Genetic Diversity in Diospyros Germplasm in the Western Caucasus Based on SSR and ISSR Polymorphism
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. Genetic Analysis
2.3. Statistical Analysis
3. Results
3.1. Efficiency and Resolving Power of SSR and ISSR Markers
3.2. Genetic Diversity and Population Structure of Diospyros Germplasm
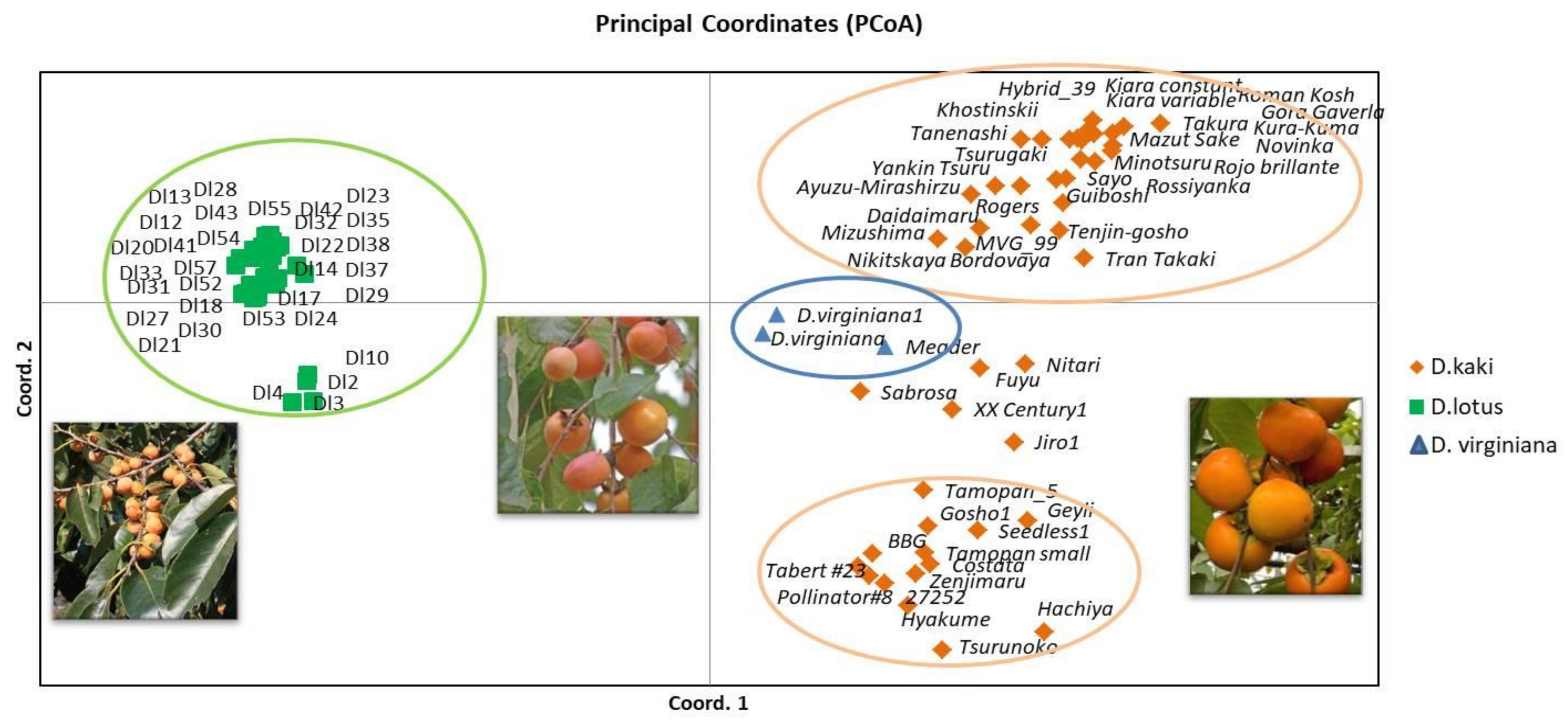
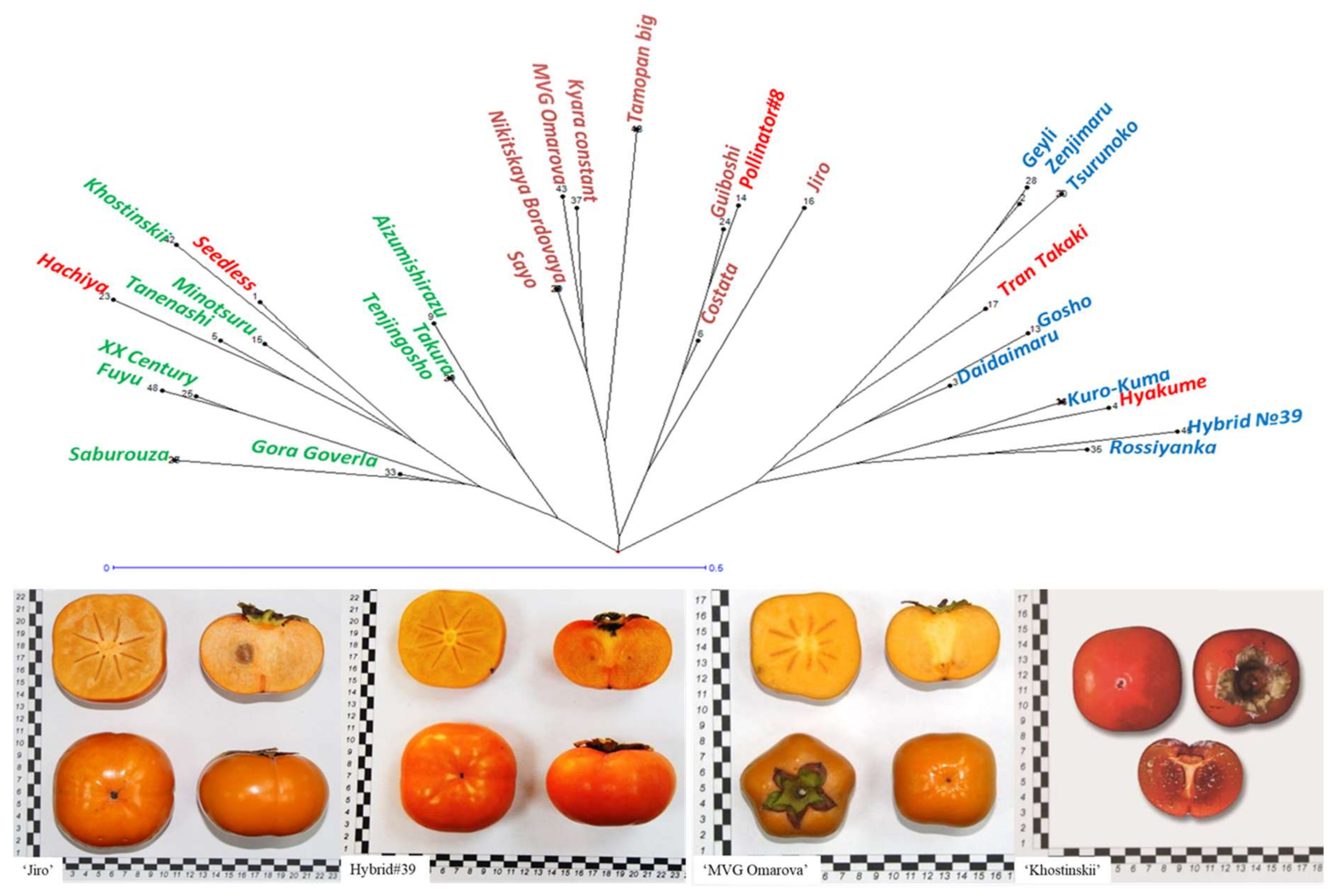
3.3. Phylogenetic Relationships and Connection with Phenotype in Diospyros Collection
4. Discussion
4.1. Efficiency and Resolving Power of SSR and ISSR Markers
4.2. Genetic Diversity and Population Structure of Diospyros Germplasm
4.3. Phylogenetic Relationships and Connection with Phenotype in Diospyros Collection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yesiloglu, T.; Çimen, B.; Incesu, M.; Yilmaz, B. Genetic Diversity and Breeding of Persimmon. In Breeding and Health Benefits of Fruit and Nut Crops; Jaya, R., Nageswara-Rao, M., Eds.; IntechOpen: London, UK, 2018. [Google Scholar]
- Intrigliolo, D.S.; Visconti, F.; Bonet, L.; Parra, M.; Besada, C.; Abrisqueta, I.; Rubio, J.S.; De Paz, J.M. Persimmon (Diospyros kaki) Trees Responses to Restrictions in Water Amount and Quality. In Water Scarcity and Sustainable Agriculture in Semiarid Environment, 1st ed.; Garcia Tejero, I.F., Duran Zuazo, V.H., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 149–177. [Google Scholar]
- Robert, H.G.; Faik, A.A.; Millson, M.; Huang, H.S.; Chuang, L.T.; Sanz, C.; Golding, J.B. Changes in sugars, acids and fatty acids in naturally parthenocarpic date plum persimmon (Diospyros lotus L.) fruit during maturation and ripening. Eur. Food Res. Technol. 2005, 221, 113–118. [Google Scholar] [CrossRef]
- Yang, Y.; Ruan, X.; Wang, R. Indigenous Persimmon Germplasm Resources in China. Acta Hortic. 2013, 89–96. [Google Scholar] [CrossRef]
- Yildirim, N.; Ercisli, S.; Agar, G.; Orhan, E.; Hizarci, Y. Genetic variation among date plum (Diospyros lotus) genotypes in Turkey. Genet. Mol. Res. 2010, 9, 981–986. [Google Scholar] [CrossRef]
