Characterization of Immune Cell Subsets of Tumor Infiltrating Lymphocytes in Brain Metastases
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. TILs Scoring
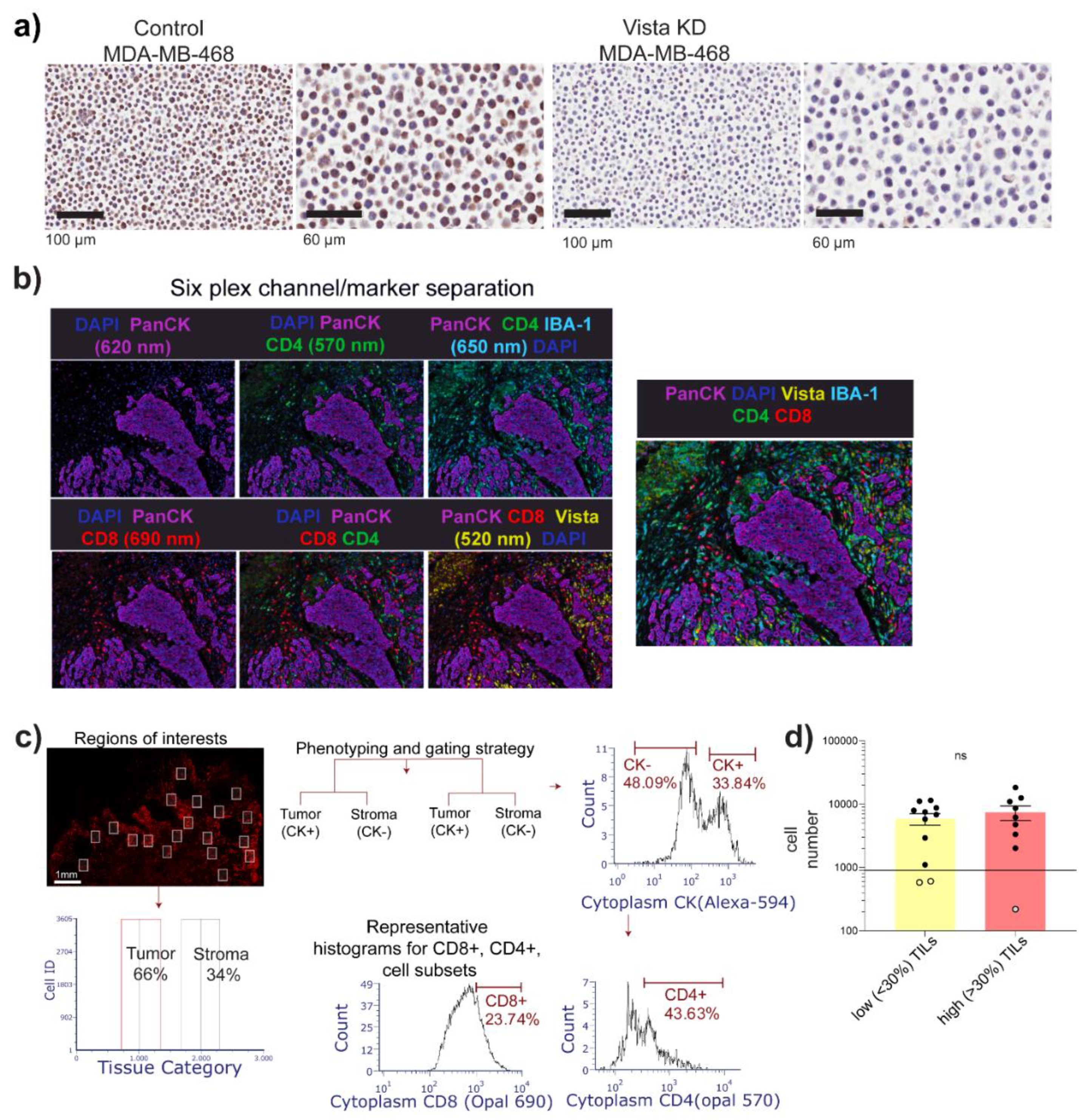
2.3. Transient Knockdown
| sense (5′-3′) | antisense (5′-3′) |
| CCGUAUUCCCUGUAUGUCUTT | AGACAUACAGGGAAUACGGT |
| GCAAAGAUGCACCAUCCAATT | UUGGAUGGUGCAUCUUUGCTT |
| GCAACAUUCAAGGGAUUGATT | UCAAUCCCUUGAAUGUUGCTT |
2.4. mIF and IHC
2.5. Dataset Analysis
2.6. Statistical Analysis
3. Results
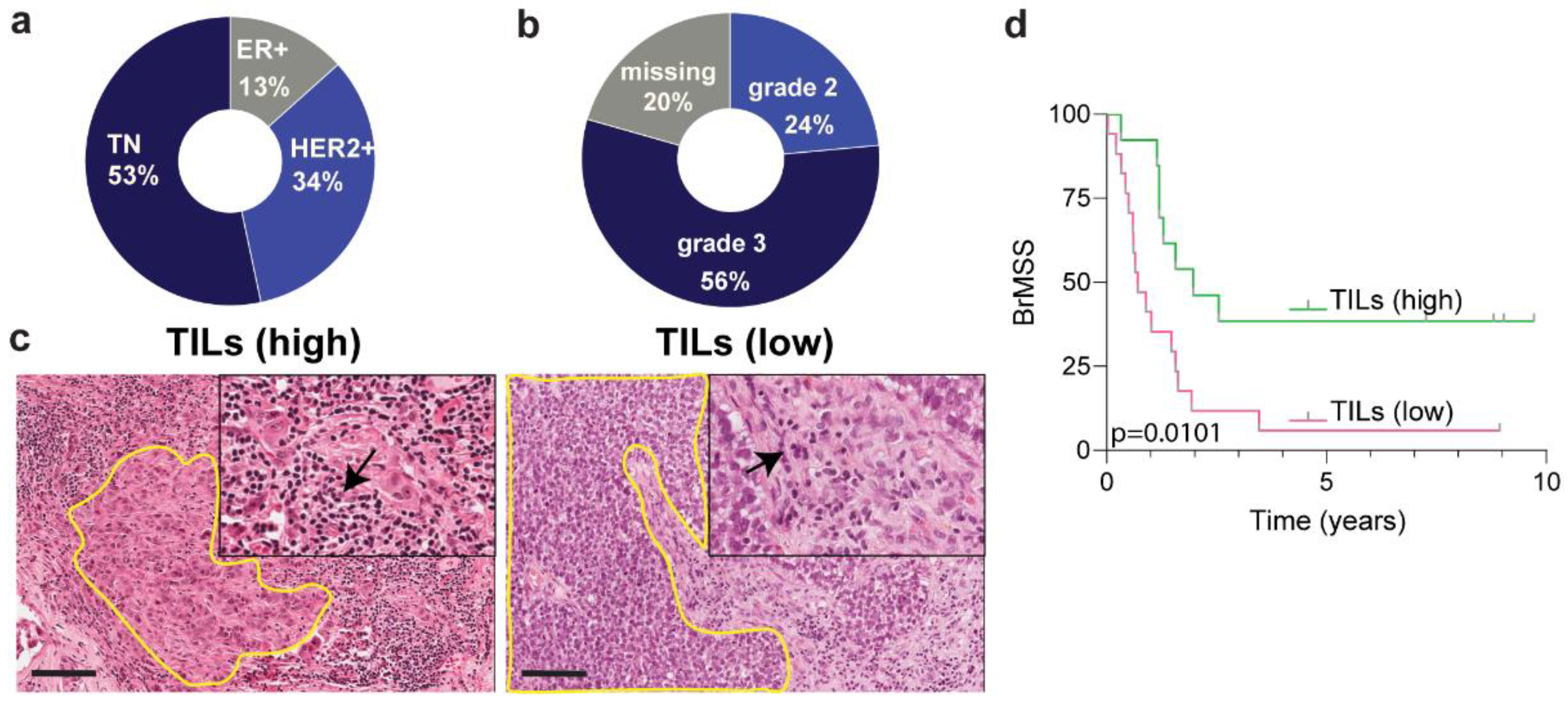
3.1. Cohort Description and Image Cytometry
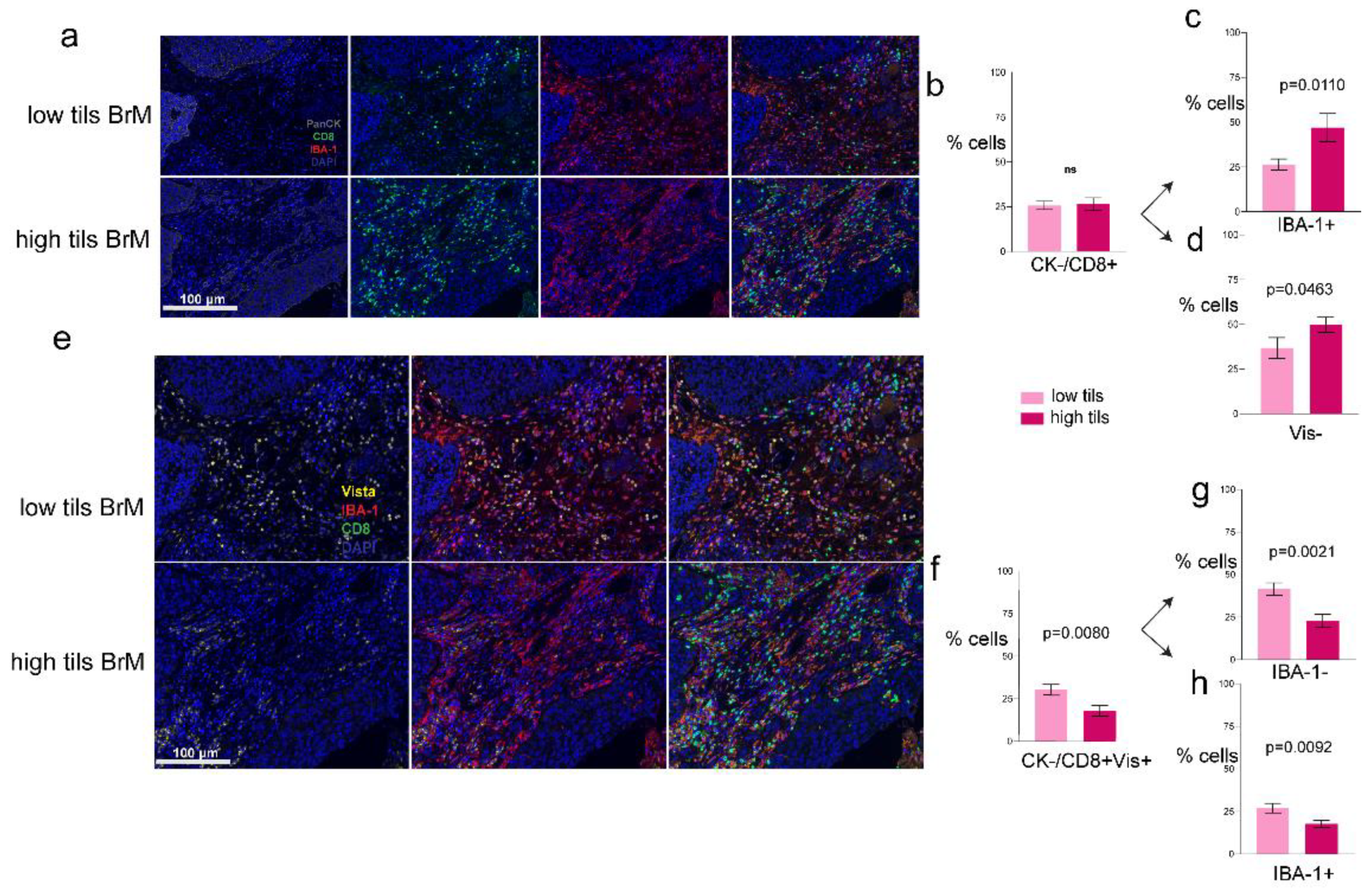
3.2. Increased VISTA and IBA-1 Expression in the TME
3.3. Phenotypes of the CD8+ T-Cells
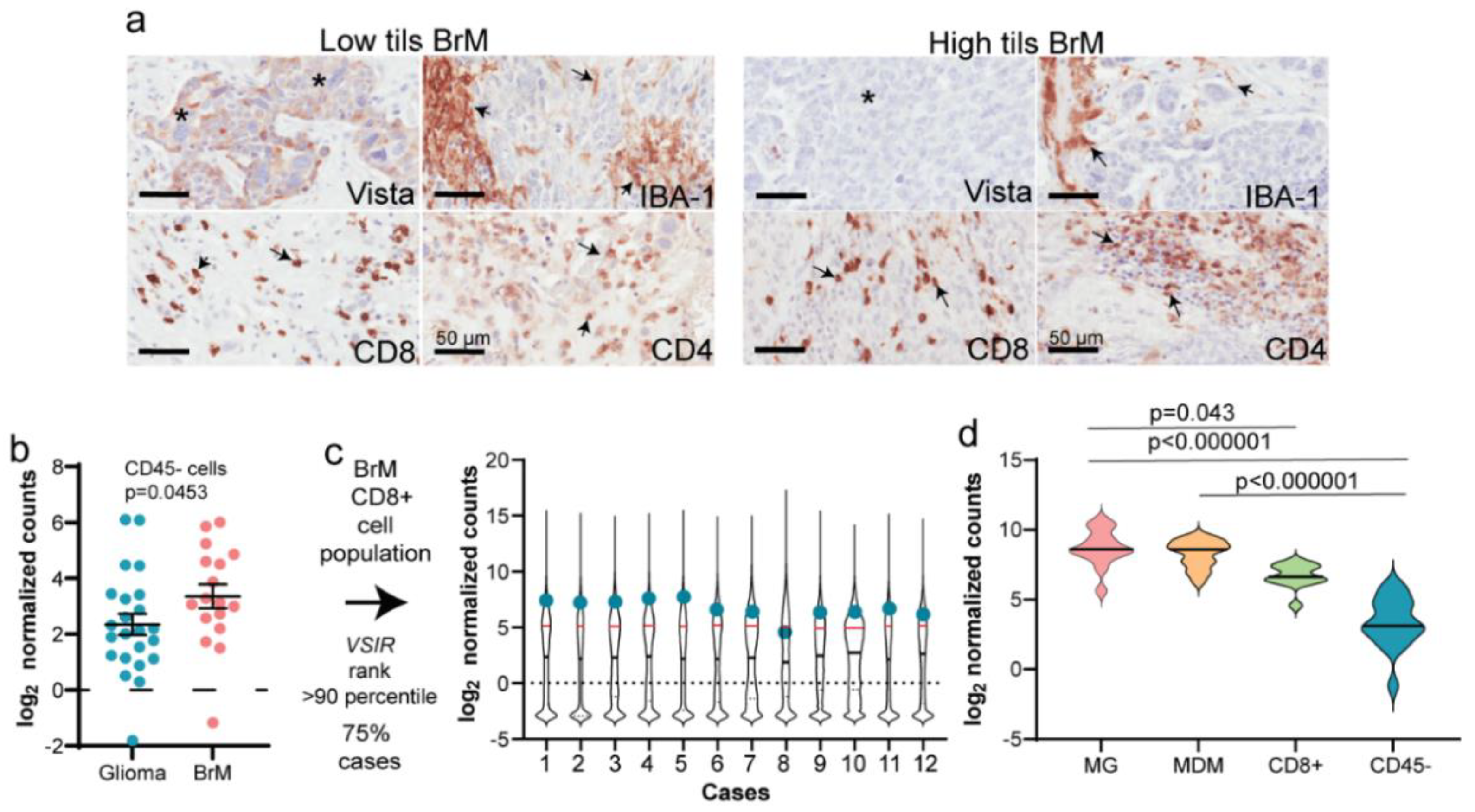
3.4. Validation of mIF Using IHC and RNA Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Cagney, D.N.; Martin, A.M.; Catalano, P.J.; Redig, A.J.; Lin, N.U.; Lee, E.Q.; Wen, P.Y.; Dunn, I.F.; Bi, W.L.; Weiss, S.E.; et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: A population-based study. Neuro-Oncol. 2017, 19, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Tawbi, H.A.; Forsyth, P.A.; Algazi, A.; Hamid, O.; Hodi, F.S.; Moschos, S.J.; Khushalani, N.I.; Lewis, K.; Lao, C.D.; Postow, M.A. Combined nivolumab and ipilimumab in melanoma metastatic to the brain. N. Engl. J. Med. 2018, 379, 722–730. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Atkinson, V.; Lo, S.; Sandhu, S.; Guminski, A.D.; Brown, M.P.; Wilmott, J.S.; Edwards, J.; Gonzalez, M.; Scolyer, R.A. Combination nivolumab and ipilimumab or nivolumab alone in melanoma brain metastases: A multicentre randomised phase 2 study. Lancet Oncol. 2018, 19, 672–681. [Google Scholar] [CrossRef]
