The Oxindole Derivatives, New Promising GSK-3β Inhibitors as One of the Potential Treatments for Alzheimer’s Disease—A Molecular Dynamics Approach
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bertrand, J.A.; Thieffine, S.; Vulpetti, A.; Cristiani, C.; Valsasina, B.; Knapp, S.; Kalisz, H.M.; Flocco, M. Structural characterization of the GSK-3β active site using selective and non-selective ATP-mimetic inhibitors. J. Mol. Biol. 2003, 333, 393–407. [Google Scholar] [CrossRef]
- Dajani, R.; Fraser, E.; Roe, S.M.; Young, N.; Good, V.; Dale, T.C.; Pearl, L.H. Crystal structure of glycogen synthase kinase 3 beta: Structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 2001, 105, 721–732. [Google Scholar] [CrossRef]
- Ter Haar, E.; Coll, J.T.; Austen, D.A.; Hsiao, H.M.; Swenson, L.; Jain, J. Structure of GSK3β reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 2001, 8, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Lovestone, S.; Killick, R.; Di Forti, M.; Murray, R. Schizophrenia as a GSK-3 dysregulation disorder. Trends Neurosci. 2007, 30, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Avila, J.; Hernández, F. GSK-3 inhibitors for Alzheimer’s disease. Expert Rev. Neurother. 2007, 7, 1527–1533. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.; Killick, R.; Lovestone, S. The GSK3 hypothesis of Alzheimer’s disease. J. Neurochem. 2008, 104, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Rowe, M.K.; Wiest, C.; Chuang, D.M. GSK-3 is a viable potential target for therapeutic intervention in bipolar disorder. Neurosci. Biobehav. Rev. 2007, 31, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Sugden, P.H.; Fuller, S.J.; Weiss, S.C.; Clerk, A. Glycogen synthase kinase 3 (GSK3) in the heart: A point of integration in hypertrophic signalling and a therapeutic target? A critical analysis. Br. J. Pharmacol. 2008, 153, S137–S153. [Google Scholar] [CrossRef] [PubMed]
- Kerkelä, R.; Woulfe, K.; Force, T. Glycogen Synthase Kinase-3β-Actively Inhibiting Hypertrophy. Trends Cardiovasc. Med. 2007, 17, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Obligado, S.; Ibraghimov-Beskrovnaya, O.; Zuk, A.; Meijer, L.; Nelson, P. CDK/GSK-3 inhibitors as therapeutic agents for parenchymal renal diseases. Kidney Int. 2008, 73, 684–690. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, K.; Liu, Z.; Patel, S.; Doble, B.W.; Li, L.; Cras-Méneur, C.; Martinez, S.C.; Welling, C.M.; White, M.F.; Bernal-Mizrachi, E.; et al. Genetic deficiency of glycogen synthase kinase-3β corrects diabetes in mouse models of insulin resistance. PLoS Biol. 2008, 6, 0307–0318. [Google Scholar] [CrossRef]
- Martinez, A.; Castro, A.; Dorronsoro, I.; Alonso, M. Glycogen synthase kinase 3 (GSK-3) inhibitors as new promising drugs for diabetes, neurodegeneration, cancer, and inflammation. Med. Res. Rev. 2002, 22, 373–384. [Google Scholar] [CrossRef]
- Ougolkov, A.V.; Billadeau, D.D. Targeting GSK-3: A promising approach for cancer therapy? Futur. Oncol. 2006, 2, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Hao, Y.; Liu, B.; Qian, L. Indirubin and Meisoindigo in the Treatment of Chronic Myelogenous Leukemia in China. Leuk. Lymphoma 2002, 43, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Meijer, L.; Hoessel, R.; Leclerc, S.; Endicott, J.A.; Nobel, M.E.M.; Lawrie, A.; Tunnah, P.; Leost, M.; Damiens, E.; Marie, D.; et al. Indirubin, the active constituent of a Chinese antileukaemia medicine, inhibits cyclin-dependent kinases. Nat. Cell Biol. 1999, 1, 60–67. [Google Scholar]
