Characteristics of the Soil Microbial Communities in Different Slope Positions along an Inverted Stone Slope in a Degraded Karst Tiankeng
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sample Collection
2.3. DNA Extraction and Sequancing
2.4. Processing of Sequencing Data
2.5. Statistical Analysis
3. Results
3.1. Characteristics of the Environmental Factors among Slope Positions
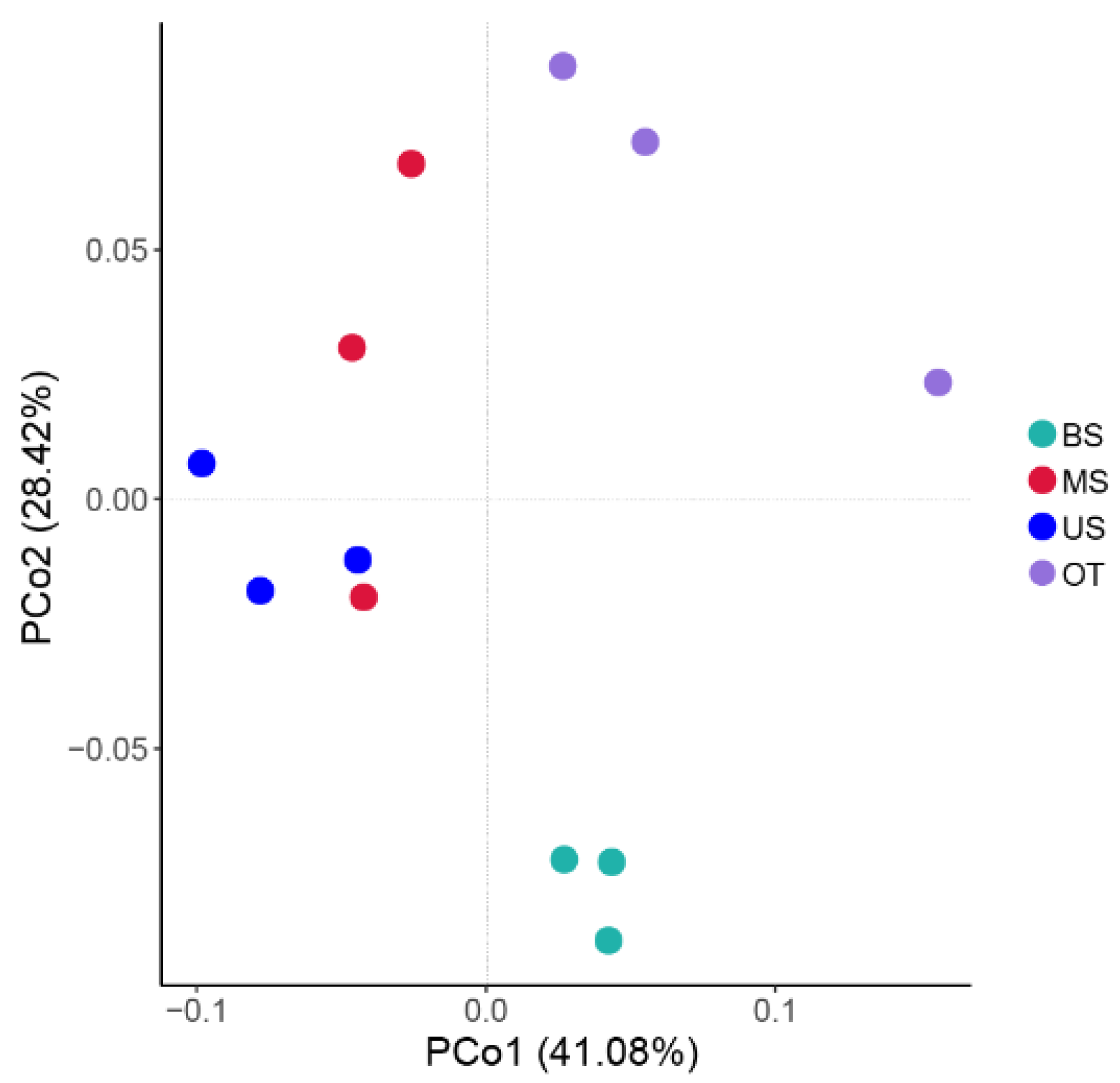
3.2. Composition and Diversity of the Microbial Community along the Slope
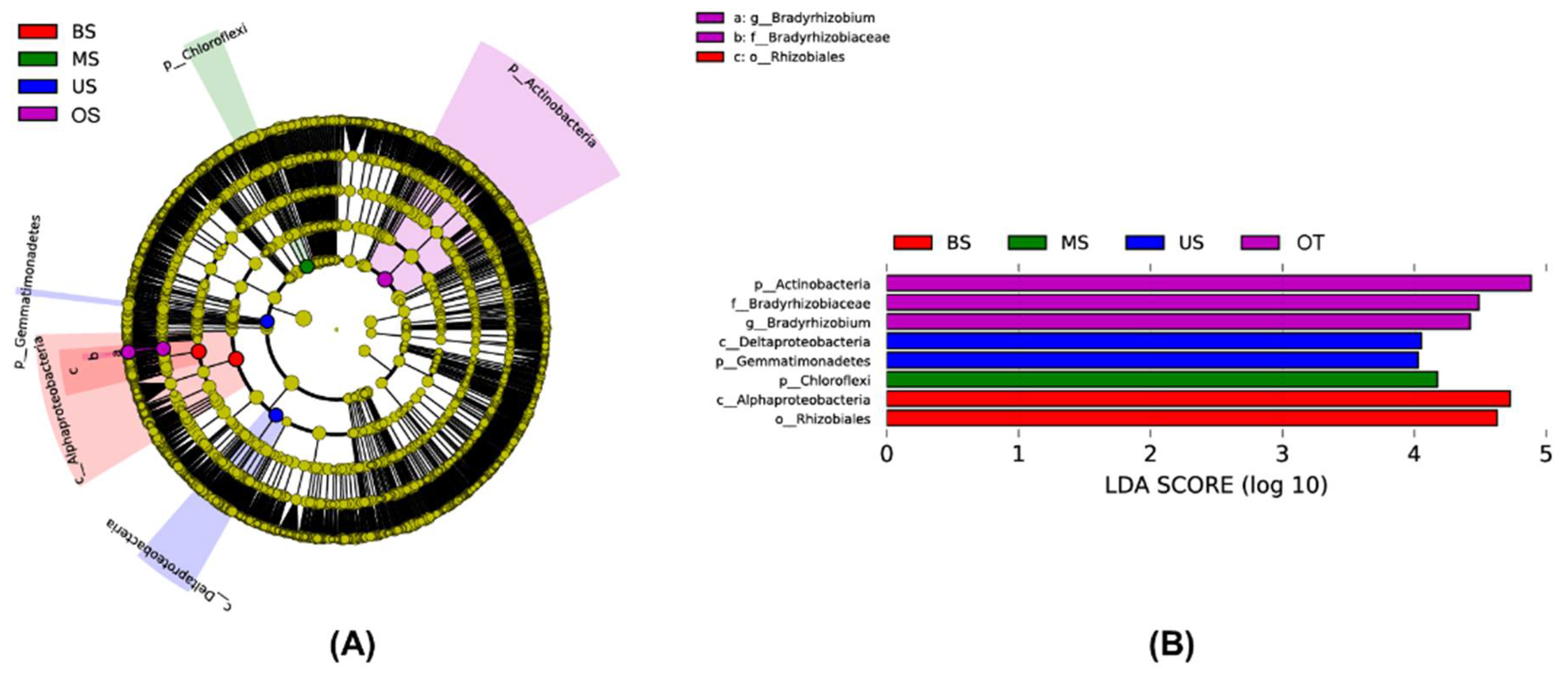
3.2.1. Composition of the Microbial Community
3.2.2. Diversity of the Microbial Community
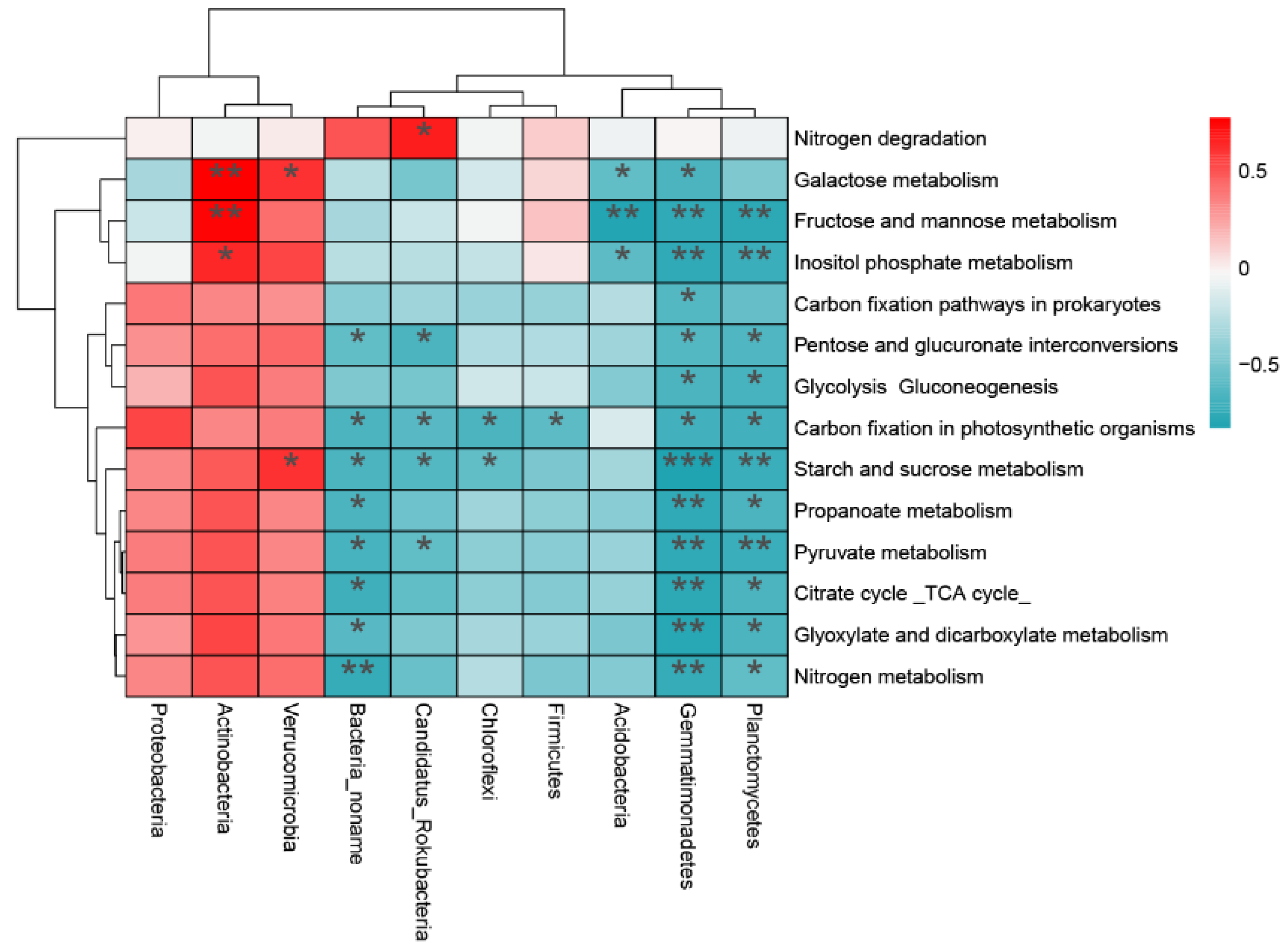
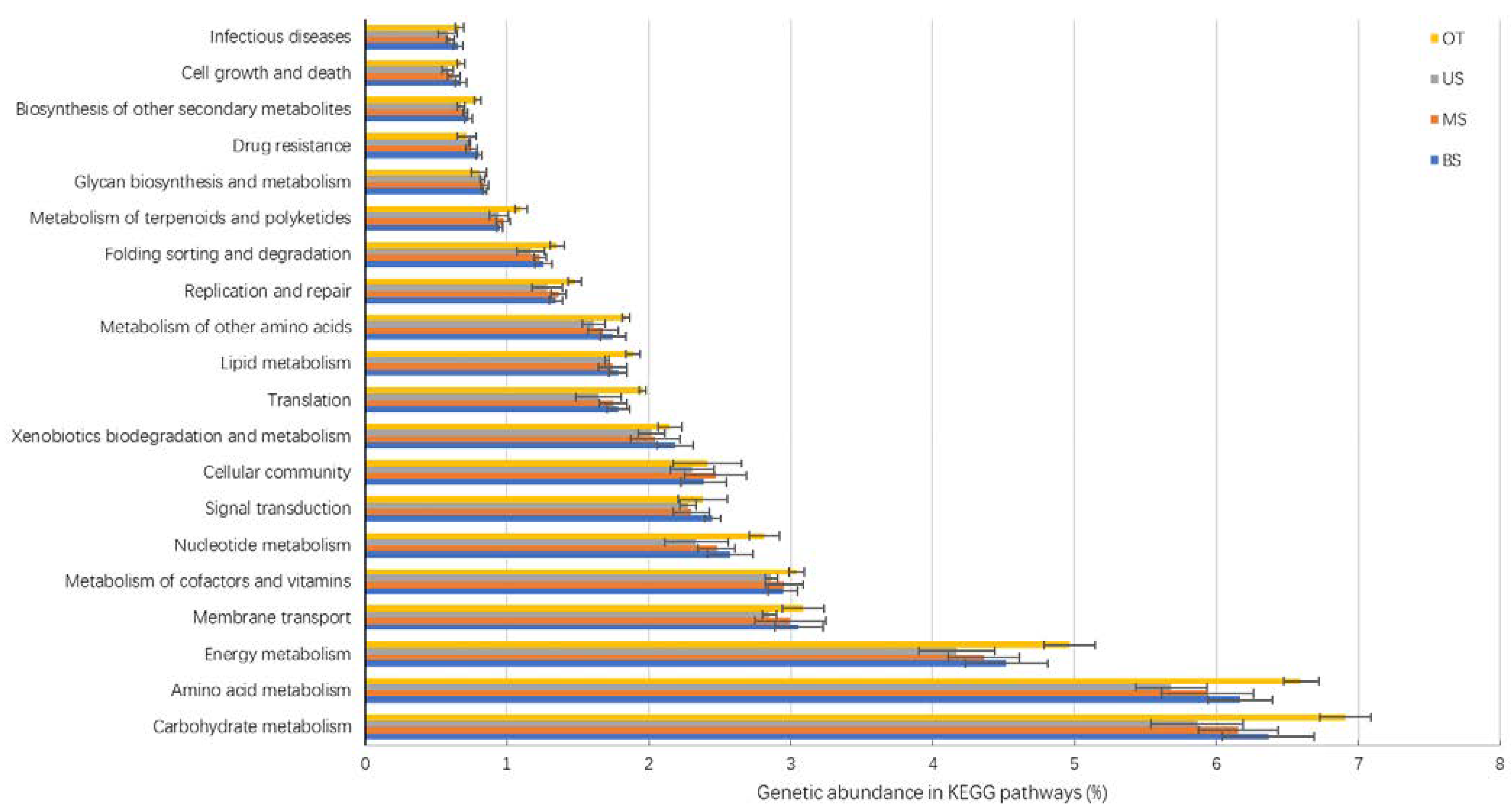
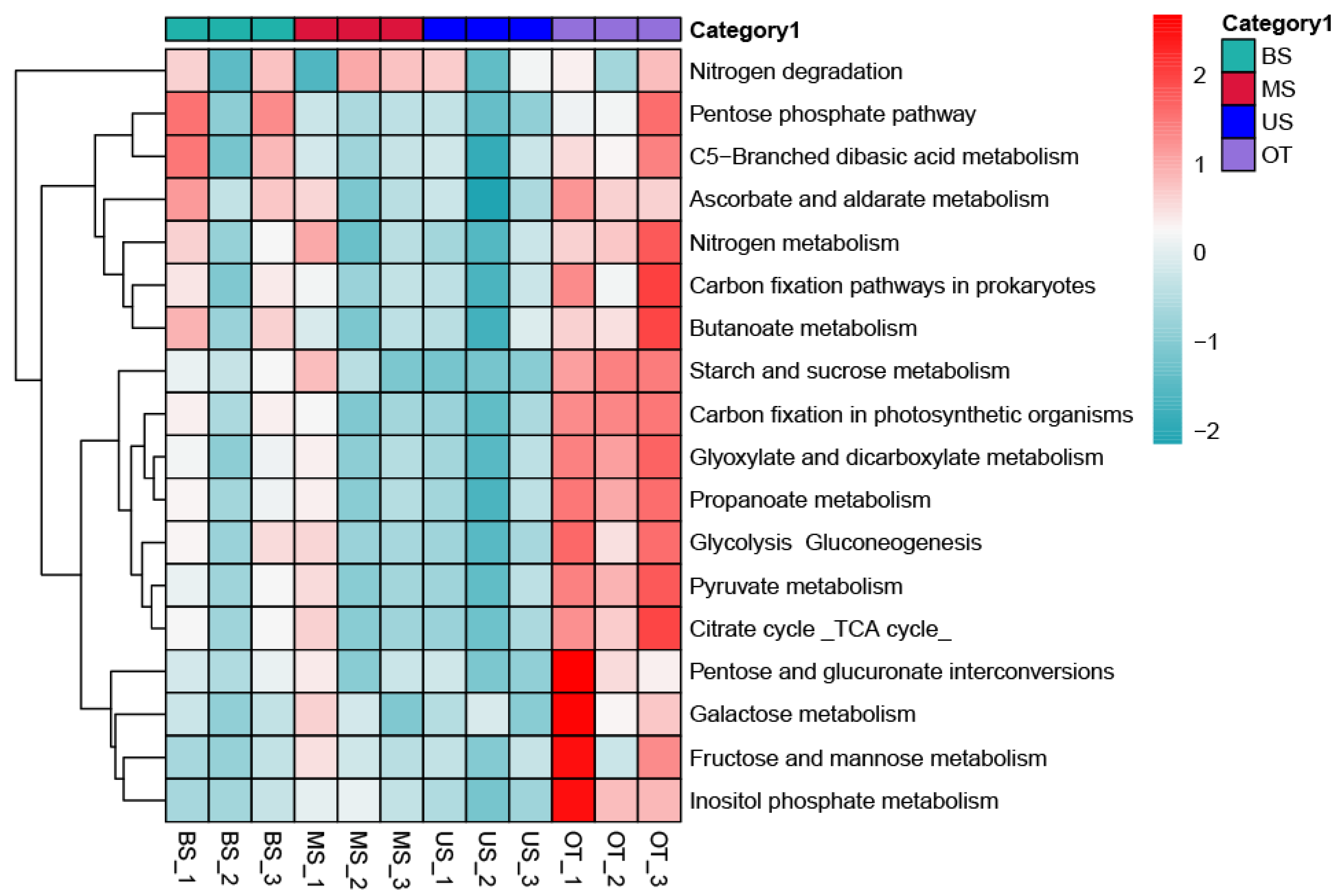
3.3. Functional Differences in Microbial Communities along the Slope
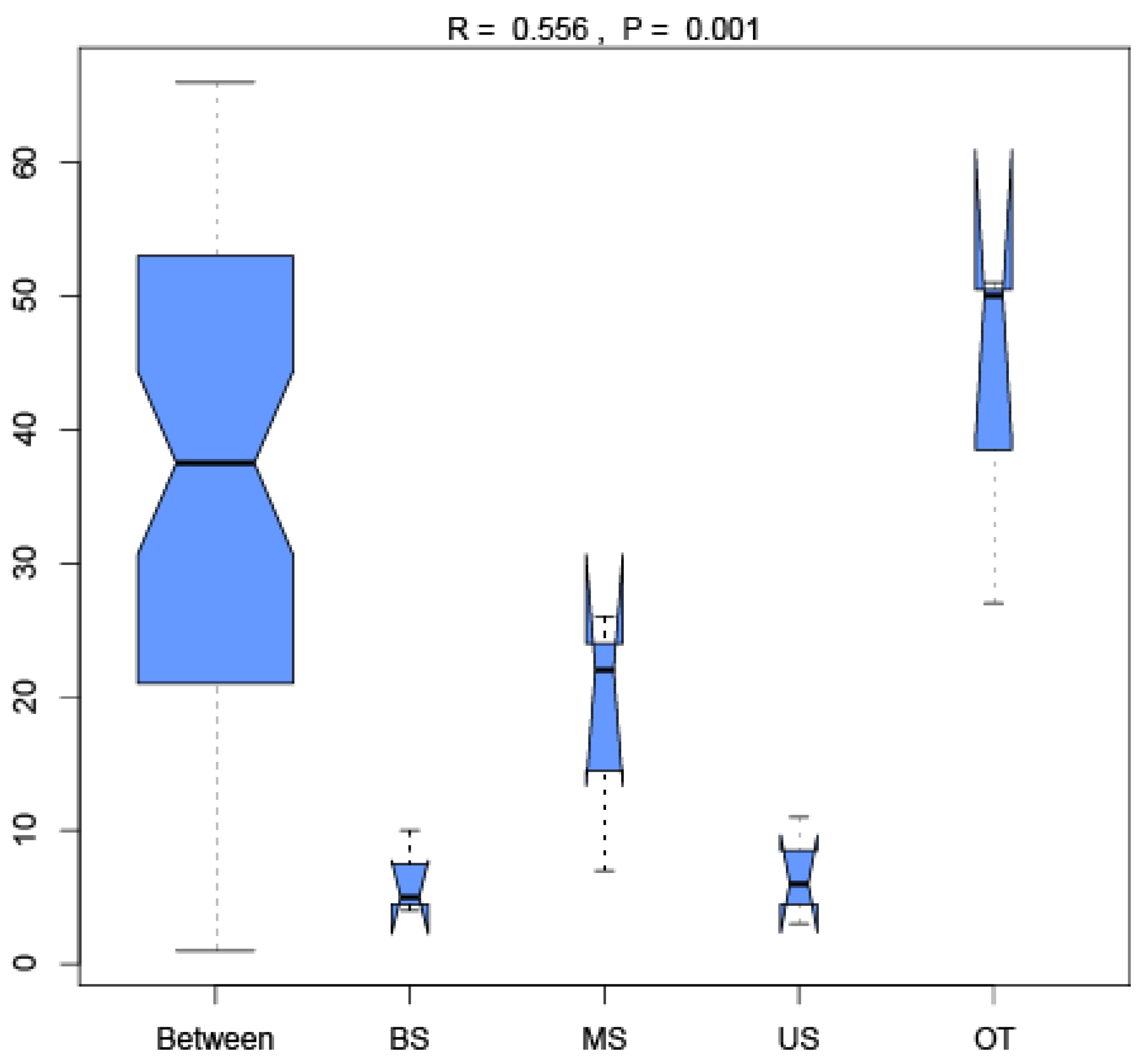
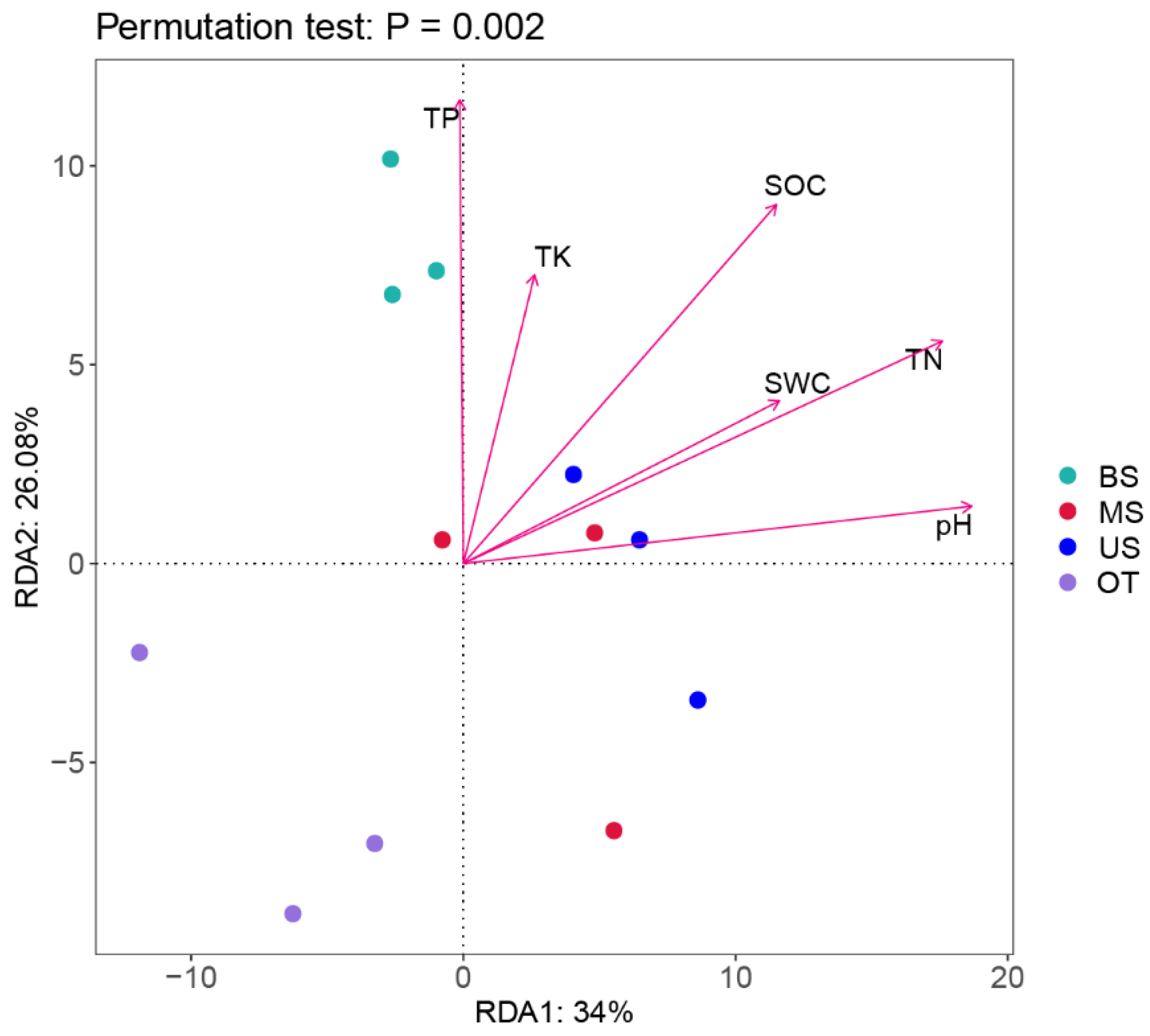
3.4. Relationship between the Microbial Community and Soil Characteristics
4. Discussion
4.1. Diversity and Composition of Microbial Communities along a Slope Gradient
4.2. Comparisons of Microbial Community Functional Groups among the Slope Gradient
4.3. Relationships between Soil Characteristics and Microbial Communities
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Item | Light Degradation | Moderate Degradation | Severe Degradation | Heavy Degradation |
|---|---|---|---|---|
| Depth-width rate | (0.45, 1) | (0.35, 0.45) | (0.1, 0.35) | (0, 0.1) |
| Damage degree of wall area | 0–20% | 21%–50% | 51%–80% | >81% |
| Damage degree of wall area | <1 | 1~2 | 3 | >4(Circularity distribution) |
| Trapping | Good trapping | General trapping | Slightly poor trapping | Poor trapping |
| Pattern of pithead | Approximately ellipse | Irregular ellipse | Irregular polygon | Approximately large doline |
| Sampl Sites | Geographic Coordinate | Altitude (m) | Plant Shannon–Wiener | |
|---|---|---|---|---|
| Longitude (E) | Latitude (N) | |||