- Hendriks, L.E.L.; Henon, C.; Auclin, E.; Mezquita, L.; Ferrara, R.; Audigier-Valette, C.; Mazieres, J.; Lefebvre, C.; Rabeau, A.; Le Moulec, S.; et al. Outcome of Patients with Non-Small Cell Lung Cancer and Brain Metastases Treated with Checkpoint Inhibitors. J. Thorac. Oncol. 2019, 14, 1244–1254. [Google Scholar] [CrossRef]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target 2020, 5, 166. [Google Scholar] [CrossRef]
- Sevenich, L.; Bowman, R.L.; Mason, S.D.; Quail, D.F.; Rapaport, F.; Elie, B.T.; Brogi, E.; Brastianos, P.K.; Hahn, W.C.; Holsinger, L.J.; et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat. Cell Biol. 2014, 16, 876–888. [Google Scholar] [CrossRef]
- Neman, J.; Choy, C.; Kowolik, C.M.; Anderson, A.; Duenas, V.J.; Waliany, S.; Chen, B.T.; Chen, M.Y.; Jandial, R. Co-evolution of breast-to-brain metastasis and neural progenitor cells. Clin. Exp. Metastasis 2013, 30, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Sartorius, C.A.; Hanna, C.T.; Gril, B.; Cruz, H.; Serkova, N.J.; Huber, K.M.; Kabos, P.; Schedin, T.B.; Borges, V.F.; Steeg, P.S.; et al. Estrogen promotes the brain metastatic colonization of triple negative breast cancer cells via an astrocyte-mediated paracrine mechanism. Oncogene 2016, 35, 2881–2892. