Identification of an 11-Autophagy-Related-Gene Signature as Promising Prognostic Biomarker for Bladder Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Data Acquisition
2.2. Functional Analysis
2.3. Construction of an ARGs-Related Prognostic Model
2.4. Statistical Analysis
3. Result
3.1. Identification of Prognostic Autophagy-Related Genes
3.2. Identification of Involved Signaling Pathways and Functional Annotation
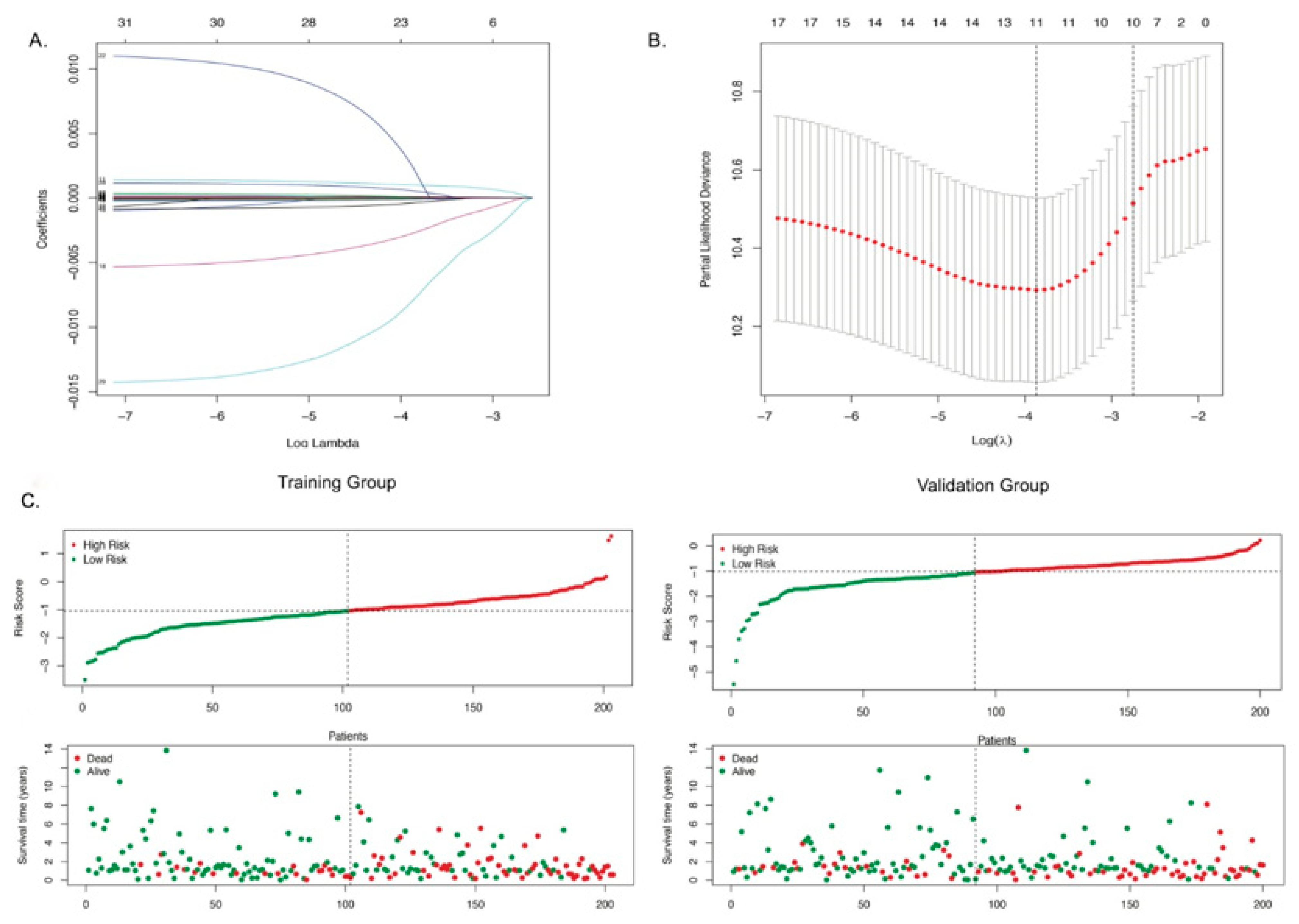
3.3. Identification of Autophagy-Related Gene Signatures
3.4. The 11-ARG Signature as an Independent Risk Factor for Bladder Cancer
3.5. Establishment of a Nomogram for Predicting Survival in Bladder Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statements
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.E.M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. 2018. Available online: https://gco.iarc.fr/today (accessed on 20 May 2020).
