Rhizospheric Communication through Mobile Genetic Element Transfers for the Regulation of Microbe–Plant Interactions
Abstract
Simple Summary
Abstract
1. Introduction
2. General Introduction to Classic MGE Transfers
2.1. MGE Transfer in Prokaryotes
2.2. MGE Transfers in Eukaryotes
2.3. MGE Transfer between Microbes and Plants
3. MGE Transfers in the Rhizosphere
3.1. MGE Transfers among Soil Bacteria
3.2. HGTs among Filamentous Eukaryotes
3.3. Cross-Kingdom HGTs
3.3.1. Cross-Kingdom MGE Transfers among Microbes in the Rhizosphere Play an Important Role in Regulating Plant Growth and Development
3.3.2. MGE Transfers from Rhizospheric Microbes to Plants
3.3.3. Plants Could Be MGE Donors in the Rhizosphere Too
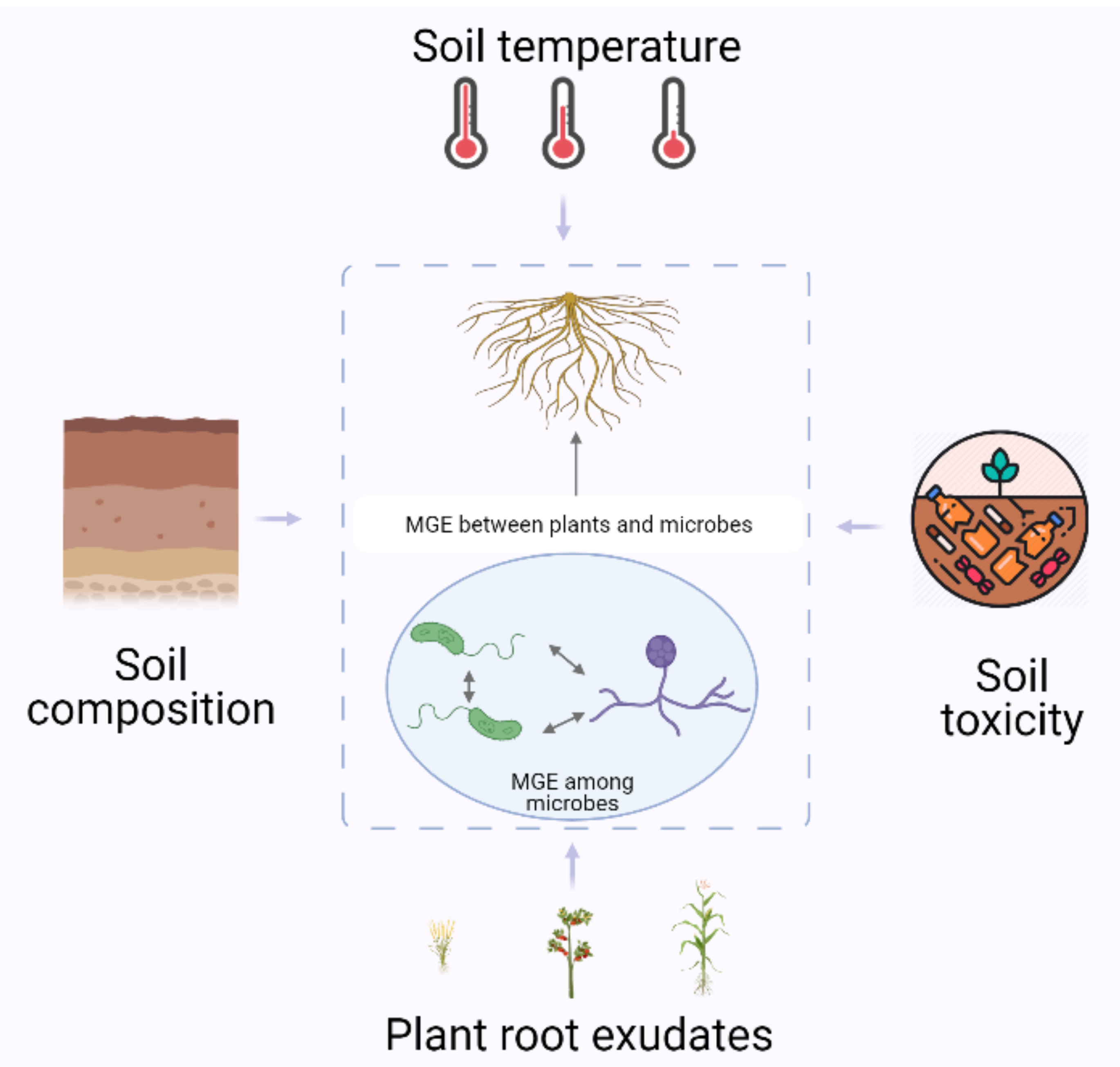
4. The Effects of Stress and Environmental Factors on the Transfer of MGEs in the Rhizosphere
4.1. Temperature
4.2. Soil Composition
4.3. Soil Toxicity
4.4. Influences by Plants
5. MGE Transfers Regulate the Adaptation to the Environment
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Venturi, V.; Keel, C. Signaling in the Rhizosphere. Trends Plant Sci. 2016, 21, 187–198. [Google Scholar] [CrossRef]
- Zhang, R.; Vivanco, J.M.; Shen, Q. The unseen rhizosphere root–soil–microbe interactions for crop production. Curr. Opin. Microbiol. 2017, 37, 8–14. [Google Scholar] [CrossRef]
- Ku, Y.-S.; Cheng, S.-S.; Gerhardt, A.; Cheung, M.-Y.; Contador, C.A.; Poon, L.-Y.W.; Lam, H.-M. Secretory peptides as bullets: Effector peptides from pathogens against antimicrobial peptides from soybean. Int. J. Mol. Sci. 2020, 21, 9294. [Google Scholar] [CrossRef] [PubMed]
- Frost, L.; Leplae, R.; Summers, A.O.; Toussaint, A. Mobile genetic elements: The agents of open source evolution. Nat. Rev. Genet. 2005, 3, 722–732. [Google Scholar] [CrossRef]
- Koonin, E.V. Viruses and mobile elements as drivers of evolutionary transitions. Philos. Trans. R. Soc. L. B Biol. Sci. 2016, 371, 20150442. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Wang, X.; Duan, J.; Ma, J. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 2019, 365, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Fauquet, C.M.; Mayo, M.A.; Maniloff, J.; Desselberger, U.; Ball, L.A. (Eds.) Virus Taxonomy; Academic Press: Cambridge, MA, USA, 2005. [Google Scholar]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A.A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Genet. 2015, 13, 641–650. [Google Scholar] [CrossRef]
- Pfeifer, E.; Sousa, J.A.M.D.; Touchon, M.; Rocha, E.P.C. Bacteria have numerous distinctive groups of phage–plasmids with conserved phage and variable plasmid gene repertoires. Nucleic Acids Res. 2021, 49, 2655–2673. [Google Scholar] [CrossRef]
- Galetti, R.; Andrade, L.N.; Varani, A.M.; Darini, A.L.C. A phage-like plasmid carrying blaKPC-2 gene in carbapenem-resistant Pseudomonas aeruginosa. Front. Microbiol. 2019, 10, 572. [Google Scholar] [CrossRef]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brüssow, H. Prophage Genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef]