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, S.; Yao, J.; Lowery, F.J.; Zhang, Q.; Huang, W.C.; Li, P.; Li, M.; Wang, X.; Zhang, C.; et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature 2015, 527, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Kalita-de Croft, P.; Lim, M.; Chittoory, H.; de Luca, X.M.; Kutasovic, J.R.; Day, B.W.; Al-Ejeh, F.; Simpson, P.T.; McCart Reed, A.E.; Lakhani, S.R.; et al. Clinicopathologic significance of nuclear HER4 and phospho-YAP(S-127) in human breast cancers and matching brain metastases. Ther. Adv. Med. Oncol. 2020, 12. [Google Scholar] [CrossRef]
- Kalita-de Croft, P.; Straube, J.; Lim, M.; Al-Ejeh, F.; Lakhani, S.R.; Saunus, J.M. Proteomic Analysis of the Breast Cancer Brain Metastasis Microenvironment. Int. J. Mol. Sci. 2019, 20, 2524. [Google Scholar] [CrossRef] [PubMed]
- Klemm, F.; Maas, R.R.; Bowman, R.L.; Kornete, M.; Soukup, K.; Nassiri, S.; Brouland, J.P.; Iacobuzio-Donahue, C.A.; Brennan, C.; Tabar, V.; et al. Interrogation of the Microenvironmental Landscape in Brain Tumors Reveals Disease-Specific Alterations of Immune Cells. Cell 2020, 181, 1643–1660 e1617. [Google Scholar] [CrossRef] [PubMed]
- Friebel, E.; Kapolou, K.; Unger, S.; Nunez, N.G.; Utz, S.; Rushing, E.J.; Regli, L.; Weller, M.; Greter, M.; Tugues, S.; et al. Single-Cell Mapping of Human Brain Cancer Reveals Tumor-Specific Instruction of Tissue-Invading Leukocytes. Cell 2020, 181, 1626–1642 e1620. [Google Scholar] [CrossRef] [PubMed]
- Kalita-de Croft, P.; Sadeghi Rad, H.; Gasper, H.; O’Byrne, K.; Lakhani, S.R.; Kulasinghe, A. Spatial profiling technologies and applications for brain cancers. Expert Rev. Mol. Diagn. 2021, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Rubinstein, R.; Lines, J.L.; Wasiuk, A.; Ahonen, C.; Guo, Y.; Lu, L.F.; Gondek, D.; Wang, Y.; Fava, R.A.; et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 2011, 208, 577–592. [Google Scholar] [CrossRef] [PubMed]
