. Res. 2021, 117, 109–122. [Google Scholar] [CrossRef] [PubMed]


| Characteristics | AD/CS− | AD/CS+ | H/CS− | H/CS+ |
|---|---|---|---|---|
| Discovery cohort | ||||
| Female, n (%) | 15 (100) | 15 (100) | 15 (100) | 15 (100) |
| Age, years, mean (SD) | 62.8 (12.4) | 68 (10) | 57.9 (6.7) | 56.6 (4.8) |
| BMI, kg/m2, mean (SD) | 25 (5.2) | 25 (3.4) | 25.3 (2) | 24.9 (3.2) |
| Cigarette smoking, n (%) | 0 (0) | 15 (100) | 0 (0) | 15 (100) |
| Pack/years, mean (SD) | NA | 21.8 (7.9) | NA | 37 (11.7) ‡ |
| Diabetes mellitus, n (%) | 2 (13) | 0 | 0 | 0 |
| Hypertension, n (%) | 7 (46.7) | 3 (20) | 0 | 0 |
| Cardiac disease, n (%) | 1 (6.7) | 2 (13.3) | 0 | 0 |
| Obesity | 2 (13.3) | 2 (13.3) | 0 | 0 |
| Histological subtype | AD | AD | NA | NA |
| Disease stage | ||||
| IIB, n (%) | 5 (33.3) | 4 (26.7) | NA | NA |
| IV, n (%) | 10 (66.7) | 11 (73.3) | NA | NA |
| Validation cohort | ||||
| Female, n | 27 (100) | 14 (100) | 39 (100) | 22 (100) |
| Age, years, mean (SD) | 61.4 (12.3) | 66.5 (10) | 61 (8.2) | 56 (5.9) |
| BMI, kg/m2, mean (SD) | 25.1 (5.9) | 25 (3.9) | 26 (3.5) | 26.5 (4.5) |
| Cigarette smoking, n (%) | 0 (0) | 14 (100) | 0 (0) | 29 (100) |
| Pack/years, mean (SD) | NA | 22.2 (8.4) | NA | 32.9 (13) ‡ |
| Diabetes mellitus, n (%) | 4 (14.8) | 1 (7) | 0 | 0 |
| Hypertension, n (%) | 5 (18.5) | 2 (14.3) | 0 | 0 |
| Cardiac disease, n (%) | 1 (3.7) | 2 (14) § | 0 | 0 |
| Obesity | 3 (11.1) | 2 (14) | 0 | 0 |
| Histological subtype | AD | AD | NA | NA |
| Disease stage | ||||
| IIB | 5 (18.5) | 4 (28.6) | NA | NA |
| IIIB | 4 (14.8) | 2 (14.3) | NA | NA |
| IV | 18 (66.7) | 8 (57.1) | NA | NA |
| MicroRNA | Study Groups | AUC | 95% CI | Sensitivity (%) | Specificity (%) | p-Value |
|---|---|---|---|---|---|---|
| miR-532-5p | H vs. AD | 0.745 | 0.641–0.849 | 76.5 | 59 | <0.0001 |
| H/CS− vs. AD/CS− | 0.762 | 0.649–0.875 | 81.5 | 56.4 | <0.0001 | |
| H/CS+ vs. AD/CS− | 0.886 | 0.79–0.981 | 77.8 | 81.8 | <0.0001 | |
| miR-25-3p | H vs. AD | 0.674 | 0.565–0.782 | 76.5 | 60.7 | 0.005 |
| H/CS+ vs. AD/CS− | 0.779 | 0.649–0.91 | 81.5 | 72.7 | 0.001 | |
| H/CS+ vs. AD/CS+ | 0.779 | 0.578–0.98 | 85.7 | 72.7 | 0.028 | |
| miR-133a-3p | H vs. AD | 0.588 | NA | NA | NA | 0.158 |
| H/CS+ vs. AD/CS+ | 0.935 | 0.836–1 | 85.7 | 95.5 | 0.001 | |
| miR-133a-3p | AD/CS− vs. AD/CS+ | 0.884 | 0.733–1 | 85.7 | 88.9 | 0.002 |
| H/CS− vs. H/CS+ | 0.765 | 0.627–0.902 | 82.1 | 68.2 | 0.001 | |
| miR-532-5p + miR-25-3p + miR-133a-3p | H vs. AD | 0.70 | 0.596–0.804 | 70.6 | 59 | 0.001 |
| miR-25-3p + miR-133a-3p | H/CS+ vs. AD/CS+ | 0.961 | 0.895–1 | 100 | 86.4 | <0.0001 |
| Variable | miR-532-5p | miR-25-3p | miR-133a-3p | |||
|---|---|---|---|---|---|---|
| rho | p-Value | rho | p-Value | rho | p-Value | |
| AD patients | ||||||
| Age (years) | 0.217 | 0.225 | 0.301 | 0.088 | 0.482 ** | 0.005 |
| BMI (kg/m2) | 0.162 | 0.377 | −0.105 | 0.567 | 0.137 | 0.453 |
| Pack/years | −0.352 * | 0.041 | −0.107 | 0.546 | 0.357 * | 0.038 |
