Transdifferentiation of Human Fibroblasts into Skeletal Muscle Cells: Optimization and Assembly into Engineered Tissue Constructs through Biological Ligands
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmid Construction and Lentivirus Preparation
2.3. Viral Transduction
2.4. Ligand Screening and Combination Experiments
2.5. PDMS Fabrication
2.6. Construction of Engineered tHFs and C2C12 Tissue Culture
2.7. Cell Immunohistochemistry and Imaging
2.8. Tissue Immunohistochemistry and Imaging
2.9. Statistical Analysis
3. Results
3.1. Skeletal Muscle Screening of Biological Ligands
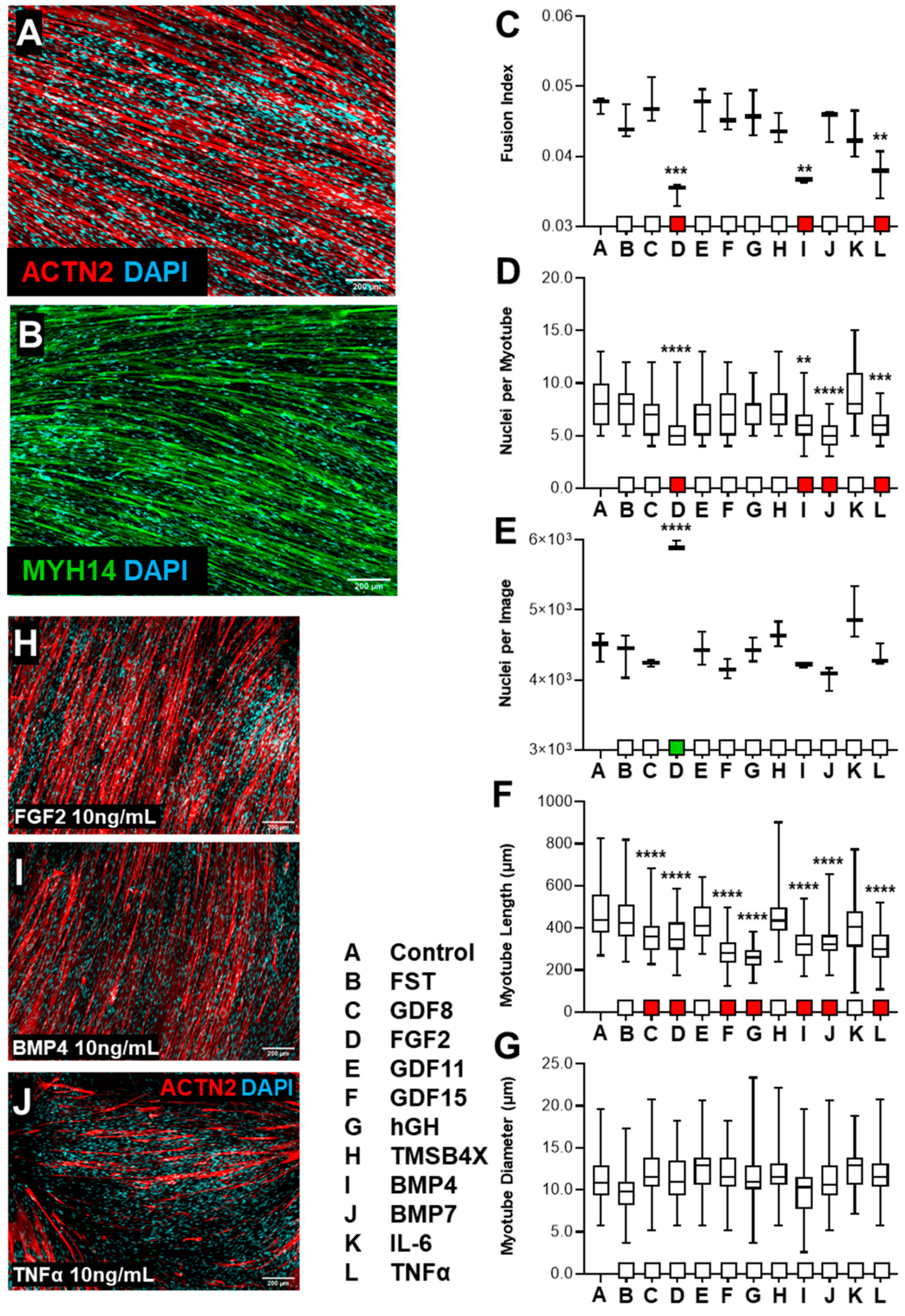
3.1.1. Effect of Screening C2C12s with Biological Ligands
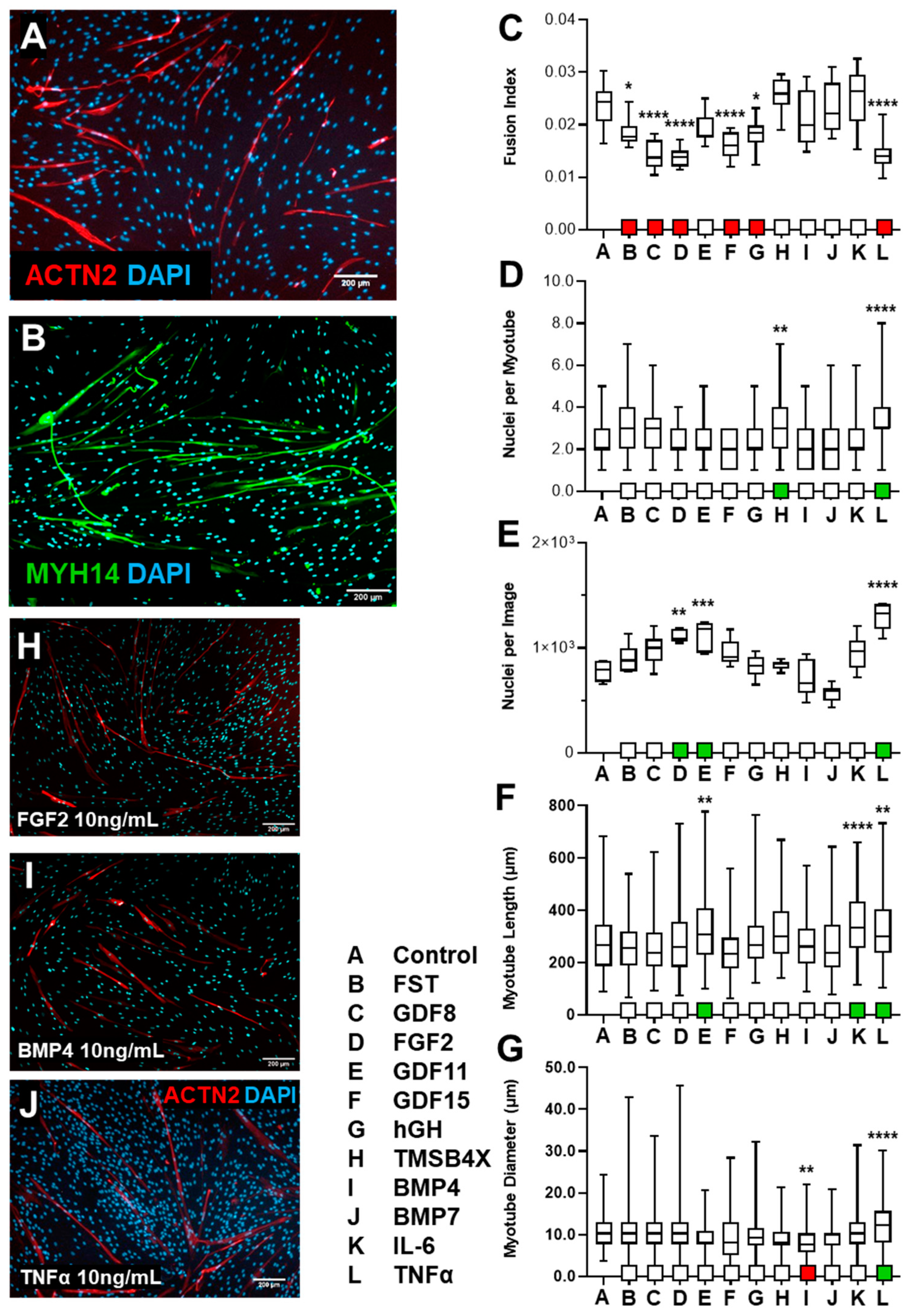
3.1.2. HSkM Response to Biological Ligand Exposure Was Limited
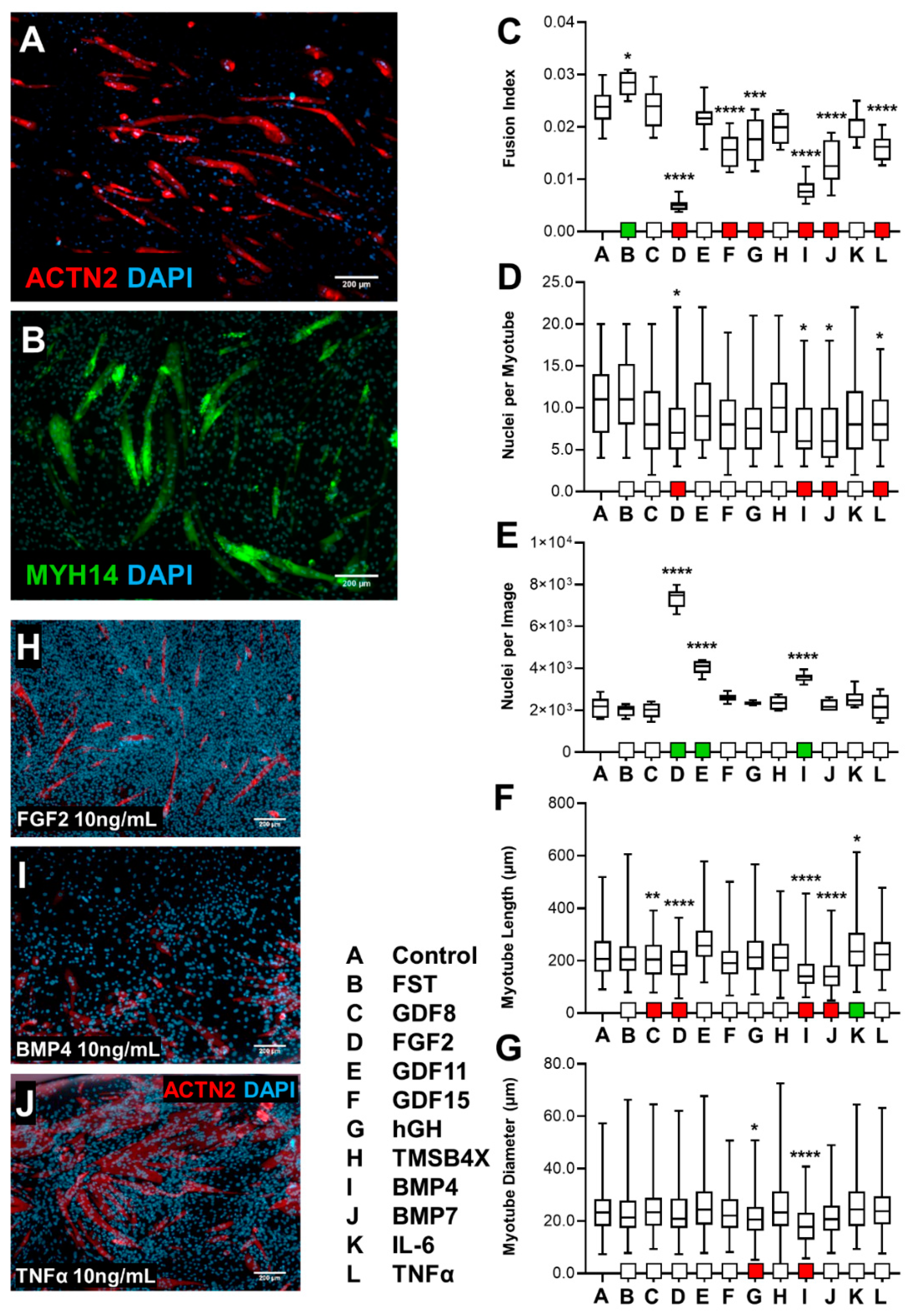
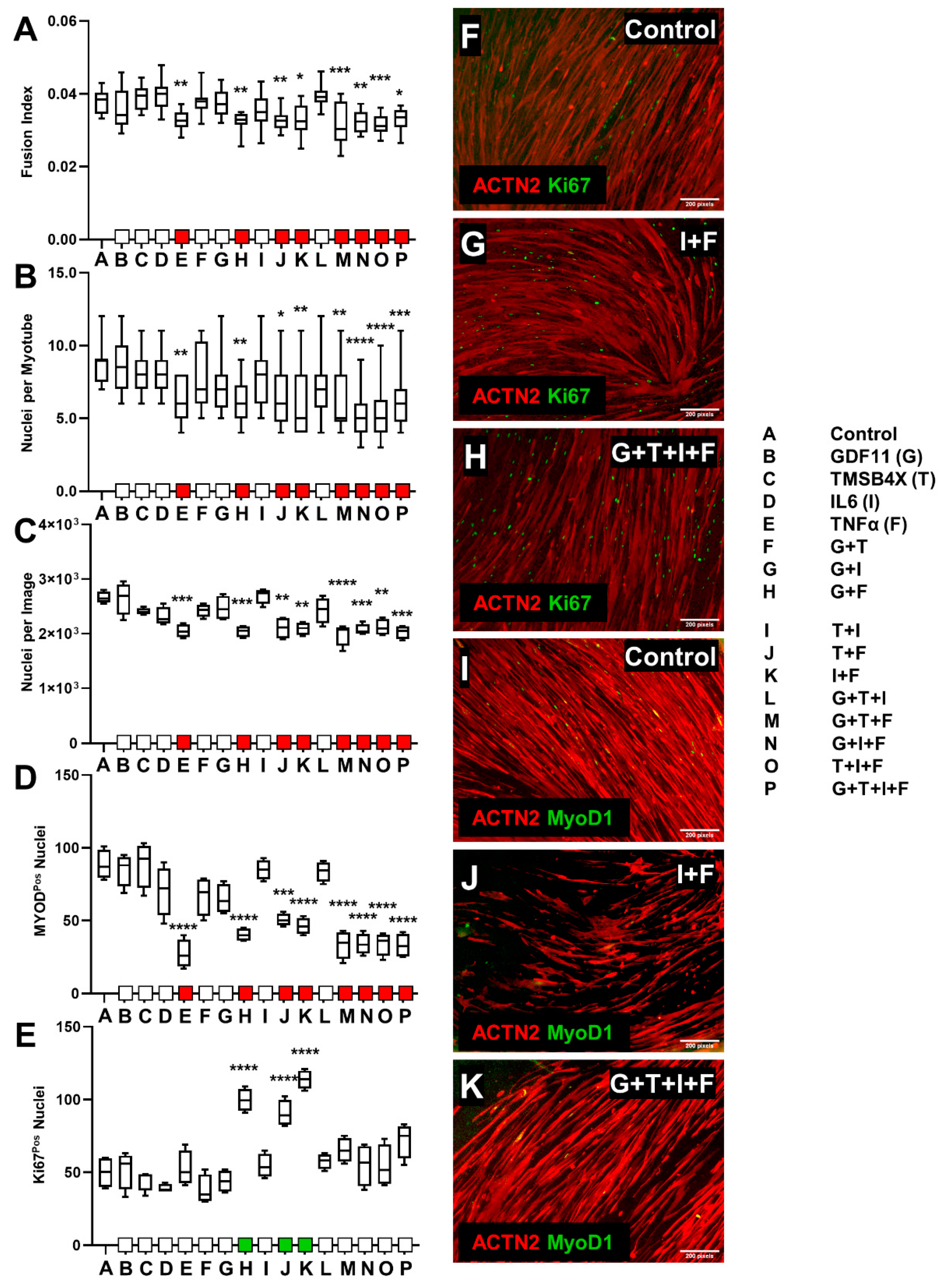
3.1.3. tHF Treatment with Biological Ligands Resulted in Decreased Differentiation with Partial Increases in Proliferation
3.2. Ligand Combination Contribution to Skeletal Muscle Differentiation
3.2.1. C2C12 Ligand Combination Did Not Improve Differentiation Response
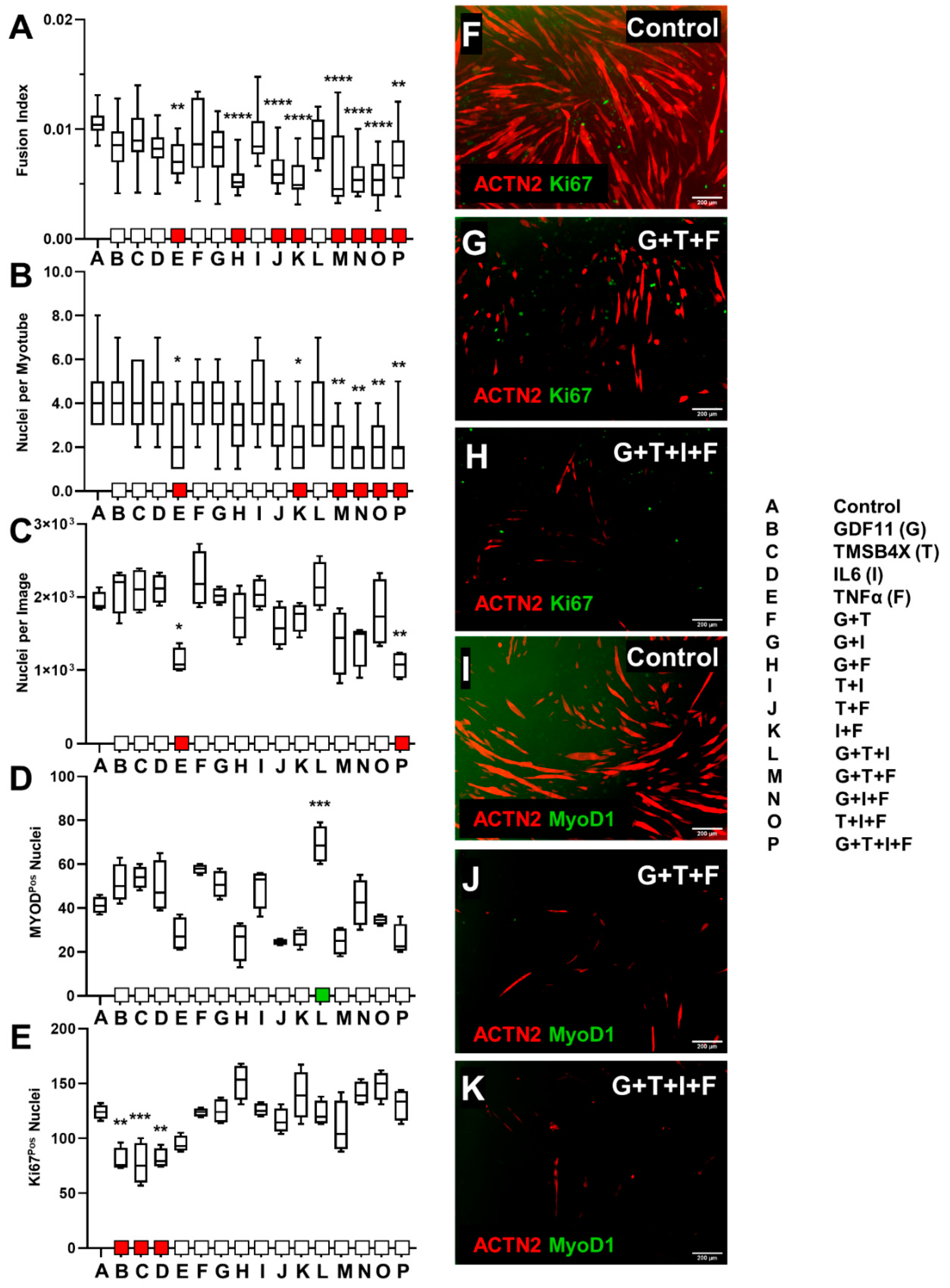
3.2.2. tHFs Ligand Combinations Were Negatively Associated with Differentiation Capacity
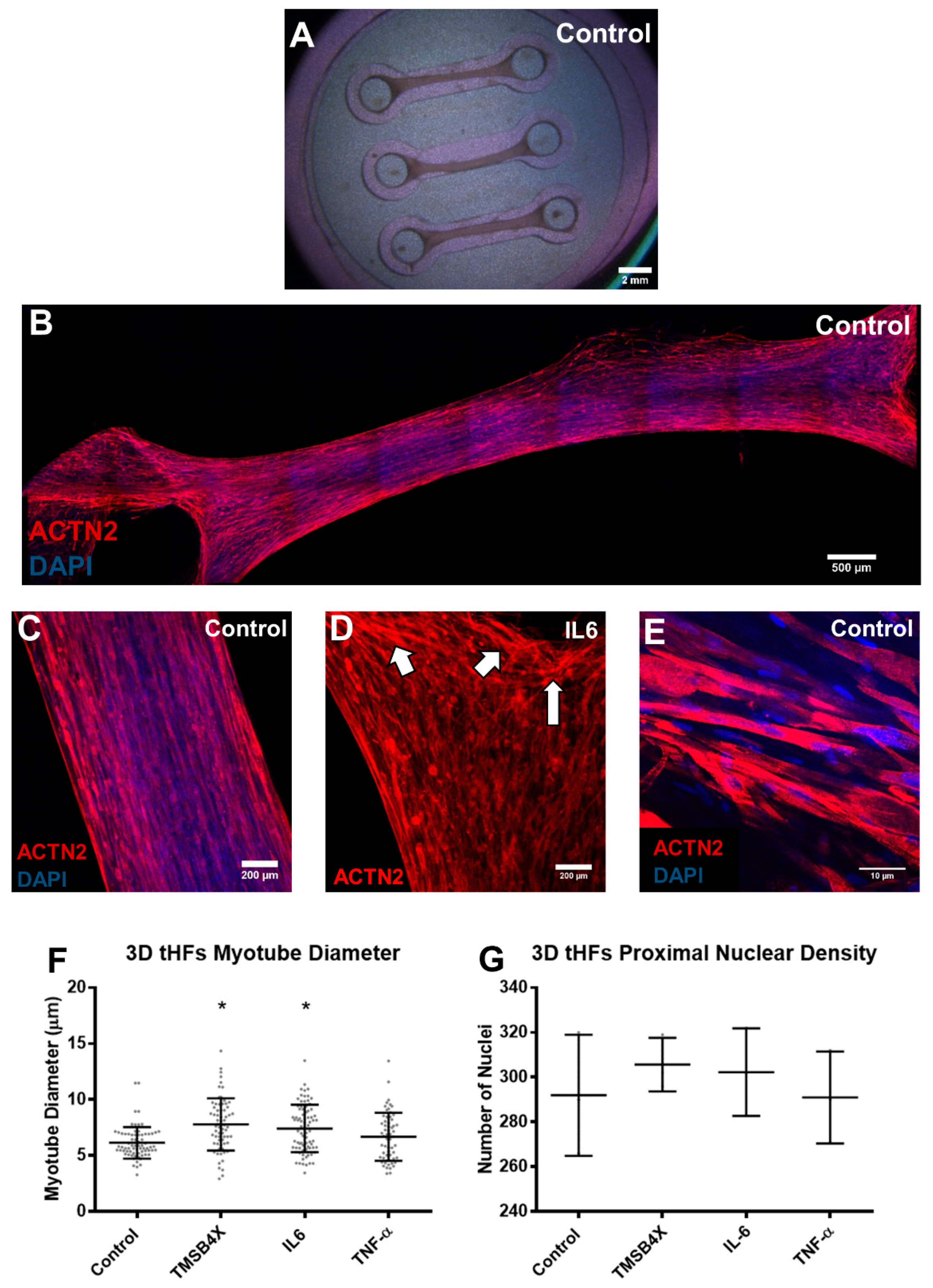
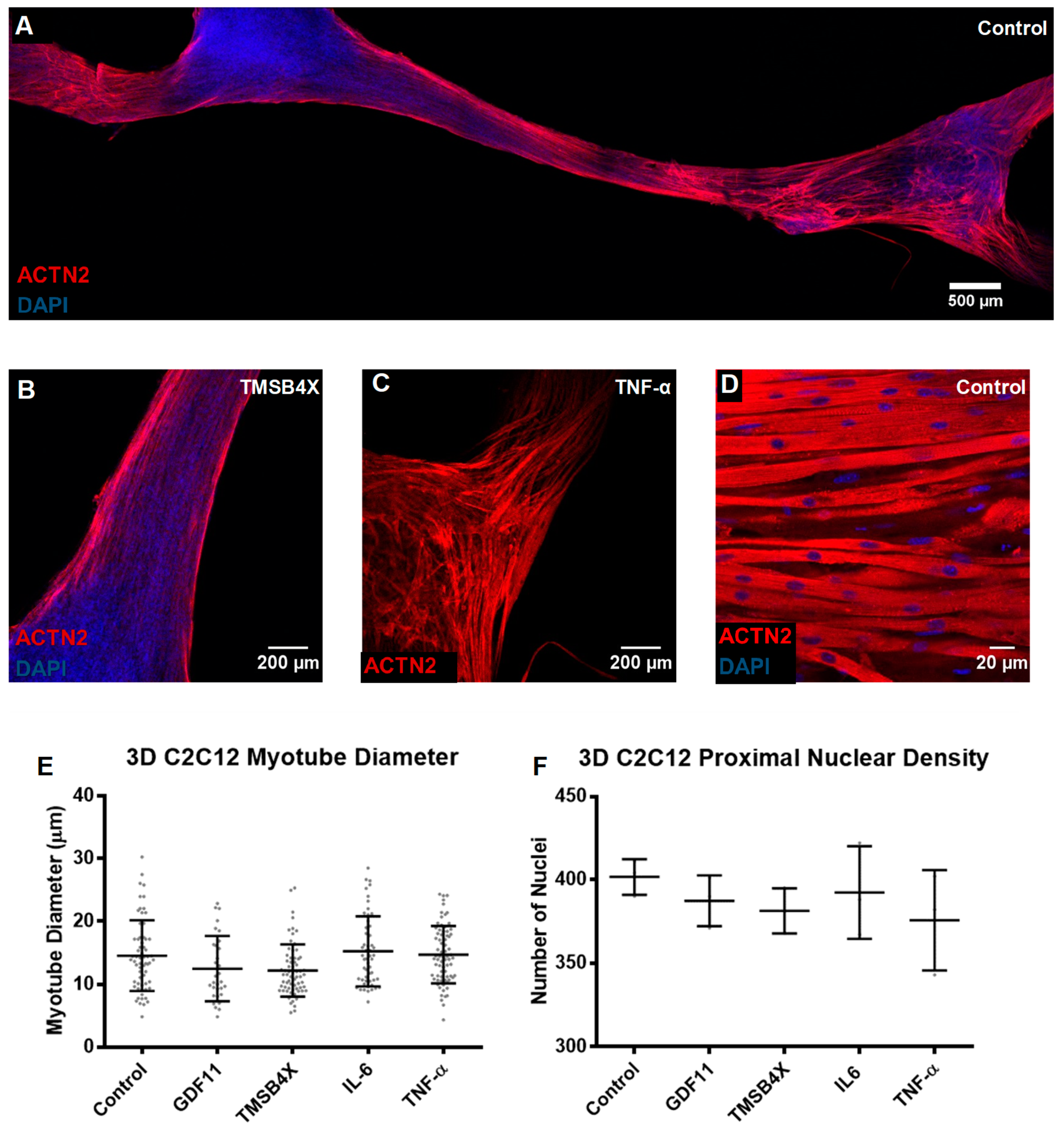
3.3. Tissue Engineering of C2C12s and tHFs Derived Skeletal Muscle Constructs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schultz, E.; McCormick, K.M. Skeletal muscle satellite cells. Rev. Physiol. Biochem. Pharmacol. 1994, 123, 213. Available online: http://www.ncbi.nlm.nih.gov/pubmed/8209136 (accessed on 7 May 2018).
- Tidball, J.G. Inflammatory processes in muscle injury and repair. Am. J. Physiol. Integr. Comp. Physiol. 2005, 288, R345–R353. [Google Scholar] [CrossRef]
- Fisher, M.B.; Mauck, R.L. Tissue Engineering and Regenerative Medicine: Recent Innovations and the Transition to Translation. Tissue Eng. Part B Rev. 2012, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chen, K.; Whaley-Connell, A.T.; Stump, C.S.; Ibdah, J.A.; Sowers, J.R. Skeletal muscle insulin resistance: Role of inflammatory cytokines and reactive oxygen species. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R673–R680. [Google Scholar] [CrossRef]
- Fairclough, R.J.; Wood, M.J.; Davies, K.E. Therapy for Duchenne muscular dystrophy: Renewed optimism from genetic approaches. Nat. Rev. Genet. 2013, 14, 373. Available online: http://www.nature.com/doifinder/10.1038/nrg3460%5Cn (accessed on 20 July 2018). [CrossRef]
- Grogan, B.F.; Hsu, J.R.; Skeletal Trauma Research Consortium. Volumetric Muscle Loss. J. Am. Acad. Orthop. Surg. 2011, 19, S35–S37. [Google Scholar] [CrossRef]
- Klumpp, D.; Horch, R.E.; Kneser, U.; Beier, J.P. Engineering skeletal muscle tissue-new perspectives in vitro and in vivo. J. Cell. Mol. Med. 2010, 14, 2622–2629. [Google Scholar] [CrossRef]
- Wang, J.; Khodabukus, A.; Rao, L.; Vandusen, K.; Abutaleb, N.; Bursac, N. Engineered skeletal muscles for disease modeling and drug discovery. Biomaterials 2019, 221, 119416. [Google Scholar] [CrossRef]
- Khodabukus, A.; Prabhu, N.; Wang, J.; Bursac, N. In Vitro Tissue-Engineered Skeletal Muscle Models for Studying Muscle Physiology and Disease. Adv. Healthc. Mater. 2018, 7, 1701498. [Google Scholar] [CrossRef]