- Rankin, D.J.; Rocha, E.; Brown, S.P. What traits are carried on mobile genetic elements, and why? Heredity 2010, 106, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Phage-phage, phage-bacteria, and phage-environment communication. In Biocommunicatiton of Phages; Witzany, G., Ed.; Springer: Cham, Switzerland, 2020; pp. 23–70. [Google Scholar]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef]
- Richards, T.A. Genome Evolution: Horizontal Movements in the Fungi. Curr. Biol. 2011, 21, R166–R168. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A.; Beardall, J.; Flynn, K.J.; Maberly, S.C. Phagotrophy in the origins of photosynthesis in eukaryotes and as a com-plementary mode of nutrition in phototrophs: Relation to Darwin’s insectivorous plants. J. Exp. Clin. Cancer Res. 2009, 60, 3975–3987. [Google Scholar]
- Roca, M.G.; Davide, L.C.; Davide, L.M.; Mendes-Costa, M.C.; Schwan, R.F.; Wheals, A.E. Conidial anastomosis fusion between Colletotrichum species. Mycol. Res. 2004, 108, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, D.A. Horizontal gene transfer in fungi. FEMS Microbiol. Lett. 2011, 329, 1–8. [Google Scholar] [CrossRef]
- Huamanquispe, D.G.Q.; Gheysen, G.; Kreuze, J.F. Horizontal gene transfer contributes to plant evolution: The case of Agrobacterium T-DNAs. Front. Plant Sci. 2017, 8, 2015. [Google Scholar] [CrossRef]
- Zupan, J.R.; Zambryski, P. Transfer of T-DNA from Agrobacterium to the Plant Cell. Plant Physiol. 1995, 107, 1041–1047. [Google Scholar] [CrossRef]
- Tzfira, T.; Li, J.; Lacroix, B.; Citovsky, V. Agrobacterium T-DNA integration: Molecules and models. Trends Genet. 2004, 20, 375–383. [Google Scholar] [CrossRef]
- Richards, T.A.; Soanes, D.M.; Foster, P.; Leonard, G.; Thornton, C.R.; Talbot, N.J. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell 2009, 21, 1897–1911. [Google Scholar] [CrossRef]
- Li, M.; Zhao, J.; Tang, N.; Sun, H.; Huang, J. Horizontal gene transfer from bacteria and plants to the arbuscular mycorrhizal fungus Rhizophagus irregularis. Front. Plant Sci. 2018, 9, 701. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.-L.; Zhao, J.-H.; Wang, S.; Jin, Y.; Chen, Z.-Q.; Fang, Y.-Y.; Jian-Hua, Z.; Ding, S.-W.; Guo, H.-S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.-M.; Palmquist, J.; Huang, S.-D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and bat-tlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Maheshwari, M.; Abulreesh, H.H.; Khan, M.S.; Ahmad, I.; Pichtel, J. Horizontal gene transfer in soil and the rhizosphere: Impact on ecological fitness of bacteria. In Agriculturally Important Microbes for Sustainable Agriculture; Meena, V.S., Mishra, P.K., Bisht, J.K., Pattanayak, A., Eds.; Springer: Singapore, 2017; pp. 111–130. [Google Scholar]