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110–3115. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Richters, A.; Aben, K.K.H.; Kiemeney, L.A.L.M. The global burden of urinary bladder cancer: An update. World J. Urol. 2020, 38, 1895–1904. [Google Scholar] [CrossRef]
- Cumberbatch, M.G.; Rota, M.; Catto, J.W.; La Vecchia, C. The Role of Tobacco Smoke in Bladder and Kidney Carcinogenesis: A Comparison of Exposures and Meta-analysis of Incidence and Mortality Risks. Eur. Urol. 2016, 70, 458–466. [Google Scholar] [CrossRef]
- Freedman, N.D. Association between Smoking and Risk of Bladder Cancer among Men and Women. JAMA 2011, 306, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Witjes, J.A.; Bruins, H.M.; Cathomas, R.; Compérat, E.M.; Cowan, N.C.; Gakis, G.; Hernández, V.; Espinós, E.L.; Lorch, A.; Neuzillet, Y.; et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol. 2020, 79, 82–104. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K. Bladder Cancer: New Insights into Its Molecular Pathology. Cancers 2018, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; McConkey, D.J. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017, 171, 540–556.e25. [Google Scholar] [CrossRef]
- Apel, A.; Herr, I.; Schwarz, H.; Rodemann, H.P.; Mayer, A. Blocked Autophagy Sensitizes Resistant Carcinoma Cells to Radiation Therapy. Cancer Res. 2008, 68, 1485–1494. [Google Scholar] [CrossRef]
- Zachari, M.; Gudmundsson, S.R.; Li, Z.; Manifava, M.; Cugliandolo, F.; Shah, R.; Smith, M.; Stronge, J.; Karanasios, E.; Piunti, C.; et al. Selective Autophagy of Mitochondria on a Ubiquitin-Endoplasmic-Reticulum Platform. Dev. Cell 2019, 50, 627–643.e5. [Google Scholar] [CrossRef]
- Dower, C.M.; Wills, C.A.; Frisch, S.M.; Wang, H.-G. Mechanisms and context underlying the role of autophagy in cancer metastasis. Autophagy 2018, 14, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- White, E.; DiPaola, R.S. The Double-Edged Sword of Autophagy Modulation in Cancer. Clin. Cancer Res. 2009, 15, 5308–5316. [Google Scholar] [CrossRef] [PubMed]
- Apel, A.; Zentgraf, H.; Büchler, M.W.; Herr, I. Autophagy-A double-edged sword in oncology. Int. J. Cancer 2009, 125, 991–995. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Karantza-Wadsworth, V.; White, E. Role of autophagy in cancer. Nat. Rev. Cancer 2007, 7, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Dickstein, R.J.; Nitti, G.; Dinney, C.P.; Davies, B.R.; Kamat, A.M.; McConkey, D.J. Autophagy limits the cytotoxic effects of the AKT inhibitor AZ7328 in human bladder cancer cells. Cancer Biol. Ther. 2012, 13, 1325–1338. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Huang, Y.; Chen, H.; Wang, Y.; Xia, L.; Chen, Y.; Liu, Y.; Qiu, F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. BioSyst. 2013, 9, 407–411. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Z.; Wang, Y.; Zhu, K.; Deng, W.; Li, Y.; Fu, B. Downregulation of HMGA1 Mediates Autophagy and Inhibits Migration and Invasion in Bladder Cancer via miRNA-221/TP53INP1/p-ERK Axis. Front. Oncol. 2020, 10, 589. [Google Scholar] [CrossRef]