- Kjær, M. Role of Extracellular Matrix in Adaptation of Tendon and Skeletal Muscle to Mechanical Loading. Physiol. Rev. 2004, 84, 649–698. [Google Scholar] [CrossRef]
- Velleman, S.G. The role of the extracellular matrix in skeletal muscle development. Poult. Sci. 1999, 78, 778–784. [Google Scholar] [CrossRef]
- Osses, N.; Brandan, E. ECM is required for skeletal muscle differentiation independently of muscle regulatory factor expression. Am. J. Physiol. Cell Physiol. 2002, 282, C383–C394. [Google Scholar] [CrossRef] [PubMed]
- Gillies, A.R.; Lieber, R.L. Structure and function of the skeletal muscle extracellular matrix. Muscle Nerve 2011, 44, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Qazi, T.H.; Mooney, D.J.; Pumberger, M.; Geiler, S.; Duda, G.N. Biomaterials based strategies for skeletal muscle tissue engineering: Existing technologies and future trends. Biomaterials 2015, 53, 502–521. [Google Scholar] [CrossRef]
- Wolf, M.T.; Dearth, C.L.; Sonnenberg, S.B.; Loboa, E.G.; Badylak, S.F. Naturally derived and synthetic scaffolds for skeletal muscle reconstruction. Adv. Drug Deliv. Rev. 2015, 84, 208–221. [Google Scholar] [CrossRef] [PubMed]
- Badylak, S.F.; Freytes, D.O.; Gilbert, T.W. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomatera. 2009, 5, 1–13. [Google Scholar] [CrossRef]
- Cezar, C.A.; Mooney, D.J. Biomaterial-based delivery for skeletal muscle repair. Adv. Drug Deliv. Rev. 2015, 84, 188–197. [Google Scholar] [CrossRef]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef]
- Cheng, C.S.; El-Abd, Y.; Bui, K.; Hyun, Y.-E.; Hughes, R.H.; Kraus, W.E.; Truskey, G.A. Conditions that promote primary human skeletal myoblast culture and muscle differentiation in vitro. Am. J. Physiol. Cell Physiol. 2014, 306, C385–C395. Available online: http://ajpcell.physiology.org.ubproxy.ub.uni-heidelberg.de/content/306/4/C385.jnl_ajpcell_tab_info (accessed on 5 August 2018). [CrossRef]
- Powell, C.A.; Smiley, B.L.; Mills, J.; Vandenburgh, H.H. Mechanical stimulation improves tissue-engineered human skeletal muscle. Am. J. Physiol. Cell Physiol. 2002, 283, C1557–C1565. Available online: http://www.ncbi.nlm.nih.gov/pubmed/12372817 (accessed on 5 August 2018). [CrossRef] [PubMed]
- Tedesco, F.S.; Dellavalle, A.; Diaz-manera, J.; Messina, G.; Cossu, G. Review series repairing skeletal muscle: Regenerative potential of skeletal muscle stem cells. J. Clin. Investig. 2010, 120, 11–19. [Google Scholar] [CrossRef]
- Pantellic, M.N.; Larkin, L.M. Stem Cells for Skeletal Muscle Tissue Engineering. Tissue Eng. Part B Rev. 2018, 24, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Abujarour, R.; Bennett, M.; Valamehr, B.; Lee, T.T.; Robinson, M.; Robbins, D.; Le, T.; Lai, K.; Flynn, P. Embryonic Stem Cells/Induced Pluripotent Stem (iPS) Cells Myogenic Differentiation of Muscular Dystrophy- Specific Induced Pluripotent Stem Cells for Use in Drug Discovery. Stem Cells Transl. Med. 2014, 3, 149. [Google Scholar] [CrossRef]
- Maffioletti, S.M.; Sarcar, S.; Henderson, A.B.; Mannhardt, I.; Pinton, L.; Moyle, L.A.; Steele-Stallard, H.; Cappellari, O.; Wells, K.E.; Ferrari, G.; et al. Three-Dimensional Human iPSC-Derived Artificial Skeletal Muscles Model Muscular Dystrophies and Enable Multilineage Tissue Engineering. Cell Rep. 2018, 23, 899–908. [Google Scholar] [CrossRef]
