Alterations in microRNA Expression during Hematopoietic Stem Cell Mobilization
Abstract
Simple Summary
Abstract
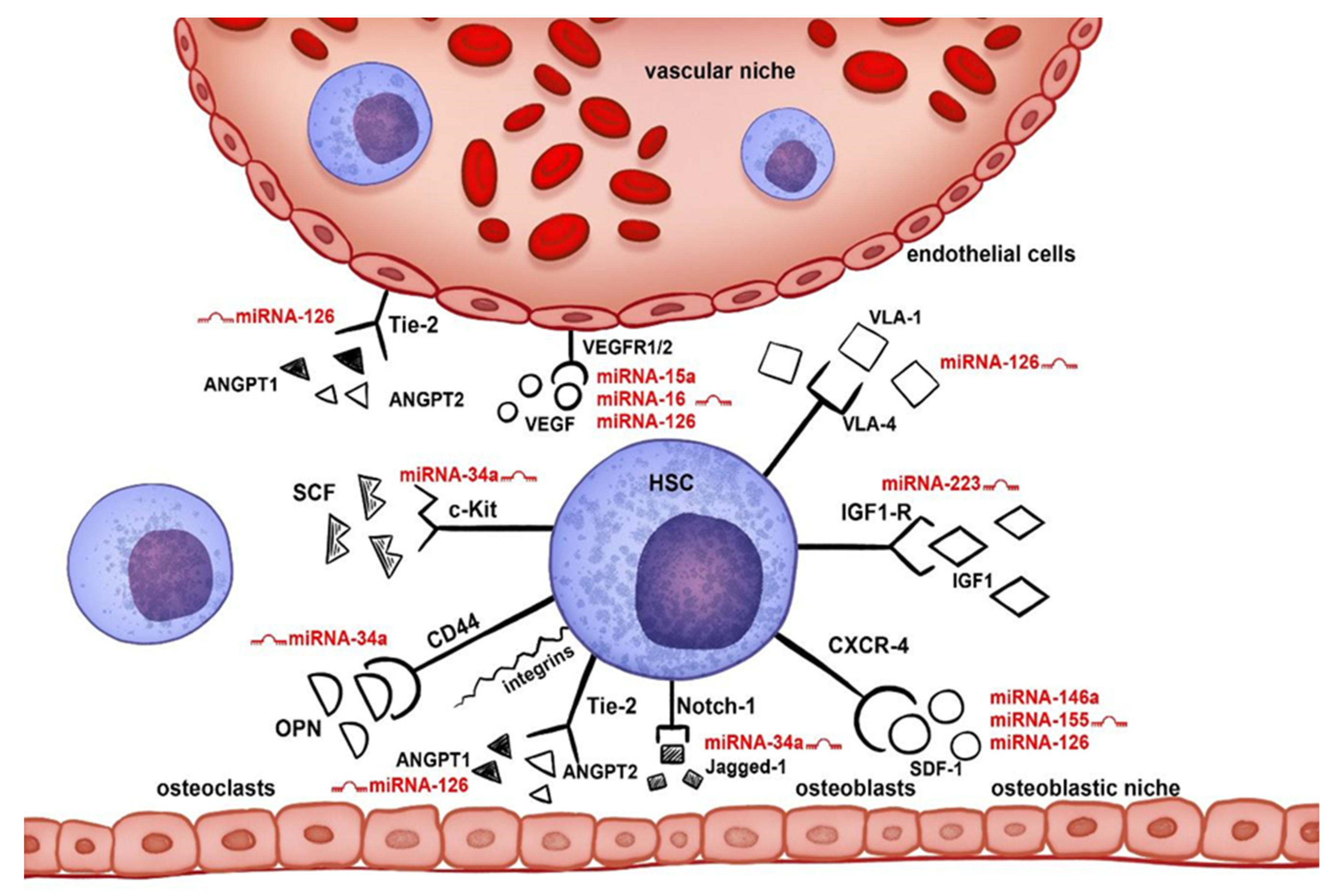
1. Introduction
2. Materials and Methods
2.1. RNA Isolation and cDNA Synthesis
2.2. qPCR miRNA Expression
2.3. Statistical Analysis
3. Results
3.1. Mobilization Data
3.2. miRNA Expression and Mobilization Efficacy
3.2.1. miRNA Expression and the Number of CD34+ Cells in Peripheral Blood at First Apheresis
3.2.2. miRNA Expression and the Number of Collected CD34+ Cells on the Day of First Apheresis
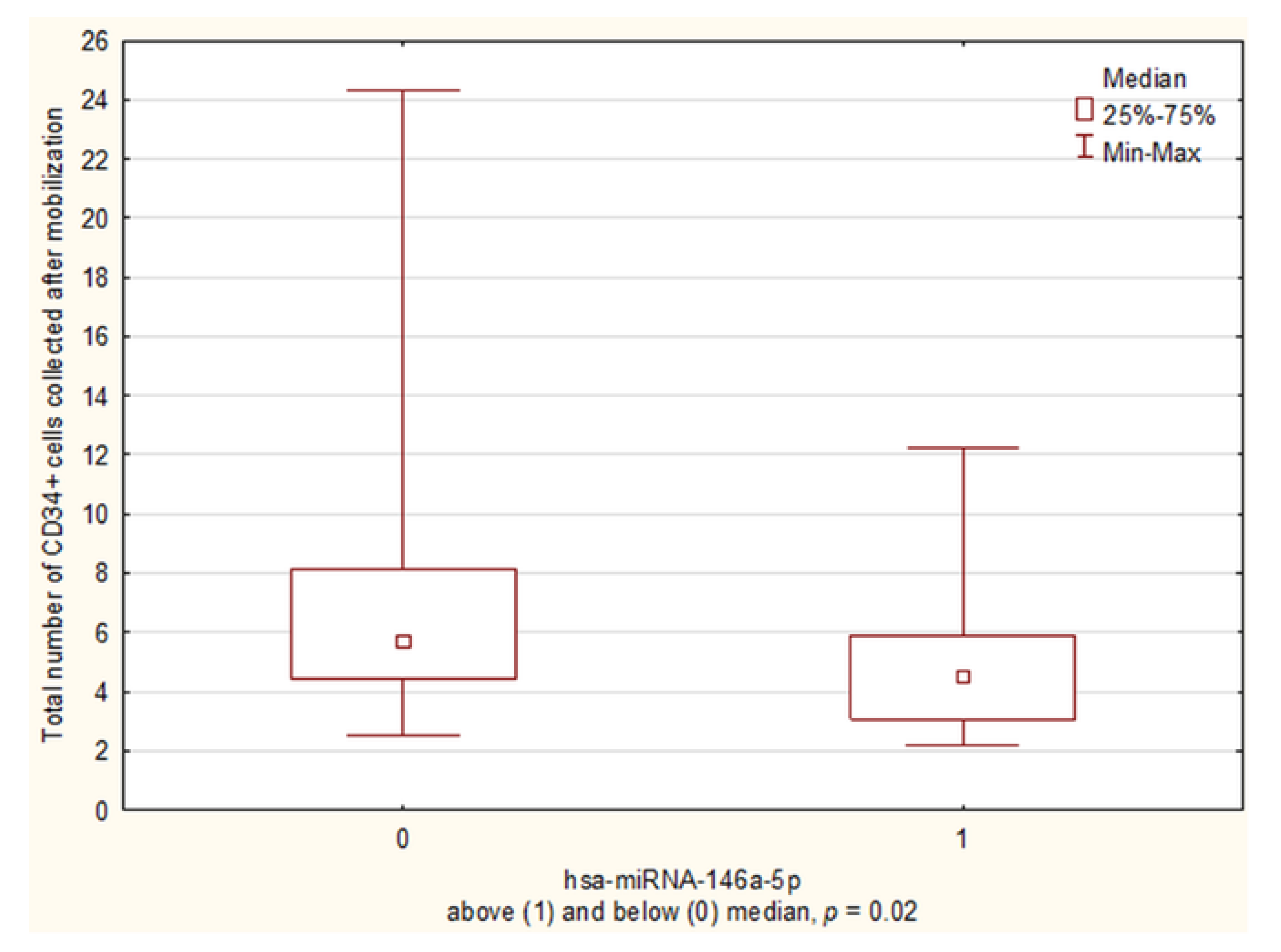
3.2.3. miRNA Expression and the Total Number of CD34+ Cells Collected during Mobilization
3.2.4. miRNA Expression and the Number of Apheresis Attempts
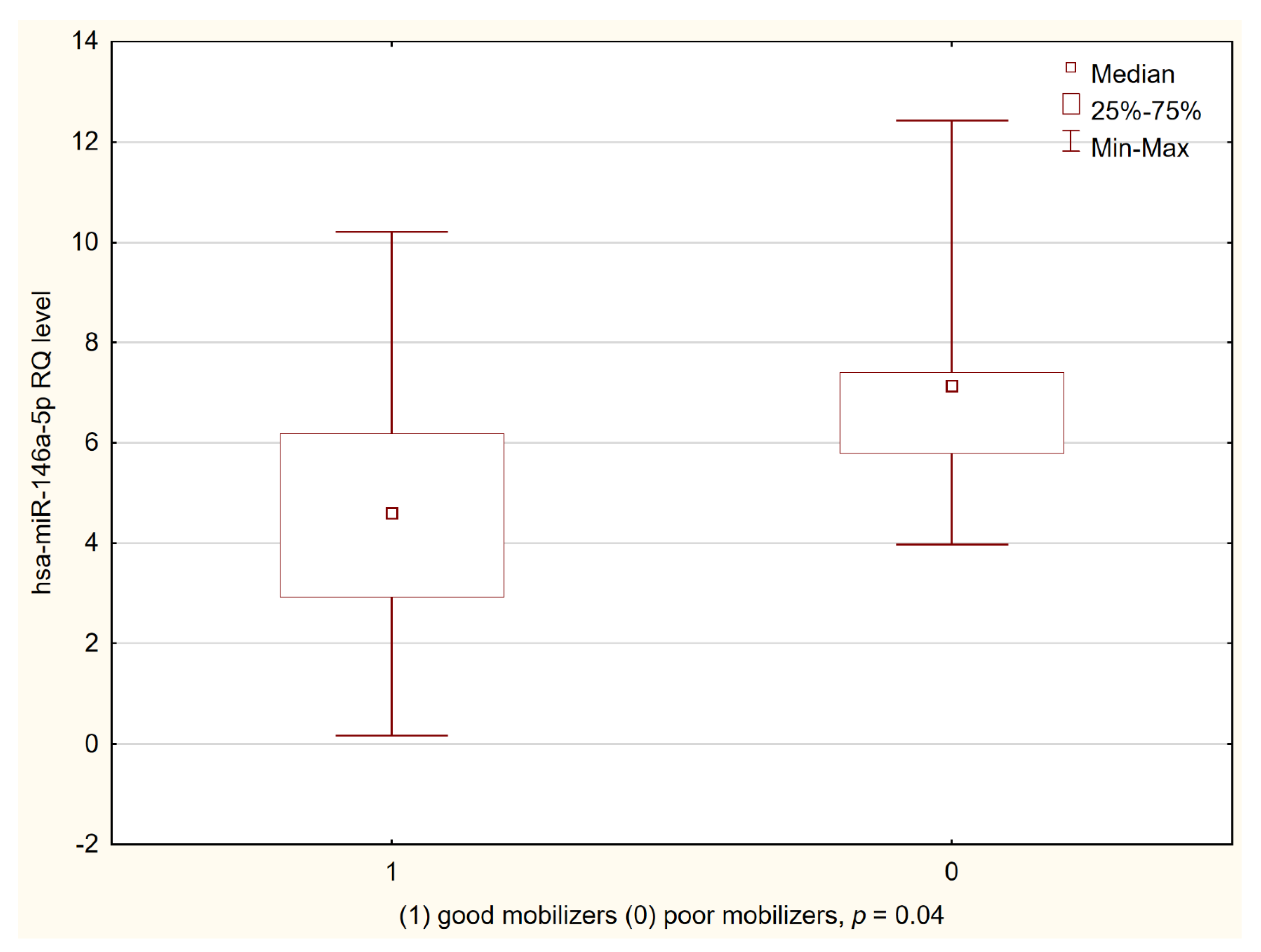
3.3. miRNA Expression in Good and Poor Mobilizers According GITMO Criteria
miRNA Expression According to CD34 Peak in Peripheral Blood
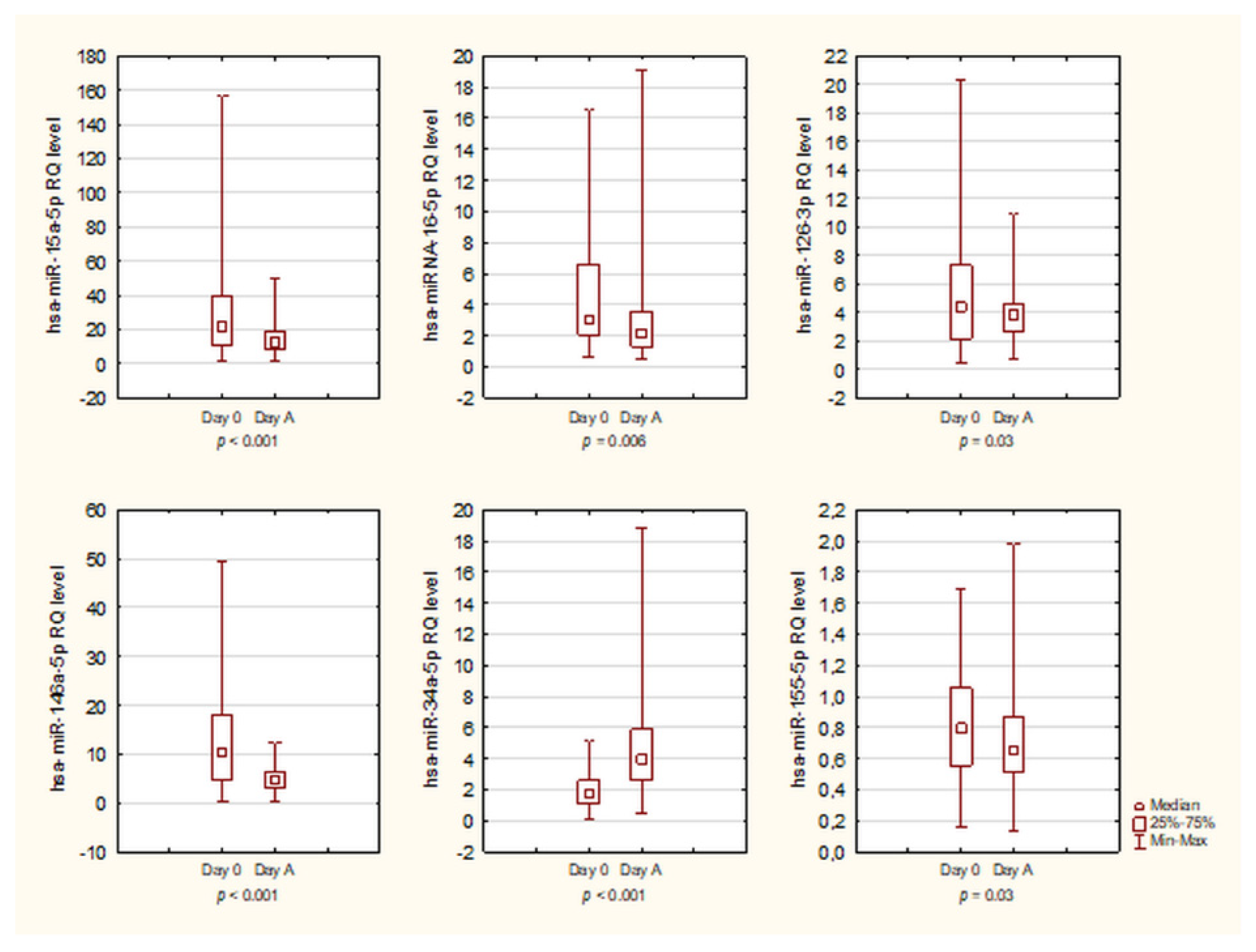
3.4. Kinetics of miRNA
3.5. Relationship between WBC Count and miRNA Expression
3.6. miRNA Expression and Remission Status
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HSC | Hematopoietic Stem Cells |
| HSCT | Hematopoietic Stem Cell Transplantation |
| LD | Lymphoproliferative Disorders |
| G-CSF | Granulocyte Colony-stimulating Factor |
| EC | Endothelial Cells |
| HPSC | Hematopoietic Progenitor and Stem Cells |
| PB | Peripheral Blood |
References
- Subramaniam, K.; D’Rozario, J.; Pavli, P. Lymphoma and other lymphoproliferative disorders in inflammatory bowel disease: A review. J. Gastroenterol. Hepatol. 2013, 28, 24–30. [Google Scholar] [CrossRef]
- Devine, S.M.; Flomenberg, N.; Vesole, D.H.; Liesveld, J.; Weisdorf, D.; Badel, K.; Calandra, G.; DiPersio, J.F. Rapid Mobilization of CD34+ Cells Following Administration of the CXCR4 Antagonist AMD3100 to Patients With Multiple Myeloma and Non-Hodgkin’s Lymphoma. J. Clin. Oncol. 2004, 22, 1095–1102. [Google Scholar] [CrossRef]
- Nowicki, M.; Wierzbowska, A.; Małachowski, R.; Robak, T.; Grzybowska-Izydorczyk, O.; Pluta, A.; Szmigielska-Kaplon, A. VEGF, ANGPT1, ANGPT2, and MMP-9 expression in the autologous hematopoietic stem cell transplantation and its impact on the time to engraftment. Ann. Hematol. 2017, 96, 2103–2112. [Google Scholar] [CrossRef]
- Nervi, B.; Link, D.C.; DiPersio, J.F. Cytokines and hematopoietic stem cell mobilization. J. Cell. Biochem. 2006, 99, 690–705. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B. New class of microRNA targets containing simultaneous 5’-UTR and 3’-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Siemerink, M.J.; Klaassen, I.; Vogels, I.M.C.; Griffioen, A.W.; Van Noorden, C.J.F.; Schlingemann, R.O. CD34 marks angiogenic tip cells in human vascular endothelial cell cultures. Angiogenesis 2012, 15, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Szmigielska-Kaplon, A.; Szemraj, J.; Hamara, K.; Robak, M.; Wolska, A.; Pluta, A.; Czemerska, M.; Krawczynska, A.; Jamroziak, K.; Szmigielska, K.; et al. Polymorphism of CD44 Influences the Efficacy of CD34+ Cells Mobilization in Patients with Hematological Malignancies. Biol. Blood Marrow Transplant. 2014, 20, 986–991. [Google Scholar] [CrossRef] [PubMed]