| BS | 103°34′46.67” | 25°48′7.75” | 1949.2 | 1.76 ± 0.24 ab |
| MS | 103°34′46.86” | 25°48′6.90” | 1972.9 | 2.04 ± 0.36 a |
| US | 103°34′46.88” | 25°48′7.34” | 1998.3 | 2.19 ± 0.18 a |
| OT | 103°34′45.93” | 25°48′3.63” | 2052.6 | 1.65 ± 0.08 b |
| Sample Sites | Plant | |
|---|---|---|
| Dominant Species | Number of Species | |
| BS | Myrsine africana Linn. (Shrub) | 145 |
| MS | Myrsine africana Linn. (Shrub) | 216 |
| Debregeasia orientalis C. J. Chen (Shrub) | 25 | |
| Ternstroemia gymnanthera (Wight et Arn.) Beddome (Shrub) | 18 | |
| Swida oblonga (Arbor) | 19 | |
| US | Cyclobalanopsis glauca (Arbor) | 41 |
| Keteleeria evelyniana Mast. (Arbor) | 40 | |
| Fraxinus griffithii C. B. Clarke (Arbor) | 10 | |
| OT | Myrsine africana Linn. (Shrub) | 132 |
| Viburnum propinquum (Shrub) | 28 | |
| BS | MS | US | OT | |
|---|---|---|---|---|
| Archaea | 0.68% | 0.82% | 0.78% | 0.50% |
| Bacteria | 98.91% | 98.94% | 98.90% | 99.29% |
| Fungi | 0.37% | 0.21% | 0.30% | 0.17% |
| Viruses | 0.03% | 0.03% | 0.02% | 0.03% |
| BS | MS | US | OT | ||
|---|---|---|---|---|---|
| Archaea | Euryarchaeota | 0.3851% | 0.4911% | 0.5203% | 0.3353% |
| Candidatus_Bathyarchaeota | 0.0439% | 0.0579% | 0.0612% | 0.0348% | |
| Archaea_noname | 0.0406% | 0.0426% | 0.0469% | 0.0250% | |
| Crenarchaeota | 0.0262% | 0.0376% | 0.0377% | 0.0235% | |
| Candidatus_Thorarchaeota | 0.0083% | 0.0107% | 0.0107% | 0.0068% | |
| Candidatus_Lokiarchaeota | 0.0043% | 0.0045% | 0.0059% | 0.0043% | |
| Candidatus_Korarchaeota | 0.0013% | 0.0015% | 0.0013% | 0.0014% | |
| Candidatus_Micrarchaeota | 0.0003% | 0.0001% | 0.0003% | 0.0002% | |
| Candidatus_Nanohaloarchaeota | 0.0003% | 0.0004% | 0.0004% | 0.0004% | |
| Candidatus_Parvarchaeota | 0.0001% | 0.0002% | 0.0001% | 0.0003% | |
| Nanoarchaeota | 0.0000% | 0.0002% | 0.0000% | 0.0001% | |
| Fungi | Basidiomycota | 0.2812% | 0.0850% | 0.0691% | 0.0969% |
| Ascomycota | 0.0992% | 0.1978% | 0.1201% | 0.1283% | |