- Rao, L.; Qian, Y.; Khodabukus, A.; Ribar, T.; Bursac, N. Engineering human pluripotent stem cells into a functional skeletal muscle tissue. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bakooshli, M.A.; Lippmann, E.S.; Mulcahy, B.; Iyer, N.; Nguyen, C.T.; Tung, K.; Stewart, B.A.; van den Dorpel, H.; Fuehrmann, T.; Shoichet, M.; et al. A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction. Elife 2019, 8, e44530. [Google Scholar] [CrossRef]
- Osaki, T.; Uzel, S.G.M.; Kamm, R.D. On-chip 3D neuromuscular model for drug screening and precision medicine in neuromuscular disease. Nat. Protoc. 2020, 15, 421–449. [Google Scholar] [CrossRef] [PubMed]
- Urciuolo, A.; Serena, E.; Ghua, R.; Zatti, S.; Giomo, M.; Mattei, N.; Vetralla, M.; Selmin, G.; Luni, C.; Vitulo, N. Engineering a 3D in vitro model of human skeletal muscle at the single fiber scale. PLoS ONE 2020, 15, e0232081. [Google Scholar] [CrossRef]
- Selman, K.; Kafatos, F.C. Transdifferentiation in the labial gland of silk moths: Is DNA required for cellular metamorphosis? Cell Differ. 1974, 3, 81–94. [Google Scholar] [CrossRef]
- Boukamp, P.; Chen, J.; Gonzales, F.; Jones, P.A.; Fusenig, N.E. Progressive stages of “transdifferentiation” from epidermal to mesenchymal phenotype induced by MyoD1 transfection, 5-aza-2-deoxycytidine treatment, and selection for reduced cell attachment in the human keratinocyte line HaCaT. J. Cell Biol. 1992, 116, 1257–1271. [Google Scholar] [CrossRef]
- Zhou, Q.; Melton, D.A. Extreme Makeover: Converting One Cell into Another. Cell Stem Cell 2008, 3, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Boue, S.; Belmonte, J.C.I. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, M.; Sebastian-Perez, R.; Cosma, M.P. Dedifferentiation, transdifferentiation and cell fusion: In vivo reprogramming strategies for regenerative medicine. FEBS J. 2018, 286, 1074–1093. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, K.; Wang, J. Exploring the Mechanisms of Differentiation, Dedifferentiation, Reprogramming and Transdifferentiation. PLoS ONE 2014, 9, e105216. [Google Scholar] [CrossRef]
- Smart, N.; Bollini, S.; Dubé, K.N.; Vieira, J.; Zhou, B.; Davidson, S.; Yellon, D.; Riegler, J.; Price, A.N.; Lythgoe, M.; et al. De novo cardiomyocytes from within the activated adult heart after injury. Nat. Cell Biol. 2011, 474, 640–644. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct Reprogramming of Fibroblasts into Functional Cardiomyocytes by Defined Factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef]
- Kalani, M.Y.S.; Martirosyan, N. Direct conversion of fibroblasts to functional neurons. World Neurosurg. 2012, 77, 7–8. [Google Scholar]
- Kaur, K.; Yang, J.; Eisenberg, C.A.; Eisenberg, L.M. 5-Azacytidine Promotes the Transdifferentiation of Cardiac Cells to Skeletal Myocytes. Cell. Reprogram. 2014, 16, 324. Available online: http://www.ncbi.nlm.nih.gov/pubmed/25090621 (accessed on 27 September 2018). [CrossRef]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- L’Ecuyer, T.J.; Tompach, P.C.; Morris, E.; Fulton, A.B. Transdifferentiation of chicken embryonic cells into muscle cells by the 3′ untranslated region of muscle tropomyosin. Proc. Natl. Acad. Sci. USA 1995, 92, 7520–7524. [Google Scholar] [CrossRef]