- Van Elsas, J.D.; Bailey, M.J. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Lett. 2002, 42, 187–197. [Google Scholar] [CrossRef]
- Haritha, A.; Sagar, K.P.; Tiwari, A.; Kiranmayi, P.; Rodrigue, A.; Mohan, P.M.; Singh, S.S. MrdH, a novel metal resistance determinant of Pseudomonas putida KT2440, is flanked by metal-inducible mobile genetic elements. J. Bacteriol. 2009, 191, 5976–5987. [Google Scholar] [CrossRef] [PubMed]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018, 31, 00088-17. [Google Scholar] [CrossRef]
- Shintani, M.; Nour, E.; Elsayed, T.; Blau, K.; Wall, I.; Jechalke, S.; Spröer, C.; Bunk, B.; Overmann, J.; Smalla, K. Plant species-dependent increased abundance and diversity of IncP-1 plasmids in the rhizosphere: New insights into their role and ecology. Front. Microbiol. 2020, 11, 590776. [Google Scholar] [CrossRef] [PubMed]
- Seiler, C.; Berendonk, T.U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Loper, J.E.; Paulsen, I.T.; Thomashow, L.S. Mobile genetic elements in the genome of the beneficial rhizobacterium Pseudomonas fluorescens Pf-5. BMC Microbiol. 2009, 9, 8. [Google Scholar] [CrossRef]
- Bruto, M.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 2014, 4, 6261. [Google Scholar] [CrossRef]
- Cai, P.; Sun, X.; Wu, Y.; Gao, C.; Mortimer, M.; Holden, P.A.; Redmile-Gordon, M.; Huang, Q. Soil biofilms: Microbial interactions, challenges, and advanced techniques for ex-situ characterization. Soil Ecol. Lett. 2019, 1, 85–93. [Google Scholar] [CrossRef]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef]
- Berthold, T.; Centler, F.; Hübschmann, T.; Remer, R.; Thullner, M.; Harms, H.; Wick, L.Y. Mycelia as a focal point for horizontal gene transfer among soil bacteria. Sci. Rep. 2016, 6, 36390. [Google Scholar] [CrossRef]
- Wang, B.; Qiu, Y.-L. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 2006, 16, 299–363. [Google Scholar] [CrossRef]
- Akyol, T.Y.; Niwa, R.; Hirakawa, H.; Maruyama, H.; Sato, T.; Suzuki, T.; Fukunaga, A.; Sato, T.; Yoshida, S.; Tawaraya, K.; et al. Impact of introduction of arbuscular mycorrhizal community in agricultural fields. Microbes Environ. 2019, 34, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Feng, G. Arbuscular mycorrhizal fungi interactions in the rhizosphere. In Rhizosphere Biology: Interaction between Microbes and Plants; Gupta, V.V.S.R., Sharma, A.K., Eds.; Springer: Singapore, 2021; pp. 217–235. [Google Scholar]
- Kuhn, G.; Hijri, M.; Sanders, I. Evidence for the evolution of multiple genomes in arbuscular mycorrhizal fungi. Nat. Cell Biol. 2001, 414, 745–748. [Google Scholar] [CrossRef]
- Hijri, M.; Sanders, I.R. Low gene copy number shows that arbuscular mycorrhizal fungi inherit genetically different nuclei. Nat. Cell Biol. 2005, 433, 160–163. [Google Scholar] [CrossRef]
- Angelard, C.; Colard, A.; Niculita-Hirzel, H.; Croll, D.; Sanders, I. Segregation in a mycorrhizal fungus alters rice growth and symbiosis-specific gene transcription. Curr. Biol. 2010, 20, 1216–1221. [Google Scholar] [CrossRef] [PubMed]