- Guan, C.; Zhang, P.; Hu, C.; Chachar, S.; Riaz, A.; Wang, R.; Yang, Y. Genetic diversity, germplasm identification and population structure of Diospyros kaki Thunb. from different geographic regions in China using SSR markers. Sci. Hortic. 2019, 251, 233–240. [Google Scholar] [CrossRef]
- Omarov, M.D. Yield of different cultivars of oriental oersimmon in the humid subtropics of Russia. Subtrop. Orna Mental Hortic. 2018, 65, 137–141. [Google Scholar] [CrossRef]
- Omarov, M.D.; Zagirov, N.G.; Omarova, Z.M.; Avidzba, M.A. Atlas of Cultivars and Hybrids of Oriental Persimmon; Ryndin, A.V., Ed.; Russian Research Institute of Floriculture and Subtropical Crops: Sochi, Russia, 2014; p. 93. [Google Scholar]
- Li, W.; Liu, Y.; Yang, Y.; Xie, X.; Lu, Y.; Yang, Z.; Jin, X.; Dong, W.; Suo, Z. Interspecific chloroplast genome sequence diversity and genomic resources in Diospyros. BMC Plant Biol. 2018, 18, 210. [Google Scholar] [CrossRef]
- Reim, S.; Lochschmidt, F.; Proft, A.; Höfer, M. Genetic integrity is still maintained in natural populations of the indigenous wild apple species Malus sylvestris (Mill.) in Saxony as demonstrated with nuclear SSR and chloroplast DNA markers. Ecol. Evol. 2020, 10, 11798–11809. [Google Scholar] [CrossRef]
- Raddová, J.; Ptáčková, H.; Čechová, J.; Ondrášek, I. Genetic analysis of the genus Diospyros ssp. using RAPD and i-PBS methods. Acta univ. Agric. Silvic. Mendel. Brun. 2012, 60, 205–216. [Google Scholar] [CrossRef]
- Liang, Y.; Han, W.; Sun, P.; Liang, J.; Wuyun, T.; Li, F.; Fu, J. Genetic diversity among germplasms of Diospyros kaki based on SSR markers. Sci. Hortic. 2015, 186, 180–189. [Google Scholar] [CrossRef]
- Guo, D.-L.; Luo, Z.R. Genetic relationships of the Japanese persimmon Diospyros kaki (Ebenaceae) and related species revealed by SSR analysis. Genet. Mol. Res. 2011, 10, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Pinar, H.; Yildiz, E.; Kaplankiran, M.; Toplu, C.; Unlu, M.; Serce, S.; Ercisli, S. Molecular characterization of some selected persimmon genotypes and cultivars by srap and ssr markers. GenetIKA 2017, 49, 693–704. [Google Scholar] [CrossRef]