- Anastasia, L.; Sampaolesi, M.; Papini, N.; Oleari, D.; Lamorte, G.; Tringali, C.; Monti, E.; Galli, D.; Tettamanti, G.; Cossu, G.; et al. Reversine-treated fibroblasts acquire myogenic competence in vitro and in regenerating skeletal muscle. Cell Death Differ. 2006, 13, 2042–2051. [Google Scholar] [CrossRef]
- Veldman, M.B.; Zhao, C.; Gomez, G.A.; Lindgren, A.G.; Huang, H.; Yang, H.; Yao, S.; Martin, B.L.; Kimelman, D.; Lin, S. Transdifferentiation of Fast Skeletal Muscle Into Functional Endothelium in Vivo by Transcription Factor Etv2. PLoS Biol. 2013, 11, e1001590. [Google Scholar] [CrossRef]
- Manandhar, D.; Song, L.; Kabadi, A.; Kwon, J.B.; Edsall, L.E.; Ehrlich, M.; Tsumagari, K.; Gersbach, C.A.; Crawford, G.E.; Gordân, R. Incomplete MyoD-induced transdifferentiation is associated with chromatin remodeling deficiencies. Nucleic Acids Res. 2017, 45, 11684. [Google Scholar] [CrossRef]
- Bar-Nur, O.; Gerli, M.F.; Di Stefano, B.; Almada, A.E.; Galvin, A.; Coffey, A.; Huebner, A.J.; Feige, P.; Verheul, C.; Cheung, P.; et al. Direct Reprogramming of Mouse Fibroblasts into Functional Skeletal Muscle Progenitors. Stem Cell Rep. 2018, 10, 1505–1521. [Google Scholar] [CrossRef] [PubMed]
- Bursac, N.; Juhas, M.; Rando, T.A. Synergizing Engineering and Biology to Treat and Model Skeletal Muscle Injury and Disease. Annu. Rev. Biomed. Eng. 2015, 17, 217–242. [Google Scholar]
- Kollias, H.D.; Mcdermott, J.C. Transforming growth factor-B and myostatin signaling in skeletal muscle. J. Appl. Physiol. 2008, 104, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; McPherron, A.C. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA. 2001, 98, 9306–9311. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=55416&tool=pmcentrez&rendertype=abstract (accessed on 27 September 2018). [CrossRef] [PubMed]
- Hunter, R.B.; Stevenson, E.; Koncarevic, A.; Mitchell-Felton, H.; Essig, D.A.; Kandarian, S.C. Activation of an alternative NF-kappaB pathway in skeletal muscle during disuse atrophy. FASEB J. 2002, 16, 529–538. [Google Scholar] [CrossRef]
- Sinha, M.; Jang, Y.C.; Oh, J.; Khong, D.; Wu, E.Y.; Manohar, R.; Miller, C.; Regalado, S.G.; Loffredo, F.S.; Pancoast, J.R.; et al. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 2014, 344, 649–652. Available online: http://www.ncbi.nlm.nih.gov/pubmed/24797481%5Cn (accessed on 27 September 2018). [CrossRef]
- Egerman, M.A.; Cadena, S.M.; Gilbert, J.A.; Meyer, A.; Nelson, H.N.; Swalley, S.E.; Mallozzi, C.; Jacobi, C.; Jennings, L.L.; Clay, I.; et al. GDF11 Increases with Age and Inhibits Skeletal Muscle Regeneration. Cell Metab. 2015, 22, 164–174. [Google Scholar] [CrossRef]
- Boularaoui, S.M.; Abdel-Raouf, K.M.A.; Alwahab, N.S.A.; Kondash, M.E.; Truskey, G.A.; Teo, J.C.M.; Christoforou, N. Efficient transdifferentiation of human dermal fibroblasts into skeletal muscle. J. Tissue Eng. Regen. Med. 2017, 12, e918–e936. [Google Scholar] [CrossRef] [PubMed]
- Christoforou, N.; Chellappan, M.; Adler, A.F.; Kirkton, R.D.; Wu, T.; Addis, R.C.; Bursac, N.; Leong, K.W. Transcription Factors MYOCD, SRF, Mesp1 and SMARCD3 Enhance the Cardio-Inducing Effect of GATA4, TBX5 and MEF2C during Direct Cellular Reprogramming. PLoS ONE 2013, 8, e63577. [Google Scholar] [CrossRef]