- Ge, M.J.; Xu, K.L.; Xu, T.; Tang, Y.N.; Li, Z.Y.; Yan, Z.L.; Sun, H.Y.; Cheng, H.; Zhu, F.; Sang, W.; et al. Expression and Significance of PD-1, TIM-3 and VISTA on T Cell of Acute Myeloid Leukemia Patients. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2020, 28, 748–752. (In Chinese) [Google Scholar] [CrossRef]
- Murga-Zamalloa, C.A.; Brown, N.A.; Wilcox, R.A. Expression of the checkpoint receptors LAG-3, TIM-3 and VISTA in peripheral T cell lymphomas. J. Clin. Pathol. 2020, 73, 197–203. [Google Scholar] [CrossRef]
- Zong, L.; Mo, S.; Yu, S.; Zhou, Y.; Zhang, M.; Chen, J.; Xiang, Y. Expression of the immune checkpoint VISTA in breast cancer. Cancer Immunol. Immunother. 2020, 69, 1437–1446. [Google Scholar] [CrossRef]
- Kato, S.; Okamura, R.; Kumaki, Y.; Ikeda, S.; Nikanjam, M.; Eskander, R.; Goodman, A.; Lee, S.; Glenn, S.T.; Dressman, D.; et al. Expression of TIM3/VISTA checkpoints and the CD68 macrophage-associated marker correlates with anti-PD1/PDL1 resistance: Implications of immunogram heterogeneity. Oncoimmunology 2020, 9, 1708065. [Google Scholar] [CrossRef]
- Liao, H.; Zhu, H.; Liu, S.; Wang, H. Expression of V-domain immunoglobulin suppressor of T cell activation is associated with the advanced stage and presence of lymph node metastasis in ovarian cancer. Oncol. Lett. 2018, 16, 3465–3472. [Google Scholar] [CrossRef]
- Guldner, I.H.; Wang, Q.; Yang, L.; Golomb, S.M.; Zhao, Z.; Lopez, J.A.; Brunory, A.; Howe, E.N.; Zhang, Y.; Palakurthi, B.; et al. CNS-Native Myeloid Cells Drive Immune Suppression in the Brain Metastatic Niche through Cxcl10. Cell 2020. [Google Scholar] [CrossRef] [PubMed]