- Soriano, J.M.; Pecchioli, S.; Romero, C.; Vilanova, S.; Llacer, G.; Giordani, E.; Badenes, M.L. Development of microsatellite markers in polyploid persimmon (Diospyros kaki L.) from an enriched genomic library. Mol. Ecol. Notes 2006, 6, 368–370. [Google Scholar] [CrossRef]
- Naval, M.; Zuriaga, E.; Pecchioli, S.; Llácer, G.; Giordani, E.; Badenes, M.L. Analysis of genetic diversity among persimmon cultivars using microsatellite markers. Tree Genet. Genomes 2010, 6, 677–687. [Google Scholar] [CrossRef][Green Version]
- Yonemori, K.; Honsho, C.; Kanzaki, S.; Ino, H.; Ikegami, A.; Kitajima, A.; Sugiura, A.; Parfitt, D.E. Sequence analyses of the ITS regions and the matK gene for determining phylogenetic relationships of Diospyros kaki (persimmon) with other wild Diospyros (Ebenaceae) species. Tree Genet. Genomes 2007, 4, 149–158. [Google Scholar] [CrossRef]
- Jing, Z.; Ruan, X.; Wang, R.; Yang, Y. Genetic diversity and relationships between and within persimmon (Diospyros L.) wild species and cultivated varieties by SRAP markers. Plant Syst. Evol. 2013, 299, 1485–1492. [Google Scholar] [CrossRef]
- Guan, C.; Chachar, S.; Zhang, P.; Hu, C.; Wang, R.; Yang, Y. Inter- and Intra-specific Genetic Diversity in Diospyros Using SCoT and IRAP Markers. Hortic. Plant J. 2020, 6, 71–80. [Google Scholar] [CrossRef]
- Deng, L.; Liang, Q.; He, X.; Luo, C.; Chen, H.; Qin, Z. Investigation and Analysis of Genetic Diversity of Diospyros Germplasms Using SCoT Molecular Markers in Guangxi. PLoS ONE 2015, 10, e0136510. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, T.; Jing, Z. Genetic diversity and taxonomic studies of date plum (Diospyros lotus L.) using morphological traits and SCoT markers. Biochem. Syst. Ecol. 2015, 61, 253–259. [Google Scholar] [CrossRef]
- Fu, J.; Liu, H.; Hu, J.; Liang, Y.; Liang, J.; Wuyun, T.; Tan, X. Five Complete Chloroplast Genome Sequences from Diospyros: Genome Organization and Comparative Analysis. PLoS ONE 2016, 11, e0159566. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Wambulwa, M.C.; Meegahakumbura, M.K.; Chalo, R.; Kamunya, S.; Muchugi, A.; Xu, J.C.; Liu, J.; Li, D.Z.; Gao, L.M. Nuclear microsatellites reveal the genetic architecture and breeding history of tea germplasm of East Africa. Tree Genet. Genomes 2016, 12, 1–10. [Google Scholar] [CrossRef]