- Liang, Y.; Zhu, J.; Huang, H.; Xiang, D.; Li, Y.; Zhang, D.; Li, J.; Wang, Y.; Jin, H.; Jiang, G.; et al. SESN2/sestrin 2 induction-mediated autophagy and inhibitory effect of isorhapontigenin (ISO) on human bladder cancers. Autophagy 2016, 12, 1229–1239. [Google Scholar] [CrossRef]
- Dyrskjøt, L.; Zieger, K.; Real, F.X.; Malats, N.; Carrato, A.; Hurst, C.; Kotwal, S.; Knowles, M.; Malmström, P.-U.; De La Torre, M.; et al. Gene Expression Signatures Predict Outcome in Non–Muscle-Invasive Bladder Carcinoma: A Multicenter Validation Study. Clin. Cancer Res. 2007, 13, 3545–3551. [Google Scholar] [CrossRef]
- von Rundstedt, F.C.; Rajapakshe, K.; Ma, J.; Arnold, J.M.; Gohlke, J.; Putluri, V.; Putluri, N. Integrative Pathway Analysis of Metabolic Signature in Bladder Cancer: A Linkage to The Cancer Genome Atlas Project and Prediction of Survival. J. Urol. 2016, 195, 1911–1919. [Google Scholar] [CrossRef] [PubMed]
- Abudurexiti, M.; Xie, H.; Jia, Z.; Zhu, Y.; Zhu, Y.; Shi, G.; Zhang, H.; Dai, B.; Wan, F.; Shen, Y.; et al. Development and External Validation of a Novel 12-Gene Signature for Prediction of Overall Survival in Muscle-Invasive Bladder Cancer. Front. Oncol. 2019, 9, 856. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Yuan, L.; Ma, B.; Wang, G.; Qiu, W.; Tian, Y. An EMT-related gene signature for the prognosis of human bladder cancer. J. Cell. Mol. Med. 2020, 24, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Matboli, M.; Awad, N.; Kotb, Y. Identification and validation of a novel autophagy gene expression signature for human bladder cancer patients. Tumor Biol. 2017, 39, 1010428317698360. [Google Scholar] [CrossRef]
- Wu, Y.; Peng, Y.; Guan, B.; He, A.; Yang, K.; He, S.; Gong, Y.; Li, X.; Zhou, L. P4HB: A novel diagnostic and prognostic biomarker for bladder carcinoma. Oncol. Lett. 2020, 21, 1. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, Y.; Zhang, J.; Cao, L.; He, M.; Liu, X.; Zhao, N.; Yin, A.; Huang, H.; Wang, L. TMEM74 promotes tumor cell survival by inducing autophagy via interactions with ATG16L1 and ATG9A. Cell Death Dis. 2017, 8, e3031. [Google Scholar] [CrossRef]
- Li, F.; Guo, H.; Yang, Y.; Feng, M.; Liu, B.; Ren, X.; Zhou, H. Autophagy modulation in bladder cancer development and treatment (Review). Oncol. Rep. 2019, 42, 1647–1655. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-F.; Hwang, T.I.S. Autophagy regulation in bladder cancer as the novel therapeutic strategy. Transl. Cancer Res. 2017, 6, S708–S719. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, N.; Shao, J.; Wang, T.; Wang, X. A novel gene signature combination improves the prediction of overall survival in urinary bladder cancer. J. Cancer 2019, 10, 5744–5753. [Google Scholar] [CrossRef] [PubMed]