| Thaumarchaeota | 0.0845% | 0.1292% | 0.1366% | 0.0657% | |
| Fungi_noname | 0.0090% | 0.0087% | 0.0095% | 0.0065% | |
| Chytridiomycota | 0.0047% | 0.0041% | 0.0041% | 0.0042% | |
| Glomeromycota | 0.0020% | 0.0050% | 0.0034% | 0.0050% | |
| Blastocladiomycota | 0.0011% | 0.0009% | 0.0008% | 0.0010% | |
| Microsporidia | 0.0004% | 0.0001% | 0.0000% | 0.0004% | |
| Entomophthoromycota | 0.0004% | 0.0003% | 0.0002% | 0.0005% | |
| Cryptomycota | 0.0003% | 0.0002% | 0.0001% | 0.0001% | |
| Viruses | Viruses_noname | 0.0306% | 0.0250% | 0.0348% | 0.0286% |
| Sample Sites | Shannon-Wiener |
|---|---|
| BS | 13.62 ± 0.04 b |
| MS | 13.69 ± 0.08 ab |
| US | 13.79 ± 0.05 a |
| OT | 13.48 ± 0.08 c |
| BS | MS | US | OT | p Value | KO Description | |
|---|---|---|---|---|---|---|
| C cycle | ||||||
| K00194 | 4.76 × 10−6 | 4.19 × 10−6 | 5.78 × 10−6 | 3.83 × 10−6 | 0.834 | acetyl-CoA decarbonylase/synthase |
| K00196 | 1.06 × 10−6 | 2.11 × 10−6 | 4.22 × 10−6 | 3.34 × 10−6 | 0.629 | anaerobic carbon-monoxide dehydrogenase iron sulfur subunit |
| K00198 | 3.29 × 10−6 | 3.99 × 10−7 | 6.46 × 10−7 | 8.42 × 10−7 | 0.015 | anaerobic carbon-monoxide dehydrogenase catalytic subunit |
| K01674 | 1.93 × 10−5 | 1.97 × 10−5 | 3.71 × 10−5 | 1.24 × 10−5 | 0.06 | carbonic anhydrase |
| K03518 | 0.001744 | 0.001950 | 0.002013 | 0.001593 | 0.072 | aerobic carbon-monoxide dehydrogenase small subunit |
| K03519 | 0.001600 | 0.001825 | 0.001950 | 0.001451 | 0.075 | aerobic carbon-monoxide dehydrogenase medium subunit |
| K03520 | 0.006649 | 0.006648 | 0.006743 | 0.006103 | 0.683 | aerobic carbon-monoxide dehydrogenase large subunit |
| K03563 | 3.76 × 10−5 | 3.88 × 10−5 | 3.377 × 10−5 | 4.24 × 10−5 | 0.867 | carbon storage regulator |
| K07537 | 0.000102 | 7.09 × 10−5 | 8.59 × 10−5 | 6.33 × 10−5 | 0.103 | cyclohexa-1,5-dienecarbonyl-CoA hydratase |
| K07539 | 1.35 × 10−6 | 1.43 × 10−5 | 4.12 × 10−7 | 9.83 × 10−7 | 0.482 | 6-oxocyclohex-1-ene-carbonyl-CoA hydrolase |