- Barkley, N.A.; Krueger, R.R.; Federici, C.T.; Roose, M.L. What phylogeny and gene genealogy analyses reveal about homoplasy in citrus microsatellite alleles. Plant Syst. Evol. 2009, 282, 71–86. [Google Scholar] [CrossRef]
- Šarhanová, P.; Pfanzelt, S.; Brandt, R.; Himmelbach, A.; Blattner, F.R. SSR-seq: Genotyping of microsatellites using next-generation sequencing reveals higher level of polymorphism as compared to traditional fragment size scoring. Ecol. Evol. 2018, 8, 10817–10833. [Google Scholar] [CrossRef]
- Reddy, P.M.; Sarla, N.; Siddiq, E. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Azhar, M.; Muhammad, H.A.; Siti, N.I.; Parween, K.S.A.S. Optimization of ISSR Markers for Molecular DNA Fingerprinting in Aquilaria sp. In Nuclear Technical Convention; IAEA: Bangi, Malaysia, 2013. [Google Scholar]
- Tsumura, Y.; Ohba, K.; Strauss, S.H. Diversity and inheritance of inter-simple sequence repeat polymorphisms in Doug-las-fir (Pseudotsuga menziesii) and sugi (Cryptomeria japonica). Theor. Appl. Genet 1996, 92, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Luz, G.C.; Strioto, D.K.; Mangolin, C.A.; Machado, M.F.P.S. ISSR markers to assess genetic diversity of cultivated populations from artificial selection of Stevia rebaudiana (Bert.) Bertoni. Breed. Sci. 2020, 70, 508–514. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1991, 12, 13–15. [Google Scholar]
- Mondal, T.K. Assessment of genetic diversity of tea (Camellia sinensis (L.) O. Kuntze) by inter-simple sequence repeat polymerase chain reaction. Euphytica 2002, 128, 307–315. [Google Scholar] [CrossRef]
- Roy, S.C.; Chakraborty, B.N. Genetic diversity and relationsips among tea (Camellia sinensis) cultivars revealed by RAPD and ISSR based fingerprinting. Indian J. Biotechnol. 2009, 8, 370–376. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Earl, D.A.; Vonholdt, B.M. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- DARWIN6.0 Software. Available online: https://darwin.cirad.fr/ (accessed on 10 January 2021).