| Healthy controls | ||||||
| Age (years) | 0.303 * | 0.019 | 0.328 * | 0.01 | −0.036 | 0.783 |
| BMI (kg/m2) | 0.085 | 0.519 | −0.043 | 0.746 | −0.007 | 0.958 |
| Pack/years | −0.283 * | 0.027 | −0.205 | 0.116 | −0.45 ** | <0.0001 |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Independent Factors | Coefficient B | 95% CI | p-Value | Coefficient B | 95% CI | p-Value |
| Age | 0.311 | −0.456–8.088 | 0.078 | |||
| BMI | 0.731 | −9.402–10.865 | 0.884 | |||
| Cigarette smoking status | 145.048 | 47.162–242.933 | 0.005 | 5.009 | −89.911–99.93 | 0.915 |
| Pack/years | 6.3 | 1.311–11.289 | 0.015 | −19.101 | −26.645–−11.556 | <0.0001 |
| Diabetes mellitus | −92.961 | −246.252–60.331 | 0.226 | |||
| Hypertension | 114.022 | 14.066–213.978 | 0.027 | |||
| Cardiac disease | 197.707 | 34.312–361.103 | 0.019 | 33.62 | −66.908–134.147 | 0.499 |
| Obesity | 91.719 | −49.547–232.986 | 0.195 | |||
| ID group (AD/CS− vs. AD/CS+) | 231.769 | 138.676–324.861 | <0.0001 | 613.271 | 422.658–803.884 | <0.0001 |
| Univariate Analysis (Smoking as the Dependent Variable) | |||
| Independent Factor | Coefficient B | 95% CI | p-Value |
| miR-133a-3p | 0.002 | 0–0.003 | 0.005 |
| Univariate Analysis (Pack/Years as the Dependent Variable) | |||
| Independent Factor | Coefficient B | 95% CI | p-Value |
| miR-133a-3p | 0.027 | 0.006–0.049 | 0.015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Salazar, E.G.; Gayosso-Gómez, L.V.; Baez-Saldaña, R.; Falfán-Valencia, R.; Pérez-Padilla, R.; Higuera-Iglesias, A.L.; Vázquez-Manríquez, M.E.; Ortiz-Quintero, B. Cigarette Smoking Alters the Expression of Circulating microRNAs and Its Potential Diagnostic Value in Female Lung Cancer Patients. Biology 2021, 10, 793. https://doi.org/10.3390/biology10080793
Ramírez-Salazar EG, Gayosso-Gómez LV, Baez-Saldaña R, Falfán-Valencia R, Pérez-Padilla R, Higuera-Iglesias AL, Vázquez-Manríquez ME, Ortiz-Quintero B. Cigarette Smoking Alters the Expression of Circulating microRNAs and Its Potential Diagnostic Value in Female Lung Cancer Patients. Biology. 2021; 10(8):793. https://doi.org/10.3390/biology10080793
Chicago/Turabian StyleRamírez-Salazar, Eric Gustavo, Luis Vicente Gayosso-Gómez, Renata Baez-Saldaña, Ramcés Falfán-Valencia, Rogelio Pérez-Padilla, Anjarath L. Higuera-Iglesias, María E. Vázquez-Manríquez, and Blanca Ortiz-Quintero. 2021. "Cigarette Smoking Alters the Expression of Circulating microRNAs and Its Potential Diagnostic Value in Female Lung Cancer Patients" Biology 10, no. 8: 793. https://doi.org/10.3390/biology10080793
APA StyleRamírez-Salazar, E. G., Gayosso-Gómez, L. V., Baez-Saldaña, R., Falfán-Valencia, R., Pérez-Padilla, R., Higuera-Iglesias, A. L., Vázquez-Manríquez, M. E., & Ortiz-Quintero, B. (2021). Cigarette Smoking Alters the Expression of Circulating microRNAs and Its Potential Diagnostic Value in Female Lung Cancer Patients. Biology, 10(8), 793. https://doi.org/10.3390/biology10080793