- Chakraborty, S.; Ji, H.; Kabadi, A.M.; Gersbach, C.A.; Christoforou, N.; Leong, K.W. A CRISPR/Cas9-Based System for Reprogramming Cell Lineage Specification. Stem Cell Rep. 2014, 3, 940–947. Available online: http://linkinghub.elsevier.com/retrieve/pii/S2213671114002987 (accessed on 2 October 2018). [CrossRef] [PubMed]
- Henry, R.R.; Abrams, L.; Nikoulina, S.; Ciaraldi, T.P. Insulin Action and Glucose Metabolism in Nondiabetic Control and NIDDM Subjects Comparison Using Human Skeletal Muscle Cell Cultures. Diabetes 1995, 44, 936–946. [Google Scholar] [CrossRef]
- Cronin, E.M.; Thurmond, F.A.; Bassel-Duby, R.; Williams, R.S.; Wright, W.E.; Nelson, K.D.; Garner, H.R. Protein-coated poly(L-lactic acid) fibers provide a substrate for differentiation of human skeletal muscle cells. J. Biomed. Mater. Res. 2004, 69, 373–381. [Google Scholar] [CrossRef]
- Kang, H.W.; Lee, S.J.; Ko, I.K.; Kengla, C.; Yoo, J.J.; Atala, A. 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 2016, 34, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Hjort, K. Surface modification of PDMS by gradient-induced migration of embedded Pluronic. Lab Chip 2009, 9, 1500–1503. [Google Scholar] [CrossRef]
- Madden, L.; Juhas, M.; Kraus, W.E.; Truskey, G.A.; Bursac, N. Bioengineered human myobundles mimic clinical responses of skeletal muscle to drugs. Elife 2015, 4, e04885. [Google Scholar] [CrossRef]
- Lee, S.J. Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS ONE 2007, 2, e789. [Google Scholar] [CrossRef]
- Loffredo, F.S.; Steinhauser, M.L.; Jay, S.M.; Gannon, J.; Pancoast, J.R.; Yalamanchi, P.; Sinha, M.; Dall’Osso, C.; Khong, D.; Shadrach, J.L.; et al. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 2013, 153, 828–839. [Google Scholar] [CrossRef]
- Pedersen, B.K.; Febbraio, M.A. Muscles, exercise and obesity: Skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012, 8, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Unsicker, K.; Spittau, B.; Krieglstein, K. The multiple facets of the TGF-β family cytokine growth/differentiation factor-15/macrophage inhibitory cytokine-1. Cytokine Growth Factor Rev. 2013, 24, 373–384. [Google Scholar] [CrossRef]
- Yarasheski, K.E.; Campbell, J.A.; Smith, K.; Rennie, M.J.; Holloszy, J.O.; Bier, D.M. Effect of growth hormone and resistance exercise on muscle growth in young men. Am. J. Physiol. 1992, 262, E261–E267. Available online: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=1550219 (accessed on 5 June 2018). [CrossRef] [PubMed]
- Tokura, Y.; Nakayama, Y.; Fukada, S.I.; Nara, N.; Yamamoto, H.; Matsuda, R.; Hara, T. Muscle injury-induced thymosin B4 acts as a chemoattractant for myoblasts. J. Biochem. 2011, 149, 43–48. [Google Scholar] [CrossRef]
- Ono, Y.; Calhabeu, F.; Morgan, J.E.; Katagiri, T.; Amthor, H.; Zammit, P.S. BMP signaling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death Differ. 2011, 18, 222–234. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3044455&tool=pmcentrez&rendertype=abstract (accessed on 3 January 2019). [CrossRef]
- Komaki, M.; Asakura, A.; Rudnicki, M.; Sodek, J.; Cheifetz, S. MyoD enhances BMP7-induced osteogenic differentiation of myogenic cell cultures. J. Cell Sci. 2004, 117, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.L.; Baeza-Raja, B.; Perdiguero, E.; Jardí, M.; Muñoz-Cánoves, P. Interleukin-6 Is an Essential Regulator of Satellite Cell-Mediated Skeletal Muscle Hypertrophy. Cell Metab. 2008, 7, 33–44. [Google Scholar] [CrossRef]