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; van de Vijver, K.; Estrada, M.V.; Gonzalez-Ericsson, P.I.; et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv. Anat. Pathol. 2017, 24, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Borggrewe, M.; Grit, C.; Den Dunnen, W.F.A.; Burm, S.M.; Bajramovic, J.J.; Noelle, R.J.; Eggen, B.J.L.; Laman, J.D. VISTA expression by microglia decreases during inflammation and is differentially regulated in CNS diseases. Glia 2018, 66, 2645–2658. [Google Scholar] [CrossRef]
- Soto, M.S.; Serres, S.; Anthony, D.C.; Sibson, N.R. Functional role of endothelial adhesion molecules in the early stages of brain metastasis. Neuro Oncol. 2014, 16, 540–551. [Google Scholar] [CrossRef]
- Berghoff, A.S.; Fuchs, E.; Ricken, G.; Mlecnik, B.; Bindea, G.; Spanberger, T.; Hackl, M.; Widhalm, G.; Dieckmann, K.; Prayer, D.; et al. Density of tumor-infiltrating lymphocytes correlates with extent of brain edema and overall survival time in patients with brain metastases. Oncoimmunology 2016, 5, e1057388. [Google Scholar] [CrossRef]
- Ogiya, R.; Niikura, N.; Kumaki, N.; Yasojima, H.; Iwasa, T.; Kanbayashi, C.; Oshitanai, R.; Tsuneizumi, M.; Watanabe, K.I.; Matsui, A.; et al. Comparison of immune microenvironments between primary tumors and brain metastases in patients with breast cancer. Oncotarget 2017, 8, 103671–103681. [Google Scholar] [CrossRef]