- Dancik, G.M.; Theodorescu, D. Robust Prognostic Gene Expression Signatures in Bladder Cancer and Lung Adenocarcinoma Depend on Cell Cycle Related Genes. PLoS ONE 2014, 9, e85249. [Google Scholar] [CrossRef] [PubMed]
- Mo, Q.; Nikolos, F.; Chen, F.; Tramel, Z.; Lee, Y.-C.; Hayashi, K.; Xiao, J.; Shen, J.; Chan, K.S. Prognostic Power of a Tumor Differentiation Gene Signature for Bladder Urothelial Carcinomas. J. Natl. Cancer Inst. 2018, 110, 448–459. [Google Scholar] [CrossRef]
- Akin, D.; Wang, S.K.; Habibzadegah-Tari, P.; Law, B.; Ostrov, D.; Li, M.; Yin, X.-M.; Kim, J.-S.; Horenstein, N.; William, A.D., Jr. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy 2014, 10, 2021–2035. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luo, Q.; Yuan, L.; Miao, C.; Mu, X.; Xiao, W.; Li, J.; Sun, T.; Ma, E. JNK-dependent Atg4 upregulation mediates asperphenamate derivative BBP-induced autophagy in MCF-7 cells. Toxicol. Appl. Pharmacol. 2012, 263, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Bakula, D.; Müller, A.J.; Zuleger, T.; Takacs, Z.; Franz-Wachtel, M.; Thost, A.K.; Proikas-Cezanne, T. WIPI3 and WIPI4 β-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat. Commun. 2017, 8, 15637. [Google Scholar] [CrossRef] [PubMed]
- Crighton, D.; Wilkinson, S.; Ryan, K.M. DRAM Links Autophagy to p53 and Programmed Cell Death. Autophagy 2007, 3, 72–74. [Google Scholar] [CrossRef] [PubMed]
- Nagata, M.; Arakawa, S.; Yamaguchi, H.; Torii, S.; Endo, H.; Tsujioka, M.; Honda, S.; Nishida, Y.; Konishi, A.; Shimizu, S. Dram1 regulates DNA damage-induced alternative autophagy. Cell Stress 2018, 2, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liang, Q.; Chen, H.; Wu, P.; Feng, Z.; Ma, X.; Wu, H.; Zhou, G. DRAM1 regulates the migration and invasion of hepatoblastoma cells via autophagy‑EMT pathway. Oncol. Lett. 2018, 16, 2427–2433. [Google Scholar] [CrossRef]
- Gurusamy, N.; Lekli, I.; Gherghiceanu, M.; Popescu, L.M.; Das, D.K. BAG-1 induces autophagy for cardiac cell survival. Autophagy 2009, 5, 120–121. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cutress, R.I.; Townsend, P.A.; Brimmell, M.; Bateman, A.C.; Hague, A.; Packham, G. BAG-1 expression and function in human cancer. Br. J. Cancer 2002, 87, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.C.; Krajewski, S.; Krajewska, M.; Takayama, S.; Gumbs, A.A.; Carter, D.; Rebbeck, T.R.; Haffty, B.G.; Reed, J.C. BAG-1: A Novel Biomarker Predicting Long-Term Survival in Early-Stage Breast Cancer. J. Clin. Oncol. 2001, 19, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Reyland, M.E. Protein kinase C isoforms: Multi-functional regulators of cell life and death. Front. Biosci. 2009, 14, 2386–2399. [Google Scholar] [CrossRef] [PubMed]