| K11952 | 0 | 2.80 × 10−6 | 4.52 × 10−6 | 7.81 × 10−8 | 0.165 | bicarbonate transport system ATP-binding protein |
| K11953 | 5.85 × 10−6 | 2.70 × 10−6 | 9.04 × 10−7 | 4.70 × 10−6 | 0.427 | bicarbonate transport system ATP-binding protein |
| K19066 | 6.86 × 10−6 | 5.36 × 10−6 | 2.53 × 10−6 | 3.66 × 10−6 | 0.686 | cyclohex-1-ene-1-carbonyl-CoA dehydrogenase |
| K19067 | 1.24 × 10−5 | 1.22 × 10−5 | 8.10 × 10−6 | 2.18 × 10−5 | 0.367 | cyclohexane-1-carbonyl-CoA dehydrogenase |
| N cycle | ||||||
| K02586 | 1.00 × 10−5 | 2.08 × 10−5 | 8.84 × 10−6 | 8.31 × 10−6 | 0.296 | nitrogenase molybdenum-iron protein alpha chain |
| K02588 | 8.15 × 10−6 | 2.00 × 10−5 | 1.26 × 10−5 | 6.73 × 10−6 | 0.161 | nitrogenase iron protein NifH |
| K02591 | 4.52 × 10−6 | 1.16 × 10−5 | 6.35 × 10−6 | 2.87 × 10−6 | 0.056 | nitrogenase molybdenum-iron protein beta chain |
| K02806 | 0.000237 | 0.000214 | 0.000219 | 0.000174 | 0.121 | nitrogen PTS system EIIA component |
| K04751 | 0.000491 | 0.000530 | 0.000529 | 0.000509 | 0.627 | nitrogen regulatory protein P-II 1 |
| K07708 | 0.000334 | 0.000287 | 0.000304 | 0.000257 | 0.202 | nitrogen regulation sensor histidine kinase GlnL |
| K07712 | 0.000741 | 0.000595 | 0.000674 | 0.000564 | 0.047 | nitrogen regulation response regulator GlnG |
| K13598 | 0.000529 | 0.000470 | 0.000568 | 0.000393 | 0.059 | nitrogen regulation sensor histidine kinase NtrY |
| K13599 | 0.0009345 | 0.000741 | 0.000909 | 0.000596 | 0.01 | nitrogen regulation response regulator NtrX |
| K15861 | 9.794 × 10−5 | 5.034 × 10−5 | 3.66129 × 10−5 | 7.785 × 10−5 | 0.040 | nitrogen fixation regulation protein |
| r | p | |
|---|---|---|
| SOC | 0.1884 | 0.1675 |
| TN | 0.6960 | 3 × 10−4 |
| TP | −0.0546 | 0.5814 |
| TK | −0.0072 | 0.4769 |
| pH | 0.5397 | 5 × 10−4 |
| SWC | 0.0102 | 0.4465 |


References
- Zhu, X.W. China’s karst tiankeng and its value for science and tourism. Sci. Technol. Rev. 2001, 10, 60–63. [Google Scholar]