- Berghoff, A.S.; Ricken, G.; Wilhelm, D.; Rajky, O.; Widhalm, G.; Dieckmann, K.; Birner, P.; Bartsch, R.; Preusser, M. Tumor infiltrating lymphocytes and PD-L1 expression in brain metastases of small cell lung cancer (SCLC). J. Neurooncol. 2016, 130, 19–29. [Google Scholar] [CrossRef]
- Mulati, K.; Hamanishi, J.; Matsumura, N.; Chamoto, K.; Mise, N.; Abiko, K.; Baba, T.; Yamaguchi, K.; Horikawa, N.; Murakami, R.; et al. VISTA expressed in tumour cells regulates T cell function. Br. J. Cancer 2019, 120, 115–127. [Google Scholar] [CrossRef]
- Villarroel-Espindola, F.; Yu, X.; Datar, I.; Mani, N.; Sanmamed, M.; Velcheti, V.; Syrigos, K.; Toki, M.; Zhao, H.; Chen, L.; et al. Spatially Resolved and Quantitative Analysis of VISTA/PD-1H as a Novel Immunotherapy Target in Human Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 1562–1573. [Google Scholar] [CrossRef]
- Rosenbaum, S.R.; Knecht, M.; Mollaee, M.; Zhong, Z.; Erkes, D.A.; McCue, P.A.; Chervoneva, I.; Berger, A.C.; Lo, J.A.; Fisher, D.E.; et al. FOXD3 Regulates VISTA Expression in Melanoma. Cell Rep. 2020, 30, 510–524.e516. [Google Scholar] [CrossRef]
- Zong, L.; Mo, S.; Yu, S.; Zhou, Y.; Zhang, M.; Chen, J.; Xiang, Y. Correction to: Expression of the immune checkpoint VISTA in breast cancer. Cancer Immunol. Immunother. 2020, 69, 1447. [Google Scholar] [CrossRef] [PubMed]
- Zong, L.; Zhou, Y.; Zhang, M.; Chen, J.; Xiang, Y. VISTA expression is associated with a favorable prognosis in patients with high-grade serous ovarian cancer. Cancer Immunol. Immunother. 2020, 69, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Hanisch, U.K.; Kettenmann, H. Microglia: Active sensor and versatile effector cells in the normal and pathologic brain. Nat. Neurosci. 