- Vougogiannopoulou, K.; Ferandin, Y.; Bettayeb, K.; Myrianthopoulos, V.; Lozach, O.; Fan, Y.; Johnson, C.H.; Magiatis, P.; Skaltsounis, A.-L.; Mikros, E.; et al. Soluble 3′,6-substituted indirubins with enhanced selectivity toward glycogen synthase kinase -3 alter circadian period. J. Med. Chem. 2008, 51, 6421–6431. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Magnaudeix, A.; Wilson, C.M.; Yardin, C.; Terro, F. The new indirubin derivative inhibitors of glycogen synthase kinase-3, 6-BIDECO and 6-BIMYEO, prevent tau phosphorylation and apoptosis induced by the inhibition of protein phosphatase-2A by okadaic acid in cultured neurons. J. Neurosci. Res. 2011, 89, 1802–1811. [Google Scholar] [CrossRef] [PubMed]
- Bramson, H.N.; Holmes, W.D.; Hunter, R.N.; Lackey, K.E.; Lovejoy, B.; Luzzio, M.J.; Montana, V.; Rocque, W.J.; Rusnak, D.; Shewchuk, L.; et al. Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): Design, synthesis, enzymatic activities, and X-ray crystallographic analysis. J. Med. Chem. 2001, 44, 4339–4358. [Google Scholar] [CrossRef]
- Dermatakis, A.; Luk, K.C.; DePinto, W. Synthesis of potent oxindole CDK2 inhibitors. Bioorg. Med. Chem. 2003, 11, 1873–1881. [Google Scholar] [CrossRef]
- Luk, K.-C.; Simcox, M.E.; Schutt, A.; Rowan, K.; Thompson, T.; Chen, Y.; Kammlott, U.; DePinto, W.; Dunten, P.; Dermatakis, A. A new series of potent oxindole inhibitors of CDK2. Bioorg. Med. Chem. Lett. 2004, 14, 913–917. [Google Scholar] [CrossRef]
- Czeleń, P. Investigation of the Inhibition Potential of New Oxindole Derivatives and Assessment of Their Usefulness for Targeted Therapy. Symmetry 2019, 11, 974. [Google Scholar] [CrossRef]
- TURBOMOLE 7.0. Available online: http://www.turbomole.com/ (accessed on 20 November 2019).
- Potemkin, V.; Grishina, M. Principles for 3D/4D QSAR classification of drugs. Drug Discov. Today 2008, 13, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Potemkin, V.A.; Grishina, M.A. A new paradigm for pattern recognition of drugs. J. Comput. Aided. Mol. Des. 2008, 22, 489–505. [Google Scholar] [CrossRef] [PubMed]
- Chemosophia. Available online: http://www.chemosophia.com/ (accessed on 20 November 2019).
- Eckert, F.; Klamt, A. Fast solvent screening via quantum chemistry: COSMO-RS approach. AIChE J. 2002, 48, 369–385. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Bartashevich, E.V.; Potemkin, V.A.; Grishina, M.A.; Belik, A.V. A Method for Multiconformational Modeling of the Three-Dimensional Shape of a Molecule. J. Struct. Chem. 2002, 43, 1033–1039. [Google Scholar] [CrossRef]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Adelman, S.A. Generalized Langevin equation approach for atom/solid-surface scattering: General formulation for classical scattering off harmonic solids. J. Chem. Phys. 1976, 64, 2375. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. Amber 14 Reference Manual; University of California: Oakland, CA, USA, 2014; Available online: http://ambermd.org/doc12/Amber14.pdf (accessed on 14 April 2021).