- So, K.-Y.; Oh, S.-H. Cadmium-induced heme-oxygenase-1 expression plays dual roles in autophagy and apoptosis and is regulated by both PKC-δ and PKB/Akt activation in NRK52E kidney cells. Toxicology 2016, 370, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Shankar, S.; Srivastava, R.K. Rottlerin-induced autophagy leads to the apoptosis in breast cancer stem cells: Molecular mechanisms. Mol. Cancer 2013, 12, 171. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Walker, A.E.; Fikaris, A.J.; Kao, G.D.; Brown, E.J.; Kazanietz, M.G.; Meinkoth, J.L. Protein kinase C delta stimulates apoptosis by initiating G1 phase cell cycle progression and S phase arrest. J. Biol. Chem. 2005, 280, 32107–32114. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.; Zheng, Y.; Lyo, D.; Shen, Y.; Nakayama, K.; Nakayama, K.I.; Foster, D.A. Suppression of cell migration by protein kinase Cdelta. Oncogene 2005, 24, 3067–3072. [Google Scholar] [CrossRef][Green Version]
- Johnson, A.; Bridgham, J. Caspase-3 and -6 expression and enzyme activity in hen granulosa cells. Biol. Reprod. 2000, 62, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, X.; Wei, P.; Zhang, J.; Niu, Y.; Kang, N.; Zhang, Y.; Zhang, W.; Xing, N. Livin, Survivin and Caspase 3 as early recurrence markers in non-muscle-invasive bladder cancer. World J. Urol. 2014, 32, 1477–1484. [Google Scholar] [CrossRef]
- Burton, P.B.; Anderson, C.J.; Corbishly, C.M. Caspase 3 and p27 as Predictors of Invasive Bladder Cancer. N. Engl. J. Med. 2000, 343, 1418–1420. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhao, L.; Liu, L.; Gao, P.; Tian, W.; Wang, X.; Jin, H.; Xu, H.; Chen, Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 2010, 1, 468–477. [Google Scholar] [CrossRef]
- Betin, V.M.; Lane, J.D. Atg4D at the interface between autophagy and apoptosis. Autophagy 2009, 5, 1057–1059. [Google Scholar] [CrossRef] [PubMed]
- Tsapras, P.; Nezis, I.P. Caspase involvement in autophagy. Cell Death Differ. 2017, 24, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Zhaorigetu, S.; Wan, G.; Kaini, R.; Jiang, Z.; Hu, C.-A.A. ApoL1, a BH3-only lipid-binding protein, induces autophagic cell death. Autophagy 2008, 4, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Duchateau, P.N.; Pullinger, C.R.; Cho, M.H.; Eng, C.; Kane, J.P. Apolipoprotein L gene family: Tissue-specific expression, splicing, promoter regions; discovery of a new gene. J. Lipid Res. 2001, 42, 620–630. [Google Scholar] [CrossRef]
- Duchateau, P.N.; Pullinger, C.R.; Orellana, R.E.; Kunitake, S.T.; Naya-Vigne, J.; O’Connor, P.M.; Kane, J.P. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas Identification, cloning, characterization, and plasma distribution of apolipoprotein L. J. Biol. Chem. 1997, 272, 25576–25582. [Google Scholar] [CrossRef] [PubMed]
- Chidiac, M.; Fayyad-Kazan, M.; Daher, J.; Poelvoorde, P.; Bar, I.; Maenhaut, C.; Delrée, P.; Badran, B.; Vanhamme, L. ApolipoproteinL1 is expressed in papillary thyroid carcinomas. Pathol. Res. Pr. 2016, 212, 631–635. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.-A.A.; Klopfer, E.I.; Ray, P.E. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012, 586, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Genovese, G.; Friedman, D.J.; Ross, M.D.; Lecordier, L.; Uzureau, P.; Freedman, B.I.; Bowden, D.W.; Langefeld, C.D.; Oleksyk, T.K.; Knob, A.L.U.; et al. Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science 2010, 329, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Hao, K.; Zhao, S.; Cui, D.; Zhang, Y.; Jiang, C.; Jing, Y.; Xia, S.; Han, B. Androgen receptor antagonist bicalutamide induces autophagy and apoptosis via ULK2 upregulation in human bladder cancer cells. Int. J. Clin. Exp. Pathol. 2017, 10, 7603–7615. [Google Scholar] [PubMed]
- Wang, J.-R.; Liu, B.; Zhou, L.; Huang, Y.-X. MicroRNA-124-3p suppresses cell migration and invasion by targeting ITGA3 signaling in bladder cancer. Cancer Biomark. 2019, 24, 159–172. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, Y.; Liu, S.; Chen, Q.; Liu, Y. ITGA3 serves as a diagnostic and prognostic biomarker for pancreatic cancer. OncoTargets Ther. 2019, 12, 4141–4152. [Google Scholar] [CrossRef]
- Zhang, H.; Cui, X.; Cao, A.; Li, X.; Li, L. ITGA3 interacts with VASP to regulate stemness and epithelial-mesenchymal transition of breast cancer cells. Gene 2020, 734, 144396. [Google Scholar] [CrossRef]
- Sa, K.-D.; Zhang, X.; Li, X.-F.; Gu, Z.-P.; Yang, A.-G.; Zhang, R.; Li, J.-P.; Sun, J.-Y. A miR-124/ITGA3 axis contributes to colorectal cancer metastasis by regulating anoikis susceptibility. Biochem. Biophys. Res. Commun. 2018, 501, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.-J.; Tournier, C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy 2011, 7, 689–695. [Google Scholar] [CrossRef]
- Qiu, L.; Zhou, G.; Cao, S. Targeted inhibition of ULK1 enhances daunorubicin sensitivity in acute myeloid leukemia. Life Sci. 2020, 243, 117234. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhu, L.; Zheng, L.-P.; Cui, Q.; Zhu, H.-H.; Zhao, H.; Shen, Z.-J.; Dong, H.-Y.; Chen, S.-S.; Wu, W.-Z.; et al. Overexpression of ULK1 Represents a Potential Diagnostic Marker for Clear Cell Renal Carcinoma and the Antitumor Effects of SBI-0206965. EBioMedicine 2018, 34, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Hu, P.; Yang, Z.; Xue, C.; Gong, J.; Sun, S.; Shi, L.; Zhang, S.; Li, Z.; Yang, C.; et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol. Rep. 2017, 37, 3449–3458. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Xiang, S.; Cao, J.; Zhu, H.; Yang, B.; He, Q.; Ying, M. Kelch-like proteins: Physiological functions and relationships with diseases. Pharmacol. Res. 2019, 148, 104404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, S.; Wu, Y.; Zheng, Z.-C.; Wang, Y.; Zhao, Y. P4HB, a Novel Hypoxia Target Gene Related to Gastric Cancer Invasion and Metastasis. BioMed. Res. Int. 2019, 2019, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Li, H.; Zhang, L.; Ma, X.; Dang, Y.; Guo, J.; Liu, J.; Ge, L.; Nan, F.; Dong, H.; et al. Autophagy-related gene P4HB: A novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging 2020, 12, 1828–1842. [Google Scholar] [CrossRef]