2007, 10, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Lorger, M.; Felding-Habermann, B. Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis. Am. J. Pathol. 2010, 176, 2958–2971. [Google Scholar] [CrossRef]
- Simon, A.; Yang, M.; Marrison, J.L.; James, A.D.; Hunt, M.J.; O’Toole, P.J.; Kaye, P.M.; Whittington, M.A.; Chawla, S.; Brackenbury, W.J. Metastatic breast cancer cells induce altered microglial morphology and electrical excitability in vivo. J. Neuroinflamm. 2020, 17, 87. [Google Scholar] [CrossRef]
- Andreou, K.E.; Soto, M.S.; Allen, D.; Economopoulos, V.; de Bernardi, A.; Larkin, J.R.; Sibson, N.R. Anti-inflammatory Microglia/Macrophages As a Potential Therapeutic Target in Brain Metastasis. Front. Oncol. 2017, 7, 251. [Google Scholar] [CrossRef]
- Boddaert, J.; Bielen, K.; Jongers, B.; Manocha, E.; Yperzeele, L.; Cras, P.; Pirici, D.; Kumar-Singh, S. CD8 signaling in microglia/macrophage M1 polarization in a rat model of cerebral ischemia. PLoS ONE 2018, 13, e0186937. [Google Scholar] [CrossRef]
- Coniglio, S.J.; Eugenin, E.; Dobrenis, K.; Stanley, E.R.; West, B.L.; Symons, M.H.; Segall, J.E. Microglial stimulation of glioblastoma invasion involves epidermal growth factor receptor (EGFR) and colony stimulating factor 1 receptor (CSF-1R) signaling. Mol. Med. 2012, 18, 519–527. [Google Scholar] [CrossRef]
- Wu, S.Y.; Watabe, K. The roles of microglia/macrophages in tumor progression of brain cancer and metastatic disease. Front. Biosci. 2017, 22, 1805–1829. [Google Scholar] [CrossRef]