- Ozkan, K.; Gulsoy, S.; Mert, A.; Ozturk, M.; Muys, B. Plant distribution—Altitude and landform relationships in karstic sinkholes of Mediterranean region of Turkey. J. Environ. Biol. 2010, 31, 51–60. [Google Scholar] [PubMed]
- Su, Y.; Tang, Q.; Mo, F.; Xue, Y. Karst tiankengs as refugia for indigenous tree flora amidst a degraded landscape in southwestern china. Sci. Rep. 2017, 7, 4249. [Google Scholar] [CrossRef]
- Pu, G.; Lv, Y.; Dong, L.; Zhou, L.; Huang, K.; Zeng, D.; Mo, L.; Xu, G. Profiling the bacterial diversity in a typical karst Tiankeng of China. Biomolecules 2019, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- Shui, W.; Chen, Y.P.; Wang, Y.W.; Su, Z.A.; Zhang, S. Origination, study progress and prospect of karst tiankeng research in China. Acta Geogr. Sin. 2015, 70, 431–446. [Google Scholar]
- Waltham, T. Collapse processes at the tiankengs of Xingwen. J. Cave Karst Stud. 2005, 32, 107. [Google Scholar]
- Strickland, M.S.; Lauber, C.; Fierer, N.; Bradford, M.A. Testing the functional significance of microbial community composition. Ecology 2009, 90, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Li, J.; Tang, Z.; Shangguan, Z. Soil bacterial community succession during desertification in a desert steppe ecosystem. Land Degrad. Dev. 2020, 31, 1662–1674. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, G.; Robinson, D.; Yang, Z.; Guo, J.; Xie, J.; Fu, S.; Zhou, L.; Yang, Y. Large amounts of easily decomposable carbon stored in subtropical forest subsoil are associated with r-strategy-dominated soil microbes. Soil Biol. Biochem. 2016, 95, 233–242. [Google Scholar] [CrossRef]
- Budge, K.; Leifeld, J.; Egli, M.; Fuhrer, J. Soil microbial communities in (sub)alpine grasslands indicate a moderate shift towards new environmental conditions 11 years after soil translocation. Soil Biol. Biochem. 2011, 43, 1148–1154. [Google Scholar] [CrossRef]