- Zhuang, D.H.; Kitajima, A.; Ishida, M.; Sobajima, Y. Chromosome Numbers of Diospyros kaki Cultivars. J. Jpn. Soc. Hortic. Sci. 1990, 59, 289–297. [Google Scholar] [CrossRef]
- Ma, K.B.; Yang, S.-J.; Jo, Y.-S.; Kang, S.S.; Nam, M. Development of Kompetitive Allele Specific PCR markers for identification of persimmon varieties using genotyping-by-sequencing. Electron. J. Biotechnol. 2021, 49, 72–81. [Google Scholar] [CrossRef]
- Parfitt, D.E.; Yonemori, K.; Honsho, C.; Nozaka, M.; Kanzaki, S.; Sato, A.; Yamada, M. Relationships among Asian persimmon cultivars, astringent and non-astringent types. Tree Genet. Genomes 2015, 11, 1–9. [Google Scholar] [CrossRef]
- Omarov, M.D.; Omarova, Z.M. Cultivars of oriental persimmon (Diospyros kaki L.) and their biological characteristics. Subtrop. Ornam. Hortic. 2016, 57, 69–73. [Google Scholar]

| Species | SSR | N | Na | Ne | I | Ho |
|---|---|---|---|---|---|---|
| D. kaki | ssrdk14 | 32 | 11.000 | 5.611 | 1.977 | 0.438 |
| ssrdk26 | 33 | 16.000 | 8.475 | 2.433 | 0.606 | |
| ssrdk30 | 30 | 7.000 | 3.666 | 1.564 | 0.700 | |
| ssrdk01 | 36 | 4.000 | 2.883 | 1.142 | 0.417 | |
| ssrdk03 | 33 | 6.000 | 2.339 | 1.096 | 0.667 | |
| ssrdk06 | 33 | 6.000 | 2.276 | 1.138 | 0.333 | |
| ssrdk09 | 31 | 10.000 | 4.319 | 1.721 | 0.871 | |
| ssrdk10 | 36 | 11.000 | 6.968 | 2.110 | 0.694 | |
| MEAN ± Standard error | 33.0 ± 0.8 | 8.9 ± 1.4 | 4.6 ± 0.8 | 1.6 ± 0.2 | 0.6 ± 0.1 | |
| D. lotus | ssrdk14 | 31 | 5.000 | 2.427 | 1.041 | 0.839 |
| ssrdk26 | 30 | 3.000 | 1.753 | 0.675 | 0.600 | |
| ssrdk30 | 28 | 3.000 | 1.332 | 0.464 | 0.071 | |
| ssrdk01 | 32 | 1.000 | 1.000 | 0.000 | 0.000 | |
| ssrdk03 | 32 | 1.000 | 1.000 | 0.000 | 0.000 | |
| ssrdk06 | 32 | 1.000 | 1.000 | 0.000 | 0.000 | |