- Reid, M.B.; Li, Y.-P. Tumor necrosis factor-α and muscle wasting: A cellular perspective. Respir. Res. 2001, 2, 269–272. [Google Scholar] [CrossRef]
- Suryawan, A.; Frank, J.W.; Nguyen, H.V.; Davis, T.A. Expression of the TGF-beta family of ligands is developmentally regulated in skeletal muscle of neonatal rats. Pediatr. Res. 2006, 59, 175–179. [Google Scholar] [CrossRef][Green Version]
- Clarke, M.S.; Feeback, D.L. Mechanical load induces sarcoplasmic wounding and FGF release in differentiated human skeletal muscle cultures. FASEB J. 1996, 10, 502–509. Available online: http://www.ncbi.nlm.nih.gov/pubmed/8647349 (accessed on 3 January 2019). [CrossRef]
- Xu, J.; Kimball, T.R.; Lorenz, J.N.; Brown, D.A.; Bauskin, A.R.; Klevitsky, R.; Hewett, T.E.; Breit, S.N.; Molkentin, J.D. GDF15/MIC-1 functions as a protective and antihypertrophic factor released from the myocardium in association with SMAD protein activation. Circ. Res. 2006, 98, 342–350. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Tsuji, K.; Cox, K.; Harfe, B.D.; Rosen, V.; Tabin, C.J. Genetic Analysis of the Roles of BMP2, BMP4, and BMP7 in Limb Patterning and Skeletogenesis. PLoS Genet. 2006, 2, e216. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, B.K.; Åkerström, T.C.A.; Nielsen, A.R.; Fischer, C. Role of myokines in exercise and metabolism. J. Appl. Physiol. 2007, 103, 1093–1098. [Google Scholar] [CrossRef]
- Schafer, M.J.; Atkinson, E.J.; Vanderboom, P.M.; Kotajarvi, B.; White, T.A.; Moore, M.M.; Bruce, C.J.; Greason, K.L.; Suri, R.M.; Khosla, S.; et al. Quantification of GDF11 and Myostatin in Human Aging and Cardiovascular Disease. Cell Metab. 2016, 23, 1207–1215. [Google Scholar] [CrossRef]
- Spurney, C.F.; Cha, H.J.; Sali, A.; Pandey, G.S.; Pistilli, E.; Guerron, A.D.; Gordish-Dressman, H.; Hoffman, E.P.; Nagaraju, K. Evaluation of skeletal and cardiac muscle function after chronic administration of thymosin beta-4 in the dystrophin deficient mouse. PLoS ONE 2010, 5, e8976. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Cánoves, P.; Scheele, C.; Pedersen, B.K.; Serrano, A.L. Interleukin-6 myokine signaling in skeletal muscle: A double-edged sword? FEBS J. 2013, 280, 4131–4148. [Google Scholar] [CrossRef] [PubMed]
- Husmann, I.; Soulet, L.; Gautron, J.; Martelly, I.; Barritault, D. Growth factors in skeletal muscle regeneration. Cytokine Growth Factor Rev. 1996, 7, 249–258. Available online: http://www.sciencedirect.com/science/article/pii/S1359610196000299 (accessed on 3 March 2019). [CrossRef]
- Olwin, B.B.; Hauschka, S.D. Cell surface fibroblast growth factor and epidermal growth factor receptors are permanently lost during skeletal muscle terminal differentiation in culture. J Cell Biol. 1988, 107, 761–769. [Google Scholar] [CrossRef]
- Juhas, M.; Bursac, N. Roles of adherent myogenic cells and dynamic culture in engineered muscle function and maintenance of satellite cells. Biomaterials 2014, 35, 9438–9446. [Google Scholar] [CrossRef]