- Du, Z.; Riveros-Iregui, D.A.; Jones, R.T.; McDermott, T.R.; Dore, J.E.; McGlynn, B.L.; Emanuel, R.E.; Li, X. Landscape position influences microbial composition and function via redistribution of soil water across a watershed. Appl. Environ. Microbiol. 2015, 81, 8457–8468. [Google Scholar] [CrossRef][Green Version]
- Helgason, B.L.; Konschuh, H.J.; Bedard-Haughn, A.; VandenBygaart, A.J. Microbial distribution in an eroded landscape: Buried A horizons support abundant and unique communities. Agric. Ecosyst. Environ. 2014, 196, 94–102. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Guo, S.L.; Sun, Q.Q.; Li, N.N.; Jiang, J.S.; Wang, R.; Zhang, Y.J.; Liu, Q.F.; Wu, D.F.; Li, R.J.; et al. Soil organic carbon sequestration potential of artificial and natural vegetation in the hilly regions of Loess Plateau. Ecol. Eng. 2015, 82, 547–554. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000; pp. 263–270. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2012, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Mohammadi, M.F.; Jalali, S.G.; Kooch, Y.; Said-Pullicino, D. The effect of landform on soil microbial activity and biomass in a Hyrcanian oriental beech stand. Catena 2017, 149, 309–317. [Google Scholar] [CrossRef]
- Sun, Q.; Hu, Y.; Wang, R.; Guo, S.; Yao, L.; Duan, P. Spatial distribution of microbial community composition along a steep slope plot of the Loess Plateau. Appl. Soil Ecol. 2018, 130, 226–236. [Google Scholar] [CrossRef]
- Moorman, T.B.; Cambardella, C.A.; James, D.E.; Karlen, D.L.; Kramer, L.A. Quantification of tillage and landscape effects on soil carbon in small Iowa watersheds. Soil Tillage Res. 2004, 78, 225–236. [Google Scholar] [CrossRef]
- Zhong, Y.; Yan, W.; Wang, R.; Wang, W.; Shangguan, Z. Decreased occurrence of carbon cycle functions in microbial communities along with long-term secondary succession. Soil Biol. Biochem. 2018, 123, 207–217. [Google Scholar] [CrossRef]