- Croll, D.; Giovannetti, M.; Koch, A.M.; Sbrana, C.; Ehinger, M.; Lammers, P.J.; Sanders, I.R. Nonself vegetative fusion and genetic exchange in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2009, 181, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Angelard, C.; Sanders, I.R. Effect of segregation and genetic exchange on arbuscular mycorrhizal fungi in colonization of roots. New Phytol. 2010, 189, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Colard, A.; Angelard, C.; Sanders, I.R. Genetic exchange in an arbuscular mycorrhizal fungus results in increased rice growth and altered mycorrhiza-specific gene transcription. Appl. Environ. Microbiol. 2011, 77, 6510–6515. [Google Scholar] [CrossRef]
- Richards, T.A.; Dacks, J.B.; Jenkinson, J.M.; Thornton, C.R.; Talbot, N.J. Evolution of filamentous plant pathogens: Gene exchange across eukaryotic kingdoms. Curr. Biol. 2006, 16, 1857–1864. [Google Scholar] [CrossRef] [PubMed]
- Richards, T.A.; Soanes, D.M.; Jones, M.D.M.; Vasieva, O.; Leonard, G.; Paszkiewicz, K.; Foster, P.; Hall, N.; Talbot, N.J. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl. Acad. Sci. USA 2011, 108, 15258–15263. [Google Scholar] [CrossRef] [PubMed]
- Danchin, E.G.J.; Rosso, M.-N.; Vieira, P.; de Almeida-Engler, J.; Coutinho, P.M.; Henrissat, B.; Abad, P. Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes. Proc. Natl. Acad. Sci. USA 2010, 107, 17651–17656. [Google Scholar] [CrossRef]
- Acuna, R.; Padilla, B.E.; Florez-Ramos, C.P.; Rubio, J.D.; Herrera, J.C.; Benavides, P.; Lee, S.-J.; Yeats, T.H.; Egan, A.N.; Doyle, J.J.; et al. Adaptive horizontal transfer of a bacterial gene to an invasive insect pest of coffee. Proc. Natl. Acad. Sci. USA 2012, 109, 4197–4202. [Google Scholar] [CrossRef]
- Yue, J.; Hu, X.; Sun, H.; Yang, Y.; Huang, J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 2012, 3, 1152. [Google Scholar] [CrossRef]
- Schaller, A.; Stintzi, A.; Graff, L. Subtilases-versatile tools for protein turnover, plant development, and interactions with the environment. Physiol. Plant. 2011, 145, 52–66. [Google Scholar] [CrossRef]
- Xia, J.; Guo, Z.; Yang, Z.; Han, H.; Wang, S.; Xu, H.; Yang, X.; Yang, F.; Wu, Q.; Xie, W.; et al. Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 2021, 184, 1693–1705. [Google Scholar] [CrossRef]
- Velmourougane, K.; Saxena, G.; Prasanna, R. Plant-microbe interactions in the rhizosphere: Mechanisms and their ecological benefits. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Singapore, 2017; pp. 193–219. [Google Scholar]
- Mohanram, S.; Kumar, P. Rhizosphere microbiome: Revisiting the synergy of plant-microbe interactions. Ann. Microbiol. 2019, 69, 307–320. [Google Scholar] [CrossRef]
- Afkhami, M.E.; Almeida, B.K.; Hernandez, D.J.; Kiesewetter, K.N.; Revillini, D.P. Tripartite mutualisms as models for understanding plant-microbial interactions. Curr. Opin. Plant Biol. 2020, 56, 28–36. [Google Scholar] [CrossRef]