| ssrdk09 | 32 | 1.000 | 1.000 | 0.000 | 0.000 | |
| ssrdk10 | 32 | 1.000 | 1.000 | 0.000 | 0.000 | |
| MEAN ± Standard error | 31.1 ± 0.5 | 2.0 ± 0.5 | 1.3 ± 0.2 | 0.3 ± 0.1 | 0.2 ± 0.1 |
| Species | Marker | No bands | Av. Band Freq. | N | Na | Ne | H |
|---|---|---|---|---|---|---|---|
| D. kaki | ISSR815 | 16 | 0.390 | 56.000 | 1.750 | 1.532 | 0.307 |
| ISSR880 | 16 | 0.259 | 56.000 | 2.000 | 1.545 | 0.323 | |
| ISSR13 | 8 | 0.500 | 56.000 | 2.000 | 1.600 | 0.360 | |
| ISSR814.1 | 17 | 0.370 | 56.000 | 2.000 | 1.705 | 0.402 | |
| ISSR15 | 15 | 0.368 | 56.000 | 1.733 | 1.241 | 0.169 | |
| MEAN ± Standard error | 14.6 ± 3.0 | 0.1 ± 0.0 | 56.0 ± 0.0 | 1.9 ± 0.1 | 1.5 ± 0.0 | 0.3 ± 0.0 | |
| D. lotus | ISSR815 | 16 | 0.408 | 32.000 | 0.938 | 1.104 | 0.073 |
| ISSR880 | 16 | 0.502 | 32.000 | 1.313 | 1.130 | 0.094 | |
| ISSR13 | 8 | 0.383 | 32.000 | 1.625 | 1.213 | 0.160 | |
| ISSR814.1 | 17 | 0.309 | 32.000 | 0.824 | 1.163 | 0.096 | |
| ISSR15 | 15 | 0.867 | 32.000 | 0.867 | 1.000 | 0.000 | |
| MEAN ± Standard error | 14.4. ± 3.0 | 0.5 ± 0.1 | 32.0 ± 0.0 | 1.1 ± 0.1 | 1.1 ± 0.0 | 0.1 ± 0.0 |
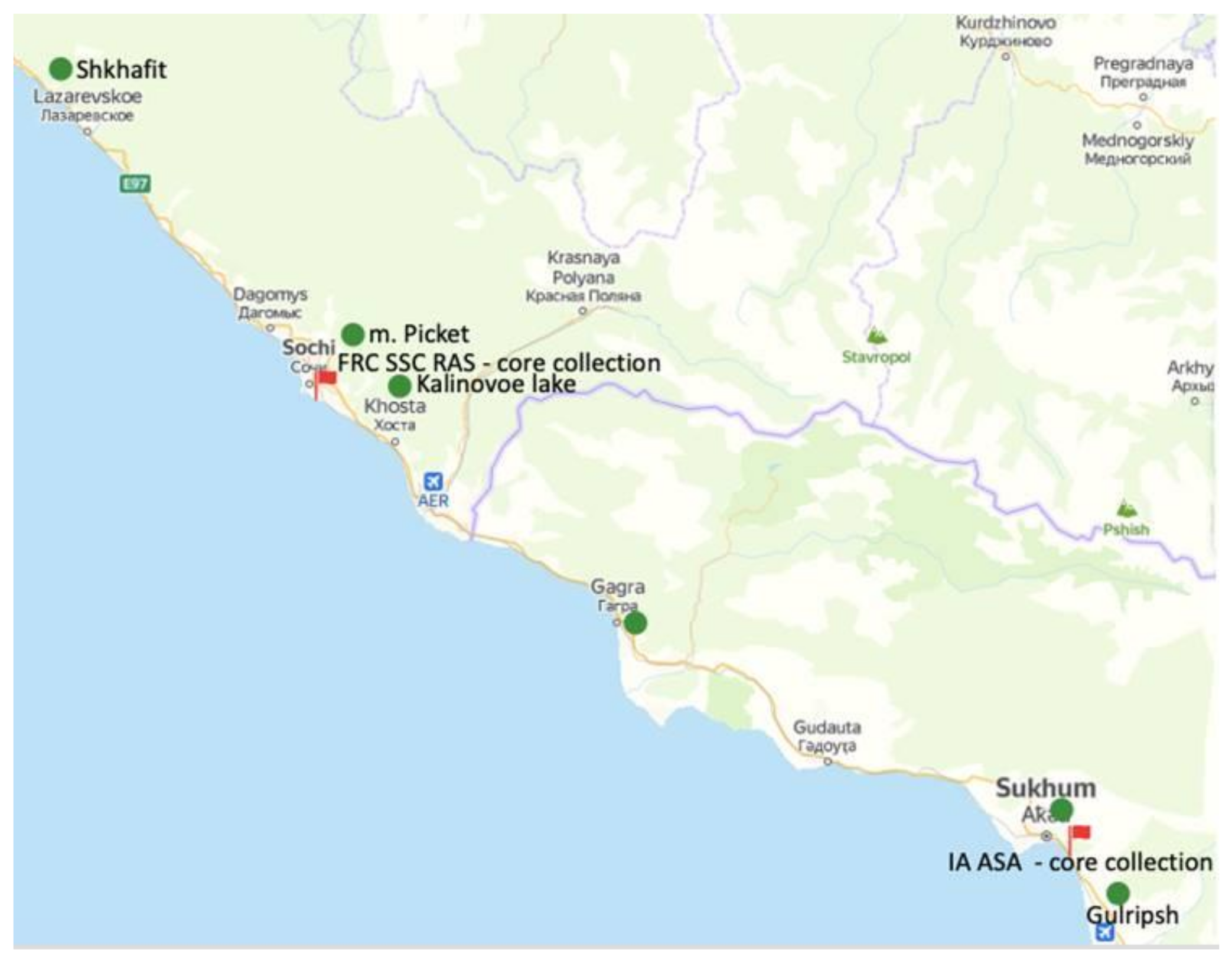
| Group | Population Location | Number of Accessions in the Population | Number of Identical Accessions Based on SSR Data | Number of Identical Accessions Based on ISSR Data |
|---|---|---|---|---|
| 1 | Shkhafit | 6 | 4 | 6 |
| 2 | Piket | 4 | 3 | 3 |
| 3 | Sochi center | 12 | 6 | 5 |
| 4 | Kalinovoe lake | 1 | 0 | 0 |
| 5 | Gagra | 19 | 12 | 7 |
| 6 | Sukhum | 10 | 4 | 8 |
| 7 | Gulripsh | 5 | 8 | 4 |
| TOTAL | 57 | 37 | 33 |
| SSR | ISSR | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cluster | N | Na | Ne | Ho | % P | Na | Ne | h | % P |
| 1 (D. lotus) | 32 | 2.00 | 1.31 | 0.20 | 37.50 | 1.06 | 1.11 | 0.08 | 33.33 |