- Pukrop, T.; Dehghani, F.; Chuang, H.N.; Lohaus, R.; Bayanga, K.; Heermann, S.; Regen, T.; Van Rossum, D.; Klemm, F.; Schulz, M.; et al. Microglia promote colonization of brain tissue by breast cancer cells in a Wnt-dependent way. Glia 2010, 58, 1477–1489. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Croft, P.K.-d.; Chittoory, H.; Nguyen, T.H.; Saunus, J.M.; Kim, W.G.; McCart Reed, A.E.; Lim, M.; De Luca, X.M.; Ferguson, K.; Niland, C.; et al. Characterization of Immune Cell Subsets of Tumor Infiltrating Lymphocytes in Brain Metastases. Biology 2021, 10, 425. https://doi.org/10.3390/biology10050425
Croft PK-d, Chittoory H, Nguyen TH, Saunus JM, Kim WG, McCart Reed AE, Lim M, De Luca XM, Ferguson K, Niland C, et al. Characterization of Immune Cell Subsets of Tumor Infiltrating Lymphocytes in Brain Metastases. Biology. 2021; 10(5):425. https://doi.org/10.3390/biology10050425
Chicago/Turabian StyleCroft, Priyakshi Kalita-de, Haarika Chittoory, Tam H. Nguyen, Jodi M. Saunus, Woo Gyeong Kim, Amy E. McCart Reed, Malcolm Lim, Xavier M. De Luca, Kaltin Ferguson, Colleen Niland, and et al. 2021. "Characterization of Immune Cell Subsets of Tumor Infiltrating Lymphocytes in Brain Metastases" Biology 10, no. 5: 425. https://doi.org/10.3390/biology10050425
APA StyleCroft, P. K.-d., Chittoory, H., Nguyen, T. H., Saunus, J. M., Kim, W. G., McCart Reed, A. E., Lim, M., De Luca, X. M., Ferguson, K., Niland, C., Mazzieri, R., Dolcetti, R., Simpson, P. T., & Lakhani, S. R. (2021). Characterization of Immune Cell Subsets of Tumor Infiltrating Lymphocytes in Brain Metastases. Biology, 10(5), 425. https://doi.org/10.3390/biology10050425