- Lee, S.-J.; Lee, Y.-S.; Zimmers, T.A.; Soleimani, A.; Matzuk, M.M.; Tsuchida, K.; Cohn, R.D.; Barton, E.R. Regulation of muscle mass by follistatin and activins. Mol. Endocrinol. 2010, 24, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Young, J.D.; Lawrence, A.J.; MacLean, A.G.; Leung, B.P.; McInnes, I.B.; Canas, B.; Pappin, D.J.C.; Stevenson, R.D. Thymosin beta 4 sulfoxide is an anti-inflammatory agent generated by monocytes in the presence of glucocorticoids. Nat. Med. 1999, 5, 1424–1427. Available online: http://www.ncbi.nlm.nih.gov/pubmed/10581087 (accessed on 6 May 2019). [CrossRef] [PubMed]
- Clever, J.L.; Sakai, Y.; Wang, R.A.; Schneider, D.B. Inefficient skeletal muscle repair in inhibitor of differentiation knockout mice suggests a crucial role for BMP signaling during adult muscle regeneration. Am. J. Physiol. Physiol. 2010, 298, C1087–C1099. [Google Scholar] [CrossRef]
- Amthor, H.; Christ, B.; Rashid-Doubell, F.; Kemp, C.; Lang, E.; Patel, K. Follistatin Regulates Bone Morphogenetic Protein-7 (BMP-7) Activity to Stimulate Embryonic Muscle Growth. Dev. Biol. 2002, 243, 115–127. Available online: http://www.ncbi.nlm.nih.gov/pubmed/11846481 (accessed on 10 June 2019). [CrossRef] [PubMed]
- Cuneo, R.; Salomon, F.; Mark Wiles, C.; Sonksen, P.H. Skeletal Muscle Performance in Adults with Growth Hormone Deficiency. Horm. Res. Paediatr. 1990, 33 (Suppl. 4), 55–60. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.E.; Allen, R.E. Activation of Skeletal Muscle Satellite Cells and the Role of Fibroblast Growth Factor Receptors. Exp. Cell Res. 1995, 219, 449–453. [Google Scholar] [CrossRef]
- Smith, S.C.; Zhang, X.; Zhang, X.; Gross, P.; Starosta, T.; Mohsin, S.; Franti, M.; Gupta, P.; Hayes, D.L.; Myzithras, M.; et al. GDF11 Does Not Rescue Aging-Related Pathological Hypertrophy. Circ. Res. 2015, 117, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Jackman, R.W.; Kandarian, S.C. The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Physiol. 2004, 287, C834–C843. Available online: http://www.ncbi.nlm.nih.gov/pubmed/15355854 (accessed on 10 June 2019). [CrossRef]
- Li, Y.P.; Schwartz, R.J.; Waddell, I.D.; Holloway, B.R.; Reid, M.B. Skeletal muscle myocytes undergo protein loss and reactive oxygen-mediated NF-kappaB activation in response to tumor necrosis factor alpha. FASEB J. 1998, 12, 871–880. [Google Scholar]
- Li, Y.P. TNF-α is a mitogen in skeletal muscle. Am. J. Physiol. Cell Physiol. 2003, 285, C370–C376. [Google Scholar] [CrossRef]
- Sen, C.K.; Khanna, S.; Reznick, A.Z.; Roy, S.; Packer, L. Glutathione regulation of tumor necrosis factor-α-induced NF-κB activation in skeletal muscle-derived L6 cells. Biochem. Biophys. Res. Commun. 1997, 237, 645–649. [Google Scholar] [CrossRef]
- Rybalko, V.; Hsieh, P.L.; Merscham-Banda, M.; Suggs, L.J.; Farrar, R.P. The development of macrophage-mediated cell therapy to improve skeletal muscle function after injury. PLoS ONE 2015, 10, e0145550. [Google Scholar] [CrossRef] [PubMed]
- Friedrichs, M.; Wirsdöerfer, F.; Flohé, S.B.; Schneider, S.; Wuelling, M.; Vortkamp, A. BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol. 2011, 12, 1–17. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3149017&tool=pmcentrez&rendertype=abstract (accessed on 28 May 2021). [CrossRef]
- Johnson, S.E.; Allen, R.E. The effects of bFGF, IGF-I, and TGF-beta on RMo skeletal muscle cell proliferation and differentiation. Exp. Cell Res. 1990, 18, 25. Available online: papers2://publication/uuid/D6949D6B-6601-4A1A-BE46-EFC76DB7FC13 (accessed on 5 May 2018).