- Song, G.-Y.; Jung, S.-H.; Ahn, S.-Y.; Jung, S.-Y.; Yang, D.-H.; Ahn, J.-S.; Kim, H.-J.; Lee, J.-J. Optimal chemo-mobilization for the collection of peripheral blood stem cells in patients with multiple myeloma. BMC Cancer 2019, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Giebel, S.; Sadus-Wojciechowska, M.; Halaburda, K.; Drozd-Sokolowska, J.; Wierzbowska, A.; Najda, J.; Mendrek, W.; Sobczyk-Kruszelnicka, M.; Nowicki, M.; Holowiecki, J.; et al. Increased efficacy of intermediate-dose cytarabine + G-CSF compared to DHAP + G-CSF for stem cell mobilization in patients with lymphoma: An analysis by the polish lymphoma research group. Ann. Hematol. 2015, 95, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Jackstadt, R.; Kaller, M.; Hermeking, H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget 2013, 4, 1399–1415. [Google Scholar] [CrossRef]
- Fish, J.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.-F.; Wythe, J.; Ivey, K.N.; Bruneau, B.; Stainier, D.; Srivastava, D. miR-126 Regulates Angiogenic Signaling and Vascular Integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef]
- Sun, C.-Y.; She, X.-M.; Qin, Y.; Chu, Z.-B.; Chen, L.; Ai, L.-S.; Zhang, L.; Hu, Y. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis 2013, 34, 426–435. [Google Scholar] [CrossRef]
- Dai, G.-H.; Ma, P.-Z.; Song, X.-B.; Liu, N.; Zhang, T.; Wu, B. MicroRNA-223-3p Inhibits the Angiogenesis of Ischemic Cardiac Microvascular Endothelial Cells via Affecting RPS6KB1/hif-1a Signal Pathway. PLoS ONE 2014, 9, e108468. [Google Scholar] [CrossRef]
- Harris, T.A.; Yamakuchi, M.; Ferlito, M.; Mendell, J.T.; Lowenstein, C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA 2008, 105, 1516–1521. [Google Scholar] [CrossRef]
- Ratajczak, J.; Shin, N.-M.; Wan, W.; Liu, R.; Masternak, M.M.; Piotrowska, K.; Wiszniewska, B.; Kucia, M.; Bartke, A.; Ratajczak, M.Z. Higher number of stem cells in the bone marrow of circulating low Igf-1 level Laron dwarf mice—novel view on Igf-1, stem cells and aging. Leukemia 2011, 25, 729–733. [Google Scholar] [CrossRef]
- Jia, C.Y.; Li, H.H.; Zhu, X.C.; Dong, Y.W.; Fu, D.; Zhao, Q.L.; Wu, W.; Wu, X.Z. MiR-223 Suppresses Cell Proliferation by Targeting IGF-1R. PLoS ONE 2011, 6, e27008. [Google Scholar] [CrossRef]