- Zou, H.; Wen, C.; Peng, Z.; Shao, Y.-Y.; ChunJie, W.; Li, S.; Li, C.; Zhou, H.-H. P4HB and PDIA3 are associated with tumor progression and therapeutic outcome of diffuse gliomas. Oncol. Rep. 2017, 39, 501–510. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, D.; Frigyesi, A.; Gudjonsson, S.; Sjödahl, G.; Hallden, C.; Chebil, G.; Veerla, S.; Ryden, T.; Månsson, W.; Liedberg, F.; et al. Combined Gene Expression and Genomic Profiling Define Two Intrinsic Molecular Subtypes of Urothelial Carcinoma and Gene Signatures for Molecular Grading and Outcome. Cancer Res. 2010, 70, 3463–3472. [Google Scholar] [CrossRef] [PubMed]
- Lyu, L.; Xiang, W.; Zheng, F.; Huang, T.; Feng, Y.; Yuan, J.; Zhang, C. Significant Prognostic Value of the Autophagy-Related Gene P4HB in Bladder Urothelial Carcinoma. Front. Oncol. 2020, 10, 1613. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Wang, W.; Wang, H.; Wang, Y.; Wang, Y.; Li, G.; Ji, C.; Ren, X.; Song, N.; Qin, C. Identification of an independent autophagy-gene prognostic index for papillary renal cell carcinoma. Transl. Androl. Urol. 2020, 9, 1945–1956. [Google Scholar] [CrossRef] [PubMed]





| Variables | TCGA (N = 403) |
|---|---|
| Status | |
| Alive | 249 |
| Dead | 154 |
| Age | 68 ± 10.57 |
| Gender | |
| Female | 102 (26.2) |
| Male | 299 (73.8) |
| Race | |
| White | 332 (82.5) |
| Asian | 43 (10.6) |
| Black or African | 28 (6.9) |
| AJCC-T | |
| T0/Ta | 1 (0.2) |
| T1 | 3 (0.8) |
| T2 | 117 (28.9) |
| T3 | 193 (47.7) |
| T4 | 58 (14.3) |
| Unknown | 31 (8.1) |
| AJCC-N | |
| N0 | 235 (58.0) |
| N1 | 46 (11.4) |
| N2 | 75 (18.5) |
| N3 | 7 (1.7) |
| Unknown | 40 (10.4) |
| AJCC-M | |
| M0 | 195 (48.1) |
| M1 | 11 (2.7) |
| Mx | 197 (49.2) |
| Pathologic_stage | |
| I&II | 130 (32.1) |
| III&IV | 271 (67.4) |
| Unknown | 2 (0.5) |
| Tumor_grade | |
| G1/G2 | 20 (5.0) |
| G3/G4 | 380 (94.3) |
| Unknown | 3 (0.7) |
| Id | HR | HR.95 L | HR.95H | p-Value |
|---|---|---|---|---|
| APOL1 | 3.68 × 10−3 | 6.33 × 10−5 | 2.14 × 10−1 | 6.87 × 10−3 |
| ATG4B | 5.23 × 10−7 | 2.63 × 10−11 | 1.04 × 10−2 | 4.18 × 10−3 |
| ATG5 | 1.06 × 10−4 | 3.86 × 10−8 | 2.90 × 10−1 | 2.34 × 10−2 |
| BAX | 5.60 × 10−1 | 5.87 × 10−1 | 9.85 × 10−1 | 3.79 × 10−2 |
| BAG1 | 3.33 × 10−4 | 2.86 × 10−7 | 3.86 × 10−1 | 2.61 × 10−2 |
| BCL2L1 | 1.17 × 10−4 | 2.79 × 10−8 | 4.93 × 10−1 | 3.35 × 10−2 |
| BID | 3.72 × 10−4 | 3.14 × 10−7 | 4.42 × 10−1 | 2.88 × 10−2 |
| CAPN10 | 2.97 × 10−5 | 1.42 × 10−8 | 6.20 × 10−2 | 7.52 × 10−3 |
| CASP3 | 1.89 × 10−4 | 6.55 × 10−8 | 5.43 × 10−1 | 3.48 × 10−2 |
| CASP8 | 1.99 × 10−3 | 1.55 × 10−5 | 2.56 × 10−1 | 1.21 × 10−2 |
| DAPK1 | 1.27 × 10−2 | 6.27 × 10−4 | 2.59 × 10−1 | 4.52 × 10−3 |
| DRAM1 | 2.98 × 10−3 | 5.88 × 10−5 | 1.51 × 10−1 | 3.70 × 10−3 |
| IKBKB | 6.90 × 10−5 | 5.82 × 10−8 | 8.18 × 10−2 | 7.97 × 10−3 |
| IL24 | 2.10 × 10−1 | 5.38 × 10−2 | 8.21 × 10−1 | 2.48 × 10−2 |
| ITGA3 | 2.35 × 10−3 | 4.75 × 10−5 | 1.16 × 10−1 | 2.34 × 10−3 |
| ITGA6 | 1.16 × 10−2 | 1.51 × 10−4 | 8.88 × 10−1 | 4.40 × 10−2 |