- Bruto, M.; Prigent-Combaret, C.; Luis, P.; Moënne-Loccoz, Y.; Muller, D. Frequent, independent transfers of a catabolic gene from bacteria to contrasted filamentous eukaryotes. Proc. R. Soc. B Boil. Sci. 2014, 281, 20140848. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Shelke, G.M.; Kumar, A.; Jha, P.N. Biochemistry and genetics of ACC deaminase: A weapon to “stress ethylene” produced in plants. Front. Microbiol. 2015, 6, 937. [Google Scholar] [PubMed]
- Lee, S.-J.; Kong, M.; Harrison, P.; Hijri, M. Conserved proteins of the RNA interference system in the arbuscular mycorrhizal fungus Rhizoglomus irregulare provide new insight into the evolutionary history of Glomeromycota. Genome Biol. Evol. 2018, 10, 328–343. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, W.; Ivanov, S.; MacLean, A.M.; Wight, H.; Ramaraj, T.; Mudge, J.; Harrison, M.J.; Fei, Z. Genome and evolution of the arbuscular mycorrhizal fungus Diversispora epigaea (formerly Glomus versiforme) and its bacterial endosymbionts. New Phytol. 2019, 221, 1556–1573. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Deng, Z.; Liu, Z.; Yan, Y.; Wang, T.; Xie, J.; Lin, M.; Cheng, Q.; Chen, S. The genome of Paenibacillus sabinae T27 provides insight into evolution, organization and functional elucidation of nif and nif-like genes. BMC Genom. 2014, 15, 723. [Google Scholar] [CrossRef]
- Swarnalakshmi, K.; Yadav, V.; Tyagi, D.; Dhar, D.W.; Kannepalli, A.; Kumar, S. Significance of plant growth promoting rhizobacteria in grain legumes: Growth promotion and crop production. Plants 2020, 9, 1596. [Google Scholar] [CrossRef]
- Checcucci, A.; Azzarello, E.; Bazzicalupo, M.; De Carlo, A.; Emiliani, G.; Mancuso, S.; Spini, G.; Viti, C.; Mengoni, A. role and regulation of ACC deaminase gene in Sinorhizobium meliloti: Is it a symbiotic, rhizospheric or endophytic gene? Front. Genet. 2017, 8, 6. [Google Scholar] [CrossRef]
- Ling, J.; Wang, H.; Wu, P.; Li, T.; Tang, Y.; Naseer, N.; Zheng, H.; Masson-Boivin, C.; Zhong, Z.; Zhu, J. Plant nodulation inducers enhance horizontal gene transfer of Azorhizobium caulinodans symbiosis island. Proc. Natl. Acad. Sci. USA 2016, 113, 13875–13880. [Google Scholar] [CrossRef]
- Kumar, A.; Verma, J.P. Does plant—Microbe interaction confer stress tolerance in plants: A review? Microbiol. Res. 2018, 207, 41–52. [Google Scholar] [CrossRef]
- Vimal, S.R.; Singh, J.S.; Arora, N.K.; Singh, S. Soil-plant-microbe interactions in stressed agriculture management: A review. Pedosphere 2017, 27, 177–192. [Google Scholar] [CrossRef]
- Shukla, N.; Awasthi, R.; Rawat, L.; Kumar, J. Biochemical and physiological responses of rice (Oryza sativa L.) as influenced by Trichoderma harzianum under drought stress. Plant Physiol. Biochem. 2012, 54, 78–88. [Google Scholar] [CrossRef]