- Chen, L.; Holmstrøm, K.; Qiu, W.; Ditzel, N.; Shi, K.; Hokland, L.; Kassem, M. MicroRNA-34a Inhibits Osteoblast Differentiation and In Vivo Bone Formation of Human Stromal Stem Cells. STEM CELLS 2014, 32, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Sessa, R.; Seano, G.; di Blasio, L.; Gagliardi, P.A.; Isella, C.; Medico, E.; Cotelli, F.; Bussolino, F.; Primo, L. The miR-126 regulates Angiopoietin-1 signaling and vessel maturation by targeting p85β. Biochim. Biophys. Acta Bioenerg. 2012, 1823, 1925–1935. [Google Scholar] [CrossRef]
- Sheng, X.; Zhong, H.; Wan, H.; Zhong, J.; Chen, F. Granulocyte colony-stimulating factor inhibits CXCR4/SDF-1α signaling and overcomes stromal-mediated drug resistance in the HL-60 cell line. Exp. Ther. Med. 2016, 12, 396–404. [Google Scholar] [CrossRef]
- Zarone, M.R.; Misso, G.; Grimaldi, A.; Zappavigna, S.; Russo, M.; Amler, E.; DI Martino, M.T.; Amodio, N.; Tagliaferri, P.; Tassone, P.; et al. Evidence of novel miR-34a-based therapeutic approaches for multiple myeloma treatment. Sci. Rep. 2017, 7, 17949. [Google Scholar] [CrossRef]
- Itkin, T.; Kumari, A.; Schneider, E.; Gur-Cohen, S.; Ludwig, C.; Brooks, R.; Kollet, O.; Golan, K.; Khatib-Massalha, E.; Russo, C.M.; et al. MicroRNA-155 promotes G-CSF-induced mobilization of murine hematopoietic stem and progenitor cells via propagation of CXCL12 signaling. Leukemia 2017, 31, 1247–1250. [Google Scholar] [CrossRef]
- Georgantas, R.W.; Hildreth, R.; Morisot, S.; Alder, J.; Liu, C.-G.; Heimfeld, S.; Calin, G.; Croce, C.M.; Civin, C.I. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA 2007, 104, 2750–2755. [Google Scholar] [CrossRef]
- Dos Santos, A.M.; Metzinger, L.; Haddad, O.; M’Baya-Moutoula, E.; Taïbi, F.; Charnaux, N.; Massy, Z.A.; Hlawaty, H.; Meuth, V.M.-L. miR-126 Is Involved in Vascular Remodeling under Laminar Shear Stress. BioMed Res. Int. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Nowicki, M.; Szemraj, J.; Wierzbowska, A.; Misiewicz, M.; Małachowski, R.; Pluta, A.; Grzybowska-Izydorczyk, O.; Robak, T.; Szmigielska-Kapłon, A. miRNA-15a, miRNA-16, miRNA-126, miRNA-146a, and miRNA-223 expressions in autologous hematopoietic stem cell transplantation and their impact on engraftment. Eur. J. Haematol. 2018, 100, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Corrigan-Cummins, M.; Barber, E.A.; Saleh, L.M.; Zingone, A.; Ghafoor, A.; Costello, R.; Zhang, Y.; Kurlander, R.J.; Korde, N.; et al. Aberrant Levels of miRNAs in Bone Marrow Microenvironment and Peripheral Blood of Myeloma Patients and Disease Progression. J. Mol. Diagn. 2015, 17, 669–678. [Google Scholar] [CrossRef]
- Fonseca, R.; Blood, E.; Rue, M.; Harrington, D.; Oken, M.M.; Kyle, R.A.; Dewald, G.W.; Van Ness, B.; Van Wier, S.A.; Henderson, K.J.; et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood 2003, 101, 4569–4575. [Google Scholar] [CrossRef]