| ITGB4 | 4.12 × 10−2 | 3.74 × 10−3 | 4.54 × 10−1 | 9.19 × 10−3 |
| IFNG | 9.21 × 10−1 | 8.58 × 10−1 | 9.88 × 10−1 | 2.22 × 10−2 |
| KLHL24 | 1.45 × 10−4 | 3.29 × 10−7 | 6.36 × 10−2 | 4.41 × 10−3 |
| MAP2K7 | 2.72 × 10−6 | 6.43 × 10−11 | 1.15 × 10−1 | 1.84 × 10−2 |
| MAPK8 | 1.91 × 10−4 | 1.44 × 10−7 | 2.53 × 10−1 | 1.95 × 10−2 |
| NFKB1 | 8.46 × 10−4 | 7.61 × 10−7 | 9.39 × 10−1 | 4.80 × 10−2 |
| PRKAB1 | 3.01 × 10−4 | 1.52 × 10−7 | 5.97 × 10−1 | 3.63 × 10−2 |
| PRKCD | 1.65 × 10−4 | 4.86 × 10−7 | 5.63 × 10−2 | 3.42 × 10−3 |
| P4HB | 1.12 × 10−1 | 1.01 × 10−1 | 1.23 × 10−1 | 2.80 × 10−2 |
| PTEN | 2.27 × 10−4 | 9.75 × 10−8 | 5.30 × 10−1 | 3.40 × 10−2 |
| PTK6 | 2.02 × 10−1 | 5.76 × 10−2 | 7.09 × 10−1 | 1.26 × 10−2 |
| TMEM74 | 1.56 × 10−1 | 1.10 × 10−1 | 2.21 × 10−1 | 1.32 × 10−2 |
| TP63 | 9.94 × 10−2 | 2.31 × 10−2 | 4.27 × 10−1 | 1.91 × 10−3 |
| ULK2 | 8.96 × 10−1 | 8.16 × 10−1 | 9.86 × 10−1 | 2.39 × 10−2 |
| VEGFA | 1.15 × 10−3 | 4.88 × 10−6 | 2.72 × 10−1 | 1.52 × 10−2 |
| WDR45 | 6.04 × 10−7 | 7.23 × 10−11 | 5.04 × 10−3 | 1.88 × 10−3 |
| Variables | Univariate Cox Regression | Multivariate Cox Regression | ||||||
|---|---|---|---|---|---|---|---|---|
| Training Group | Validation Group | Training Group | Validation Group | |||||
| HR | p-Value | HR | p-Value | HR | p-Value | HR | p-Value | |
| Age | 1.90599 | 0.00480 | 1.87244 | 0.00712 | 2.01716 | 0.00923 | 1.89282 | 0.00942 |
| Gender | 1.33404 | 0.28459 | 0.89624 | 0.68090 | 1.21995 | 0.49531 | 0.90684 | 0.72394 |
| Stage | 2.57129 | 0.00077 | 2.31364 | 0.00483 | 1.70227 | 0.08188 | 2.02544 | 0.02310 |
| TTN | 0.91655 | 0.69983 | 0.88862 | 0.61304 | 1.02461 | 0.92293 | 0.81568 | 0.43476 |
| TP53 | 0.96076 | 0.85924 | 0.75083 | 0.22132 | 1.17030 | 0.54285 | 0.61782 | 0.04981 |
| MUC16 | 0.91318 | 0.73605 | 0.99522 | 0.98484 | 1.27489 | 0.43391 | 1.48957 | 0.14201 |
| Risk Score | 4.32931 | 0.00000 | 2.24823 | 0.00035 | 4.02659 | 0.00000 | 1.67008 | 0.02760 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Li, A.H.; Liu, S.; Sun, H. Identification of an 11-Autophagy-Related-Gene Signature as Promising Prognostic Biomarker for Bladder Cancer Patients. Biology 2021, 10, 375. https://doi.org/10.3390/biology10050375
Zhou C, Li AH, Liu S, Sun H. Identification of an 11-Autophagy-Related-Gene Signature as Promising Prognostic Biomarker for Bladder Cancer Patients. Biology. 2021; 10(5):375. https://doi.org/10.3390/biology10050375
Chicago/Turabian StyleZhou, Chaoting, Alex Heng Li, Shan Liu, and Hong Sun. 2021. "Identification of an 11-Autophagy-Related-Gene Signature as Promising Prognostic Biomarker for Bladder Cancer Patients" Biology 10, no. 5: 375. https://doi.org/10.3390/biology10050375
APA StyleZhou, C., Li, A. H., Liu, S., & Sun, H. (2021). Identification of an 11-Autophagy-Related-Gene Signature as Promising Prognostic Biomarker for Bladder Cancer Patients. Biology, 10(5), 375. https://doi.org/10.3390/biology10050375