- Hill, K.E.; Top, E.M. Gene transfer in soil systems using microcosms. FEMS Microbiol. Lett. 1998, 25, 319–329. [Google Scholar] [CrossRef]
- Miransari, M. Soil microbes and the availability of soil nutrients. Acta Physiol. Plant. 2013, 35, 3075–3084. [Google Scholar] [CrossRef]
- Wang, Y.; Kou, S.; Jiang, Q.; Xu, B.; Liu, X.; Xiao, J.; Tian, Y.; Zhou, C.; Zhang, C.; Xiao, M. Factors affecting transfer of degradative plasmids between bacteria in soils. Appl. Soil Ecol. 2014, 84, 254–261. [Google Scholar] [CrossRef]
- Banerjee, G.; Ray, A.K.; Kumar, R. Effect of temperature on lateral gene transfer efficiency of multi-antibiotics resistant bacterium, Alcaligenes faecalis. Sains Malays. 2016, 45, 909–914. [Google Scholar]
- Hashimoto, M.; Hasegawa, H.; Maeda, S. High temperatures promote cell-to-cell plasmid transformation in Escherichia coli. Biochem. Biophys. Res. Commun. 2019, 515, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Peng, K.; Jiang, L.; Zhang, D.; Song, D.; Chen, G.; Xu, H.; Li, Y.; Luo, C. Alleviated antibiotic-resistant genes in the rhizosphere of agricultural soils with low antibiotic concentration. J. Agric. Food Chem. 2020, 68, 2457–2466. [Google Scholar] [CrossRef] [PubMed]
- Czekalski, N.; Berthold, T.; Caucci, S.; Egli, A.; Bürgmann, H. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 2012, 3, 106. [Google Scholar] [CrossRef] [PubMed]
- Parra, B.; Tortella, G.R.; Cuozzo, S.; Martínez, M. Negative effect of copper nanoparticles on the conjugation frequency of conjugative catabolic plasmids. Ecotoxicol. Environ. Saf. 2019, 169, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xu, H.; Liu, G.; Wei, Y.; Zhang, J.; Shi, H. Physiological and metagenomic strategies uncover the rhizosphere bacterial microbiome succession underlying three common environmental stresses in cassava. J. Hazard. Mater. 2021, 411, 125143. [Google Scholar] [CrossRef]
- Hui, N.; Jumpponen, A.; Francini, G.; Kotze, D.J.; Liu, X.; Romantschuck, M.; Strömmer, R.; Setälä, H. Soil microbial communities are shaped by vegetation type and park age in cities under cold climate. Environ. Microbiol. 2017, 19, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Yang, C.; Mei, X.; Shen, S.; Raza, W.; Shen, Q.; Xu, Y. The effect of organic acids from tomato root exudates on rhizosphere colonization of Bacillus amyloliquefaciens T-5. Appl. Soil Ecol. 2013, 64, 15–22. [Google Scholar] [CrossRef]
- Jin, J.; Wang, M.; Lu, W.; Zhang, L.; Jiang, Q.; Jin, Y.; Lu, K.; Sun, S.; Cao, Q.; Wang, Y.; et al. Effect of plants and their root exudate on bacterial activities during rhizobacterium-plant remediation of phenol from water. Environ. Int. 2019, 127, 114–124. [Google Scholar] [CrossRef]
- Miguel, A.B.R.; Jetten, M.S.; Welte, C.U. The role of mobile genetic elements in organic micropollutant degradation during biological wastewater treatment. Water Res. X 2020, 9, 100065. [Google Scholar] [CrossRef] [PubMed]