- Artemaki, P.; Letsos, P.; Zoupa, I.; Katsaraki, K.; Karousi, P.; Papageorgiou, S.; Pappa, V.; Scorilas, A.; Kontos, C. The Multifaceted Role and Utility of MicroRNAs in Indolent B-Cell Non-Hodgkin Lymphomas. Biomedicines 2021, 9, 333. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zhang, L.; An, G.; Sui, W.; Yu, Z.; Zou, D.; Xu, Y.; Chang, H.; Qiu, L. Suppressing miRNA-15a/-16 expression by interleukin-6 enhances drug-resistance in myeloma cells. J. Hematol. Oncol. 2011, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Roccaro, A.; Sacco, A.; Thompson, B.; Leleu, X.; Azab, A.K.; Azab, F.; Runnels, J.; Jia, X.; Ngo, H.T.; Melhem, M.R.; et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood 2009, 113, 6669–6680. [Google Scholar] [CrossRef]
- Xie, L.; Jing, R.; Qi, J.; Lin, Z.; Ju, S. Drug resistance-related microRNAs in hematological malignancies: Translating basic evidence into therapeutic strategies. Blood Rev. 2015, 29, 33–44. [Google Scholar] [CrossRef]
- Papanota, A.-M.; Karousi, P.; Kontos, C.; Ntanasis-Stathopoulos, I.; Scorilas, A.; Terpos, E. Multiple Myeloma Bone Disease: Implication of MicroRNAs in Its Molecular Background. Int. J. Mol. Sci. 2021, 22, 2375. [Google Scholar] [CrossRef]
- Lei, W.; Wang, S.; Yang, C.; Huang, X.; Chen, Z.; He, W.; Shen, J.; Liu, X.; Qian, W. Combined expression of miR-34a and Smac mediated by oncolytic vaccinia virus synergistically promote anti-tumor effects in Multiple Myeloma. Sci. Rep. 2016, 6, 32174. [Google Scholar] [CrossRef]
- Han, B.; Wang, S.; Zhao, H. MicroRNA-21 and microRNA-155 promote the progression of Burkitt’s lymphoma by the PI3K/AKT signaling pathway. Int. J. Clin. Exp. Pathol. 2020, 13, 89–98. [Google Scholar]
- DI Martino, M.T.; Campani, V.; Misso, G.; Cantafio, M.E.G.; Gullà, A.; Foresta, U.; Guzzi, P.H.; Castellano, M.; Grimaldi, A.; Gigantino, V.; et al. In Vivo Activity of MiR-34a Mimics Delivered by Stable Nucleic Acid Lipid Particles (SNALPs) against Multiple Myeloma. PLoS ONE 2014, 9, e90005. [Google Scholar] [CrossRef] [PubMed]
- Donahue, R.E.; Jin, P.; Bonifacino, A.C.; Metzger, M.E.; Ren, J.; Wang, E.; Stroncek, D.F. Plerixafor (AMD3100) and granulocyte colony-stimulating factor (G-CSF) mobilize different CD34+ cell populations based on global gene and microRNA expression signatures. Blood 2009, 114, 2530–2541. [Google Scholar] [CrossRef]
- Shen, W.-F.; Hu, Y.-L.; Uttarwar, L.; Passegue, E.; Largman, C. MicroRNA-126 Regulates HOXA9 by Binding to the Homeobox. Mol. Cell. Biol. 2008, 28, 4609–4619. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, O.; Jiang, K.; Gasperini, P.; Maric, D.; Zhu, J.; Sakakibara, S.; Espigol-Frigole, G.; Wang, S.; Tosato, G. MicroRNA126 contributes to granulocyte colony-stimulating factor-induced hematopoietic progenitor cell mobilization by reducing the expression of vascular cell adhesion molecule 1. Haematologica 2012, 97, 818–826. [Google Scholar] [CrossRef]