- Cheng, J.M.; Han, Z.J.; Cong, J.; Yu, J.J.; Zhou, J.Z.; Zhao, M.X.; Zhang, Y.G. Edaphic variables are better indicators of soil microbial functional structure than plant-related ones in subtropical broad-leaved forests. Sci. Total Environ. 2021, 773, 145630. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Trojan, D.; Roux, S.; Herbold, C.; Rattei, T.; Woebken, D. Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 2018, 20, 1041–1063. [Google Scholar] [CrossRef] [PubMed]
- Salles, J.F.; Poly, F.; Schmid, B.; Roux, X.L. Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology 2009, 90, 3324–3332. [Google Scholar] [CrossRef]
- Zhang, K.; Cheng, X.; Dang, H.; Ye, C.; Zhang, Y.; Zhang, Q. Linking litter production, quality and decomposition to vegetation succession following agricultural abandonment. Soil Biol. Biochem. 2013, 57, 803–813. [Google Scholar] [CrossRef]
- Zak, D.R.; Holmes, W.E.; White, D.C.; Peacock, A.D.; Tilman, D. Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology 2003, 84, 2042–2050. [Google Scholar] [CrossRef]
- Cline, L.C.; Zak, D.R. Soil microbial communities are shaped by plant-driven changes in resource availability during secondary succession. Ecology 2015, 96, 3374–3385. [Google Scholar] [CrossRef]
- Thomson, B.C.; Tisserant, E.; Plassart, P.; Uroz, S.; Griffiths, R.I.; Hannula, S.E.; Buee, M.; Mougel, C.; Ranjard, L.; Van Veen, J.A.; et al. Soil conditions and land use intensification effects on soil microbial communities across a range of European field sites. Soil Biol. Biochem. 2015, 88, 403–413. [Google Scholar] [CrossRef]
- Liu, J.J.; Sui, Y.Y.; Yu, Z.H.; Shi, Y.; Chu, H.Y.; Jin, J.; Liu, X.B.; Wang, G.H. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Shen, C.C.; Xiong, J.B.; Zhang, H.Y.; Feng, Y.Z.; Lin, X.G.; Li, X.Y.; Liang, W.J.; Chu, H.Y. Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 2013, 57, 204–211. [Google Scholar] [CrossRef]