- Allen, R.E.; Boxhorn, L.K. Regulation of skeletal muscle satellite cell proliferation and differentiation by transforming growth factor-beta, insulin-like growth factor I, and fibroblast growth factor. J. Cell. Physiol. 1989, 138, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Cassimeris, L.; Safer, D.; Nachmias, V.T.; Zigmond, S.H. Thymosin beta 4 sequesters the majority of G-actin in resting human polymorphonuclear leukocytes. J. Cell Biol. 1992, 119, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, M.; Bishop, N.C.; Stensel, D.J.; Lindley, M.R.; Mastana, S.S.; Nimmo, M.A. The anti-inflammatory effects of exercise: Mechanisms and implications for the prevention and treatment of disease. Nat. Rev. Immunol. 2011, 11, 607–610. [Google Scholar] [CrossRef]
- Bertholdt, L.; Gudiksen, A.; Schwartz, C.L.; Knudsen, J.G.; Pilegaard, H. Lack of skeletal muscle IL-6 influences hepatic glucose metabolism in mice during prolonged exercise. Am. J. Physiol. Integr. Comp. Physiol. 2017, 312, R626–R636. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Liu, D.; Huang, Q.; Cai, G.; Cui, S.; Sun, X.; Chen, X. GDF11 improves tubular regeneration after acute kidney injury in elderly mice. Sci. Rep. 2016, 6, 34624. Available online: http://www.nature.com/articles/srep34624 (accessed on 21 June 2019). [CrossRef]
- Walker, R.G.; Poggioli, T.; Katsimpardi, L.; Buchanan, S.M.; Oh, J.; Wattrus, S.; Heidecker, B.; Fong, Y.W.; Rubin, L.L.; Ganz, P.; et al. Biochemistry and Biology of GDF11 and Myostatin: Similarities, Differences, and Questions for Future Investigation. Circ. Res. 2016, 118, 1125–1142. [Google Scholar] [CrossRef] [PubMed]
- Hammers, D.W.; Merscham-Banda, M.; Hsiao, J.Y.; Engst, S.; Hartman, J.J.; Sweeney, H.L. Supraphysiological levels of GDF 11 induce striated muscle atrophy. EMBO Mol. Med. 2017, 9, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Matzuk, M.M.; Lu, N.; Vogel, H.; Sellheyer, K.; Roop, D.R.; Bradley, A. Multiple defects and perinatal death in mice deficient in follistatin. Nature 1995, 374, 360–363. Available online: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=7885475%5Cn (accessed on 7 May 2018). [CrossRef] [PubMed]
- Grant, D.S.; Rose, W.; Yaen, C.; Goldstein a Martinez, J.; Kleinman, H. Thymosin beta4 enhances endothelial cell differentiation and angiogenesis. Angiogenesis 1999, 3, 125–135. Available online: http://www.ncbi.nlm.nih.gov/pubmed/14517430 (accessed on 8 June 2019). [CrossRef]
- Winbanks, C.E.; Weeks, K.L.; Thomson, R.E.; Sepulveda, P.V.; Beyer, C.; Qian, H.; Chen, J.L.; Allen, J.M.; Lancaster, G.I.; Febbraio, M.A.; et al. Follistatin-mediated skeletal muscle hypertrophy is regulated by Smad3 and mTOR independently of myostatin. J. Cell Biol. 2012, 197, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Nixon, B.T.; Green, H. Growth hormone promotes the differentiation of myoblasts and preadipocytes generated by azacytidine treatment of 10T1/2 cells. Proc. Natl. Acad. Sci. USA 1984, 81, 3429–3432. Available online: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=345521&tool=pmcentrez&rendertype=abstract (accessed on 18 July 2019). [CrossRef]
- Isgaard, J.; Nilsson, A.; Vikman, K.; Isaksson, O.G.P. Growth hormone regulates the level of insulin-like growth factor-I mRNA in rat skeletal muscle. J. Endocrinol. 1989, 120, 107–112. Available online: http://www.ncbi.nlm.nih.gov/pubmed/2918262 (accessed on 29 July 2019). [CrossRef]
- Timper, K.; Denson, J.L.; Steculorum, S.M.; Heilinger, C.; Engström-Ruud, L.; Wunderlich, C.M.; Rose-John, S.; Wunderlich, F.T.; Brüning, J.C. IL-6 Improves Energy and Glucose Homeostasis in Obesity via Enhanced Central IL-6 trans-Signaling. Cell Rep. 2017, 19, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Ding, V.M.; Ling, L.; Natarajan, S.; Yap, M.G.; Cool, S.M.; Choo, A.B. FGF-2 modulates Wnt signaling in undifferentiated hESC and iPS cells through activated PI3-K/GSK3β signaling. J. Cell. Physiol. 2010, 225, 417–428. [Google Scholar] [CrossRef]
- Borchin, B.; Chen, J.; Barberi, T. Derivation and FACS-Mediated Purification of PAX3+/PAX7+ Skeletal Muscle Precursors from Human Pluripotent Stem Cells. Stem Cell Rep. 2013, 1, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Almada, A.E.; Wagers, A.E.A.A.J. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat. Rev. Mol. Cell Biol. 2016, 17, 267–279. [Google Scholar] [CrossRef]
- Glass, D.J. PI3 Kinase Regulation of Skeletal Muscle Hypertrophy and Atrophy. In Current Topics in Microbiology and Immunology; Springer: Berlin, Germany, 2010; Volume 346, pp. 267–278. [Google Scholar] [CrossRef]
- De Kretser, D.M.; O’Hehir, R.E.; Hardy, C.L.; Hedger, M.P. The roles of activin A and its binding protein, follistatin, in inflammation and tissue repair. Mol. Cell. Endocrinol. 2012, 359, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Kaji, H.; Ishibashi, T.; Nagamine, K.; Kanzaki, M.; Nishizawa, M. Electrically induced contraction of C2C12 myotubes cultured on a porous membrane-based substrate with muscle tissue-like stiffness. Biomaterials 2010, 31, 6981–6986. [Google Scholar] [CrossRef]
- Khodabukus, A.; Madden, L.; Prabhu, N.K.; Koves, T.R.; Jackman, C.P.; Muoio, D.M.; Bursac, N. Electrical stimulation increases hypertrophy and metabolic flux in tissue-engineered human skeletal muscle. Biomaterials 2019, 198, 259–269. [Google Scholar] [CrossRef]
- Gilbert-Honick, J.; Iyer, S.R.; Somers, S.M.; Takasuka, H.; Lovering, R.M.; Wagner, K.R.; Mao, H.-Q.; Grayson, W.L. Engineering 3D skeletal muscle primed for neuromuscular regeneration following volumetric muscle loss. Biomaterials 2020, 255, 120154. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, I.; Seol, Y.-J.; Ko, I.K.; Yoo, J.J.; Atala, A.; Lee, S.J. Neural cell integration into 3D bioprinted skeletal muscle constructs accelerates restoration of muscle function. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Seol, Y.-J.; Ko, I.K.; Kang, H.-W.; Lee, Y.K.; Yoo, J.J.; Atala, A.; Lee, S.J. 3D Bioprinted Human Skeletal Muscle Constructs for Muscle Function Restoration. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Gholobova, D.; Gerard, M.; Decroix, L.; Desender, L.; Callewaert, N.; Annaert, P.; Thorrez, L. Human tissue-engineered skeletal muscle: A novel 3D in vitro model for drug disposition and toxicity after intramuscular injection. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Nie, J.; Gao, Q.; Fu, J.; He, Y. Grafting of 3D Bioprinting to In Vitro Drug Screening: A Review. Adv. Healthc. Mater. 2020, 9, e1901773. [Google Scholar] [CrossRef]
- Nam, K.-H.; Smith, A.S.T.; Lone, S.; Kwon, S.; Kim, D.-H. Biomimetic 3D Tissue Models for Advanced High-Throughput Drug Screening. J. Lab. Autom. 2015, 20, 201–215. Available online: http://www.ncbi.nlm.nih.gov/pubmed/25385716%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4459652 (accessed on 23 May 2021). [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdel-Raouf, K.M.A.; Rezgui, R.; Stefanini, C.; Teo, J.C.M.; Christoforou, N. Transdifferentiation of Human Fibroblasts into Skeletal Muscle Cells: Optimization and Assembly into Engineered Tissue Constructs through Biological Ligands. Biology 2021, 10, 539. https://doi.org/10.3390/biology10060539
Abdel-Raouf KMA, Rezgui R, Stefanini C, Teo JCM, Christoforou N. Transdifferentiation of Human Fibroblasts into Skeletal Muscle Cells: Optimization and Assembly into Engineered Tissue Constructs through Biological Ligands. Biology. 2021; 10(6):539. https://doi.org/10.3390/biology10060539
Chicago/Turabian StyleAbdel-Raouf, Khaled M. A., Rachid Rezgui, Cesare Stefanini, Jeremy C. M. Teo, and Nicolas Christoforou. 2021. "Transdifferentiation of Human Fibroblasts into Skeletal Muscle Cells: Optimization and Assembly into Engineered Tissue Constructs through Biological Ligands" Biology 10, no. 6: 539. https://doi.org/10.3390/biology10060539
APA StyleAbdel-Raouf, K. M. A., Rezgui, R., Stefanini, C., Teo, J. C. M., & Christoforou, N. (2021). Transdifferentiation of Human Fibroblasts into Skeletal Muscle Cells: Optimization and Assembly into Engineered Tissue Constructs through Biological Ligands. Biology, 10(6), 539. https://doi.org/10.3390/biology10060539