- Lechman, E.; Gentner, B.; van Galen, P.; Giustacchini, A.; Saini, M.; Boccalatte, F.; Hiramatsu, H.; Restuccia, U.; Bachi, A.; Voisin, V.; et al. Attenuation of miR-126 Activity Expands HSC In Vivo without Exhaustion. Cell Stem Cell 2012, 11, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.L.; Rao, D.S.; O’Connell, R.M.; Garcia-Flores, Y.; Baltimore, D.; Morrison, S.J. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. eLife 2013, 2, e00537. [Google Scholar] [CrossRef]
- Zhao, J.L.; Starczynowski, D.T. Role of microRNA-146a in normal and malignant hematopoietic stem cell function. Front. Genet. 2014, 5, 219. [Google Scholar] [CrossRef] [PubMed]
- Magilnick, N.; Reyes, E.Y.; Wang, W.-L.; Vonderfecht, S.L.; Gohda, J.; Inoue, J.-I.; Boldin, M.P. miR-146a–Traf6 regulatory axis controls autoimmunity and myelopoiesis, but is dispensable for hematopoietic stem cell homeostasis and tumor suppression. Proc. Natl. Acad. Sci. USA 2017, 114, E7140–E7149. [Google Scholar] [CrossRef] [PubMed]
- Labbaye, C.; Spinello, I.; Quaranta, M.T.; Pelosi, E.; Pasquini, L.; Petrucci, E.; Biffoni, M.; Nuzzolo, E.R.; Billi, M.; Foà, R.; et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat. Cell Biol. 2008, 10, 788–801. [Google Scholar] [CrossRef]
- Bissels, U.; Wild, S.; Tomiuk, S.; Hafner, M.; Scheel, H.; Mihailovic, A.; Choi, Y.-H.; Tuschl, T.; Bosio, A. Combined Characterization of microRNA and mRNA Profiles Delineates Early Differentiation Pathways of CD133+and CD34+Hematopoietic Stem and Progenitor Cells. Stem Cells 2011, 29, 847–857. [Google Scholar] [CrossRef]
- Bi, C.; Chung, T.-H.; Huang, G.; Zhou, J.; Yan, J.J.; Ahmann, G.J.; Fonseca, R.; Chng, W.J. Genome-wide pharmacologic unmasking identifies tumor suppressive microRNAs in multiple myeloma. Oncotarget 2015, 6, 26508–26518. [Google Scholar] [CrossRef]
- Due, H.; Svendsen, P.; Bødker, J.S.; Schmitz, A.; Bøgsted, M.; Johnsen, H.E.; El-Galaly, T.C.; Roug, A.S.; Dybkær, K. miR-155 as a Biomarker in B-Cell Malignancies. BioMed Res. Int. 2016, 2016, 1–14. [Google Scholar] [CrossRef]
- Zhang, H.; Goudeva, L.; Immenschuh, S.; Schambach, A.; Skokowa, J.; Eiz-Vesper, B.; Blasczyk, R.; Figueiredo, C. miR-155 Is Associated with the Leukemogenic Potential of the Class IV Granulocyte Colony-Stimulating Factor Receptor in CD34+ Progenitor Cells. Mol. Med. 2014, 20, 736–746. [Google Scholar] [CrossRef]
- Li, X.-D.; Gu, J.-W.; Sun, X.-C. MiR-155 regulates lymphoma cell proliferation and apoptosis through targeting SOCS3/JAK-STAT3 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5153–5159. [Google Scholar]
- Ahmadvand, M.; Eskandari, M.; Pashaiefar, H.; Yaghmaie, M.; Manoochehrabadi, S.; Khakpour, G.; Sheikhsaran, F.; Zohour, M.M. Over expression of circulating miR-155 predicts prognosis in diffuse large B-cell lymphoma. Leuk. Res. 2018, 70, 45–48. [Google Scholar] [CrossRef]