- Edwards, I.P.; Buergmann, H.; Miniaci, C.; Zeyer, J. Variation in microbial community composition and culturability in the rhizosphere of Leucanthemopsis alpina (L.) heywood and adjacent bare soil along an alpine chronosequence. Microb. Ecol. 2006, 52, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; lzak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Blaszczyk, M. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]

| Characteristics | BS | MS | US | OT |
|---|---|---|---|---|
| Soil water content (%) | 43.21% ± 0.03 ab | 42.45% ± 0.02 ab | 48.46% ± 0.02 a | 39.47% ± 0.02 b |
| Total organic carbon (g/kg) | 44.43 ± 3.05 ab | 42.67 ± 2.94 ab | 48.90 ± 4.76 a | 35.50 ± 3.25 b |
| Total nitrogen (g/kg) | 2.69 ± 0.14 ab | 2.77 ± 0.34 a | 2.73 ± 0.11 a | 2.13 ± 0.23 b |
| Total phosphorus (g/kg) | 0.64 ± 0.09 a | 0.43 ± 0.17 a | 0.54 ± 0.11 a | 0.39 ± 0.07 a |
| Total potassium (g/kg) | 17.53 ± 0.93 a | 15.97 ± 3.02 a | 14.37 ± 4.43 a | 13.51 ± 2.08 a |
| pH | 6.57 ± 0.15 ab | 6.72 ± 0.21 ab | 6.79 ± 0.14 a | 6.30 ± 0.15 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, C.; Feng, J.; Zhu, S.-F.; Shui, W. Characteristics of the Soil Microbial Communities in Different Slope Positions along an Inverted Stone Slope in a Degraded Karst Tiankeng. Biology 2021, 10, 474. https://doi.org/10.3390/biology10060474
Jiang C, Feng J, Zhu S-F, Shui W. Characteristics of the Soil Microbial Communities in Different Slope Positions along an Inverted Stone Slope in a Degraded Karst Tiankeng. Biology. 2021; 10(6):474. https://doi.org/10.3390/biology10060474
Chicago/Turabian StyleJiang, Cong, Jie Feng, Su-Feng Zhu, and Wei Shui. 2021. "Characteristics of the Soil Microbial Communities in Different Slope Positions along an Inverted Stone Slope in a Degraded Karst Tiankeng" Biology 10, no. 6: 474. https://doi.org/10.3390/biology10060474
APA StyleJiang, C., Feng, J., Zhu, S.-F., & Shui, W. (2021). Characteristics of the Soil Microbial Communities in Different Slope Positions along an Inverted Stone Slope in a Degraded Karst Tiankeng. Biology, 10(6), 474. https://doi.org/10.3390/biology10060474