| MGE-Borne Gene(s)/MGE | Name of MGE-Borne Gene/MGE and Description | Nature | Direction | Significance | Reference |
|---|---|---|---|---|---|
| nifHDK | nifHDK is the operon that comprises the genes nifH, nifD and nifK. nifH and nifD encode the α subunit and the β subunit of dinitrogenase respectively, while nifH encodes the γ subunit of dinitrogenase reductase. Nitrogenase converts atmospheric nitrogen into the ammoniacal form of nitrogen to be used by plants. | DNA | Between bacteria | Encodes nitrogenase in PGPR; the transfer helped shape a taxonomic subgroup of PGPR | [34,61,62] |
| ACC deaminase structural gene | acdS encodes ACC deaminase which degrades ACC in root exudates and in turn inhibits the synthesis of ethylene. The result in the promotion of plant growth. | DNA | Between bacteria | The transfer helped shape a taxonomic subgroup of PGPR | [34,63] |
| From bacterium to filamentous eukaryotes. including oomycetes and fungi | Promotes plant growth | [57] | |||
| Class I ribonuclease III protein-coding gene | rirnc 2 encodes a class I ribonuclease III protein. | DNA | From cyanobacteria to Glomeromycota | Possible ancient symbiosis history between cyanobacteria and arbuscular mycorrhizae | [59] |
| Genes related to bacterial methylation defense system | Ribonuclease IIIs, Uma2 endonucleases, HNH endonuclease and methyltransferase. These genes are involved in the defense system against foreign DNA. | DNA | From Mycoplasma-related endobacteria to Diversispora epigaea | Facilitates the symbiosis of endobacteria and arbuscular mycorrhizae | [60] |
| Subtilase gene | It was suggested that genes of land plant subtilase family were derived from a single HGT event from bacteria. After that, rapid gene duplication occurred to give rise to the subtiliase family. | DNA | From bacterium to P. patens | Facilitates the colonization of plants on land | [51] |
| Gene for L-fucose uptake | FucP. FucP refers to L-fucose permease transporter family protein for L-fucose uptake. | DNA | From fungus to plant | Facilitates plant adaptation to growing in soil | [22] |
| Gene for membrane transporter | Protein sequence and domain analyses suggested that the gene belongs to the Major Facilitator Superfamily (MFS_1). | DNA | From fungus to plant | Facilitates plant adaptation to growing in soil | [22] |
| Gene for phospholipase/carboxylesterase family protein | Sequence analysis suggested that this gene has sequence similarity to the phospholipase/carboxylesterase protein family. Members of the phospholipase/carboxylesterase protein family have broad substrate specificity and the capacity of hydrolyzing carboxylic ester bonds. | DNA | From fungus to plant | Facilitates plant adaptation to growing in soil | [22] |
| Gene for siderophore biosynthesis | The gene for siderophore biosynthesis encodes a protein containing two domains: iucA and iucC. These two domains are involved in the sequential conversion of N epsilon-acetyl-N epsilon-hydroxylysine to the siderophore aerobactin. | DNA | From fungus to plant | Facilitates plant adaptation to growing in soil | [22] |