- Krzeminski, P.; Sarasquete, M.E.; Misiewicz-Krzeminska, I.; Corral, R.; Corchete, L.A.; Martín, A.A.; Garcia-Sanz, R.; Miguel, J.S.; Gutiérrez, N.C. Insights into epigenetic regulation of microRNA-155 expression in multiple myeloma. Biochim. Biophys. Acta Bioenerg. 2015, 1849, 353–366. [Google Scholar] [CrossRef]
- Amodio, N.; Cantafio, M.E.G.; Botta, C.; Agosti, V.; Federico, C.; Caracciolo, D.; Ronchetti, D.; Rossi, M.; Driessen, C.; Neri, A.; et al. Replacement of miR-155 Elicits Tumor Suppressive Activity and Antagonizes Bortezomib Resistance in Multiple Myeloma. Cancers 2019, 11, 236. [Google Scholar] [CrossRef]
- Duan, W.; Chen, Y.; Wang, X.R. MicroRNA-155 contributes to the occurrence of epilepsy through the PI3K/Akt/mTOR signaling pathway. Int. J. Mol. Med. 2018, 42, 1577–1584. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef]
- Xiao, Y.; Su, C.; Deng, T. miR-223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7. Oncol. Lett. 2016, 12, 3531–3536. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; O’Neill, L.A.J.; Masters, S.L. miR-223: Infection, inflammation and cancer. J. Intern. Med. 2013, 274, 215–226. [Google Scholar] [CrossRef]
- Gentner, B.; Pochert, N.; Rouhi, A.; Boccalatte, F.; Plati, T.; Berg, T.; Sun, S.M.; Mah, S.M.; Mirkovic-Hösle, M.; Ruschmann, J.; et al. MicroRNA-223 dose levels fine tune proliferation and differentiation in human cord blood progenitors and acute myeloid leukemia. Exp. Hematol. 2015, 43, 858–868.e7. [Google Scholar] [CrossRef]
- Yuan, X.; Berg, N.; Lee, J.W.; Le, T.-T.; Neudecker, V.; Jing, N.; Eltzschig, H. MicroRNA miR-223 as regulator of innate immunity. J. Leukoc. Biol. 2018, 104, 515–524. [Google Scholar] [CrossRef]
- Vian, L.; Di Carlo, M.; Pelosi, E.; Fazi, F.; Santoro, S.; Cerio, A.M.; Boe, A.; Rotilio, V.; Billi, M.; Racanicchi, S.; et al. Transcriptional fine-tuning of microRNA-223 levels directs lineage choice of human hematopoietic progenitors. Cell Death Differ. 2014, 21, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhao, F.; Zhang, J.; Zhu, H.; Ma, H.; Li, X.; Peng, L.; Sun, J.; Chen, Z. miR-223 reverses the resistance of EGFR-TKIs through IGF1R/PI3K/Akt signaling pathway. Int. J. Oncol. 2016, 48, 1855–1867. [Google Scholar] [CrossRef]
- Trissal, M.C.; DeMoya, R.A.; Schmidt, A.P.; Link, D.C. MicroRNA-223 Regulates Granulopoiesis but Is Not Required for HSC Maintenance in Mice. PLoS ONE 2015, 10, e0119304. [Google Scholar] [CrossRef]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nat. Cell Biol. 2008, 451, 1125–1129. [Google Scholar] [CrossRef]