| 2 (D. kaki) | 26 | 7.13 | 4.13 | 0.62 | 100.00 | 1.81 | 1.44 | 0.27 | 90.28 |
| 3 (D. kaki and hybrids) | 23 | 5.13 | 3.44 | 0.52 | 100.00 | 1.71 | 1.51 | 0.29 | 84.72 |
| mean | 4.75 | 2.96 | 0.44 | 79.17 | 1.52 | 1.35 | 0.21 | 69.44 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarina, L.S.; Malyarovskaya, V.I.; Reim, S.; Koninskaya, N.G.; Matskiv, A.O.; Tsaturyan, G.A.; Rakhmangulov, R.S.; Shkhalakhova, R.M.; Shurkina, E.S.; Kulyan, R.V.; et al. Genetic Diversity in Diospyros Germplasm in the Western Caucasus Based on SSR and ISSR Polymorphism. Biology 2021, 10, 341. https://doi.org/10.3390/biology10040341
Samarina LS, Malyarovskaya VI, Reim S, Koninskaya NG, Matskiv AO, Tsaturyan GA, Rakhmangulov RS, Shkhalakhova RM, Shurkina ES, Kulyan RV, et al. Genetic Diversity in Diospyros Germplasm in the Western Caucasus Based on SSR and ISSR Polymorphism. Biology. 2021; 10(4):341. https://doi.org/10.3390/biology10040341
Chicago/Turabian StyleSamarina, Lidia S., Valentina I. Malyarovskaya, Stefanie Reim, Natalia G. Koninskaya, Alexandra O. Matskiv, Gregory A. Tsaturyan, Ruslan S. Rakhmangulov, Ruset M. Shkhalakhova, Ekaterina S. Shurkina, Raisa V. Kulyan, and et al. 2021. "Genetic Diversity in Diospyros Germplasm in the Western Caucasus Based on SSR and ISSR Polymorphism" Biology 10, no. 4: 341. https://doi.org/10.3390/biology10040341
APA StyleSamarina, L. S., Malyarovskaya, V. I., Reim, S., Koninskaya, N. G., Matskiv, A. O., Tsaturyan, G. A., Rakhmangulov, R. S., Shkhalakhova, R. M., Shurkina, E. S., Kulyan, R. V., Omarova, Z. M., Omarov, M. D., & Ryndin, A. V. (2021). Genetic Diversity in Diospyros Germplasm in the Western Caucasus Based on SSR and ISSR Polymorphism. Biology, 10(4), 341. https://doi.org/10.3390/biology10040341