| Transfer RNA (tRNA)-derived small RNA fragments (tRFs) | Bj-tRF001, Bj-tRG002 and Bj-tRF003. Bj-tRF001, Bj-tRG002 and Bj-tRF003 target GmRHD3a/3b, GmHAM4a/4b and GmLRX5 respectively. in soybean. The tRFs regulate nodulation of the soybean plant. | RNA | From rhizobium to soybean | These tRFs regulate nodulation related genes in soybean plant. | [6] |
| Cytotoxin gene family | Members of cytotoxin gene family encode proteins having conserved cytotoxic domians. | DNA | From bacterium to fungus | Promotes bacterium-fungus symbiosis | [23] |
| Protein kinase family | Members of protein kinase gene family encode proteins having conserved protein kinase domians. | DNA | From plant to fungus | Promotes fungus–plant symbiosis | [23] |
| miRNAs | miR166 and miR159. miR166 targets Clp-1 transcripts, reduces virulence of pathogenic fungus. miR159 targets Hic-15 transcripts, reduces virulence of pathogenic fungus | RNA | From plant to fungus | These miRNAs target fungal transcripts which are related to the fungal virulence. | [24] |
| sRNAs | TAS1c-siR483, TAS2-siR453 and IGN-siR1. TAS1c-siR483 target the B. cinerea genes BC1G_10728 and BC1G_10508. TAS2-siR453 targets the B. cinerea gene BC1G_08464. IGN-siR1 targets the B. cinerea gene BC1G_05327. These sRNAs target fungal genes which are related to the fungal virulence. | RNA | From plant to fungus | Silence fungal virulence genes, reduce virulence of pathogenic fungus. | [25] |
| Integrative and conjugative element (ICE) | ICEAc. ICEAc is able to excise, form a circular DNA and conjugatively transfer to the gly-tRNA gene of other rhizobial genera. Such integration expands the host range of the reciepent rhizobia. | DNA | Between bacteria | Expand the host-range specificity of rhizobia for interacting with plants | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, Y.-S.; Wang, Z.; Duan, S.; Lam, H.-M. Rhizospheric Communication through Mobile Genetic Element Transfers for the Regulation of Microbe–Plant Interactions. Biology 2021, 10, 477. https://doi.org/10.3390/biology10060477
Ku Y-S, Wang Z, Duan S, Lam H-M. Rhizospheric Communication through Mobile Genetic Element Transfers for the Regulation of Microbe–Plant Interactions. Biology. 2021; 10(6):477. https://doi.org/10.3390/biology10060477
Chicago/Turabian StyleKu, Yee-Shan, Zhili Wang, Shaowei Duan, and Hon-Ming Lam. 2021. "Rhizospheric Communication through Mobile Genetic Element Transfers for the Regulation of Microbe–Plant Interactions" Biology 10, no. 6: 477. https://doi.org/10.3390/biology10060477
APA StyleKu, Y.-S., Wang, Z., Duan, S., & Lam, H.-M. (2021). Rhizospheric Communication through Mobile Genetic Element Transfers for the Regulation of Microbe–Plant Interactions. Biology, 10(6), 477. https://doi.org/10.3390/biology10060477