- Olivieri, A.; on behalf of the Italian Group for Stem Cell Transplantation (GITMO); Marchetti, M.; Lemoli, R.; Tarella, C.; Iacone, A.; Lanza, F.; Rambaldi, A.; Bosi, A. Proposed definition of ‘poor mobilizer’ in lymphoma and multiple myeloma: An analytic hierarchy process by ad hoc working group Gruppo ItalianoTrapianto di Midollo Osseo. Bone Marrow Transplant. 2011, 47, 342–351. [Google Scholar] [CrossRef]
- Báez, A.; Martin-Antonio, B.; Piruat, J.I.; Barbado, M.V.; Prats, C.; Álvarez-Laderas, I.; Carmona, M.; Perez-Simón, J.A.; Urbano-Ispizua, A. Gene and miRNA Expression Profiles of Hematopoietic Progenitor Cells Vary Depending on Their Origin. Biol. Blood Marrow Transplant. 2014, 20, 630–639. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Blix, E.S.; Husebekk, A. Raiders of the lost mark–endothelial cells and their role in transplantation for hematologic malignancies. Leuk. Lymphoma 2016, 57, 2752–2762. [Google Scholar] [CrossRef]
- Zhong, H.; Xu, L.; Zhong, J.-H.; Xiao, F.; Liu, Q.; Huang, H.-H.; Chen, F.-Y. Clinical and prognostic significance of miR-155 and miR-146a expression levels in formalin-fixed/paraffin-embedded tissue of patients with diffuse large B-cell lymphoma. Exp. Ther. Med. 2012, 3, 763–770. [Google Scholar] [CrossRef]
- Spinello, I.; Quaranta, M.T.; Riccioni, R.; Riti, V.; Pasquini, L.; Boe, A.; Pelosi, E.; Vitale, A.; Foà, R.; Testa, U.; et al. MicroRNA-146a and AMD3100, two ways to control CXCR4 expression in acute myeloid leukemias. Blood Cancer J. 2011, 1, e26. [Google Scholar] [CrossRef]
- Labbaye, C.; Testa, U. The emerging role of MIR-146A in the control of hematopoiesis, immune function and cancer. J. Hematol. Oncol. 2012, 5, 13. [Google Scholar] [CrossRef]
- Hattori, K.; Dias, S.; Heissig, B.; Hackett, N.R.; Lyden, D.; Tateno, M.; Hicklin, D.J.; Zhu, Z.; Witte, L.; Crystal, R.G.; et al. Vascular Endothelial Growth Factor and Angiopoietin-1 Stimulate Postnatal Hematopoiesis by Recruitment of Vasculogenic and Hematopoietic Stem Cells. J. Exp. Med. 2001, 193, 1005–1014. [Google Scholar] [CrossRef]
| Characteristics | Numbers |
|---|---|
| Age (years) | Median 60 (range 44–69) |
| Sex (female/male) | 25/25 |
| Multiple myeloma | 39 (7 CR, 24 VGPR, 8 PR) |
| Hodgkin lymphoma | 4 (1 CR, 3 PR) |
| non-Hodgkin lymphoma: | 7 |
| Diffuse large B-cell lymphoma | 3 (1 CR, 2 PR) |
| Mantle cell lymphoma | 2 (CR) |
| Anaplastic large-cell lymphoma | 1 (PR) |
| Hepatosplenic T-cell lymphoma | 1 (PR) |
| CD34+ cells collected during mobilization (total number) [×106/kg] | Median 5.07 (range 2.2–21) |
| CD34+ collected on Day A [×106/kg] | Median 3.0 (range 0.3–21) |
| Number of apheresis needed to collect at least 2 × 106/kg CD34+ | Median 2 (range 1–6) |
| WBC count on Day A [×103/µL] | Median 16.67 (range 2.68–47.42) |
| Mobilization chemotherapy: | |
| Multiple myeloma | |
| Endoxan (Cyclophosphamide) | 25 |
| DCEP (Dexamethasone, Cyclophosphamide, | 7 |
| Cisplatin, Etoposide) | |
| Alexan (Cytarabine) | 3 |
| only G-CSF in monotherapy | 5 |
| Hodgkin and non-Hodgkin lymphoma | |
| ICE (Ifosfamide, Carboplatin, Etoposide) | 4 |
| R-ICE (with rituximab) | 1 |
| DHAP (Dexamethasone, Cytarabine, Cisplatin) | 1 |
| R-DHAP (with rituximab) | 2 |
| Endoxan (Cyclophosphamide) | 1 |
| Alexan (Cytarabine) | 1 |
| Mobilization efficacy | |
| Good mobilizers | 44 |
| Poor mobilizers | 6 |
| Factor | Odds Ratio (95% CI) | p Value |
|---|---|---|
| Male vs. female | 1.64 (0.26–10.21) | 0.59 |
| Age (continuous) | 1.03 (0.94–1.13) | 0.46 |
| CR vs. not CR | 1.17 (0.13–10.89) | 0.89 |
| hsa-miR-15a-5p (Day 0) | 1.01 (0.96–1.07) | 0.69 |
| hsa-miR-146a-5p (Day 0) | 0.99 (0.86–1.13) | 0.89 |
| hsa-miR-15a-5p (Day A) | 0.89 (0.74–1.08) | 0.23 |
| hsa-miR-146a-5p (Day A) | 1.88 (1.06–3.33) | 0.03 |
| miRNA | Day 0 | Day A | p Value |
|---|---|---|---|
| hsa-miR-15a-5p | Me = 21.92 range: 1–156.83 | Me = 12.67 range: 1.24–50.35 | p < 0.001 |
| hsa-miR-16-5p | Me = 3.03 range: 0.64–16.53 | Me = 2.14 range: 0.52–19.07 | p = 0.006 |
| hsa-miR-126-3p | Me = 4.39 range: 0.46–20.38 | Me = 3.83 range: 0.69–10.9 | p = 0.03 |
| hsa-miR-146a-5p | Me = 10.42 range: 0.40–49.52 | Me = 4.74 range: 0.16–12.42 | p < 0.001 |
| hsa-miR-223-3p | Me = 14.51 range: 0.69–105.31 | Me = 15.69 range: 1.9–55.84 | p = 0.66 |
| hsa-miR-34a-5p | Me = 1.74 range: 0.04–5.13 | Me = 3.95 range: 0.48–18.78 | p < 0.001 |
| hsa-miR-155-5p | Me = 0.80 range: 0.16–1.69 | Me = 0.66 range: 0.13–1.98 | p = 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowicki, M.; Szemraj, J.; Wierzbowska, A.; Pluta, A.; Grzybowska-Izydorczyk, O.; Nowicka, A.; Stelmach, P.; Czemerska, M.; Szmigielska-Kapłon, A. Alterations in microRNA Expression during Hematopoietic Stem Cell Mobilization. Biology 2021, 10, 668. https://doi.org/10.3390/biology10070668
Nowicki M, Szemraj J, Wierzbowska A, Pluta A, Grzybowska-Izydorczyk O, Nowicka A, Stelmach P, Czemerska M, Szmigielska-Kapłon A. Alterations in microRNA Expression during Hematopoietic Stem Cell Mobilization. Biology. 2021; 10(7):668. https://doi.org/10.3390/biology10070668
Chicago/Turabian StyleNowicki, Mateusz, Janusz Szemraj, Agnieszka Wierzbowska, Agnieszka Pluta, Olga Grzybowska-Izydorczyk, Aleksandra Nowicka, Piotr Stelmach, Magdalena Czemerska, and Anna Szmigielska-Kapłon. 2021. "Alterations in microRNA Expression during Hematopoietic Stem Cell Mobilization" Biology 10, no. 7: 668. https://doi.org/10.3390/biology10070668
APA StyleNowicki, M., Szemraj, J., Wierzbowska, A., Pluta, A., Grzybowska-Izydorczyk, O., Nowicka, A., Stelmach, P., Czemerska, M., & Szmigielska-Kapłon, A. (2021). Alterations in microRNA Expression during Hematopoietic Stem Cell Mobilization. Biology, 10(7), 668. https://doi.org/10.3390/biology10070668