- Czeleń, P. Inhibition mechanism of CDK-2 and GSK-3β by a sulfamoylphenyl derivative of indoline—a molecular dynamics study. J. Mol. Model. 2017, 23, 230. [Google Scholar] [CrossRef] [PubMed]
- Czeleń, P.; Szefler, B. Molecular dynamics study of the inhibitory effects of ChEMBL474807 on the enzymes GSK-3β and CDK-2. J. Mol. Model. 2015, 21, 74. [Google Scholar] [CrossRef]
- Czeleń, P. Molecular dynamics study on inhibition mechanism of CDK-2 and GSK-3β by CHEMBL272026 molecule. Struct. Chem. 2016, 27, 1807–1818. [Google Scholar] [CrossRef]
- Lozinskaya, N.A.; Babkov, D.A.; Zaryanova, E.V.; Bezsonova, E.N.; Efremov, A.M.; Tsymlyakov, M.D.; Anikina, L.V.; Zakharyascheva, O.Y.; Borisov, A.V.; Perfilova, V.N.; et al. Synthesis and biological evaluation of 3-substituted 2-oxindole derivatives as new glycogen synthase kinase 3β inhibitors. Bioorg. Med. Chem. 2019, 27, 1804–1817. [Google Scholar] [CrossRef] [PubMed]
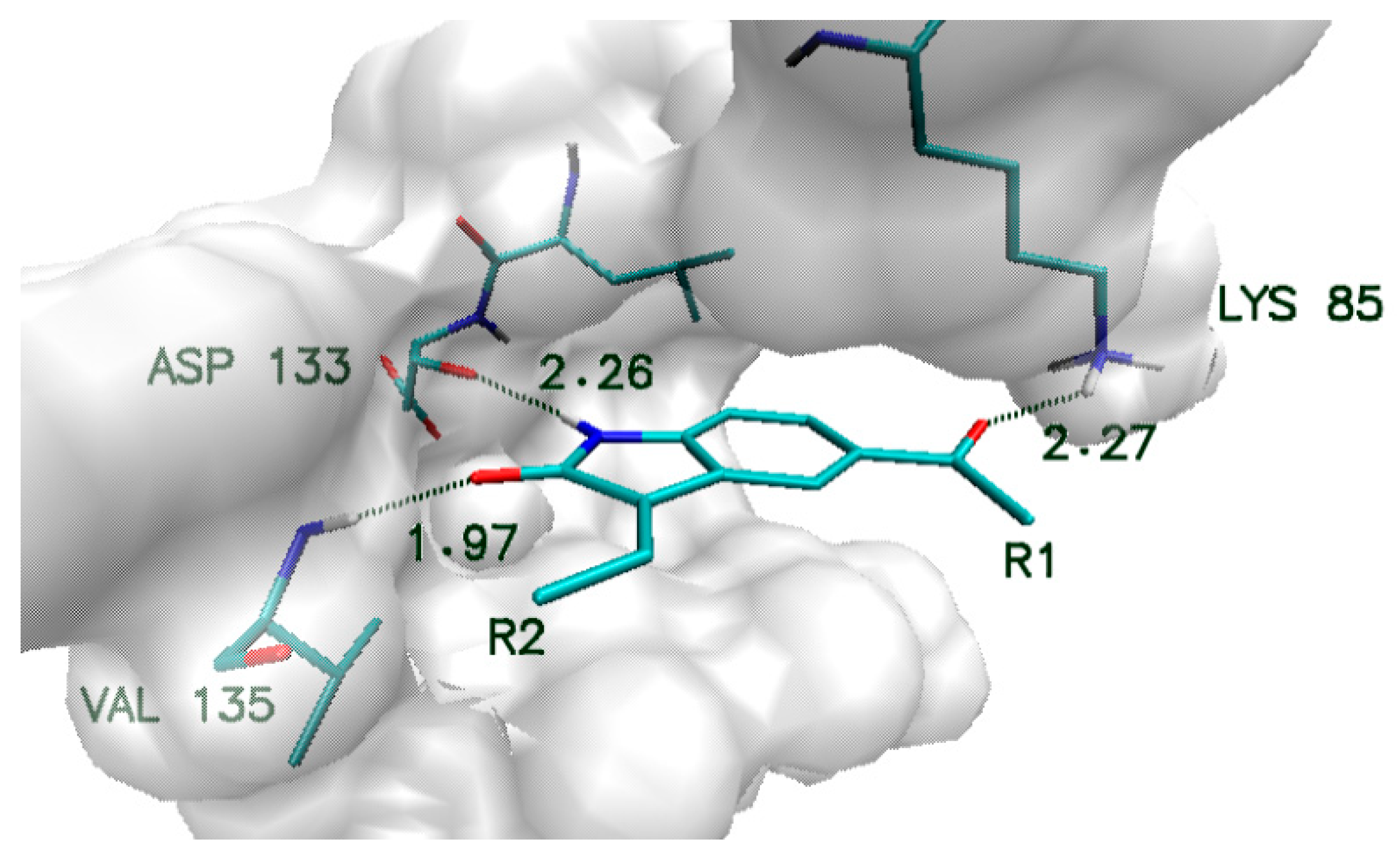
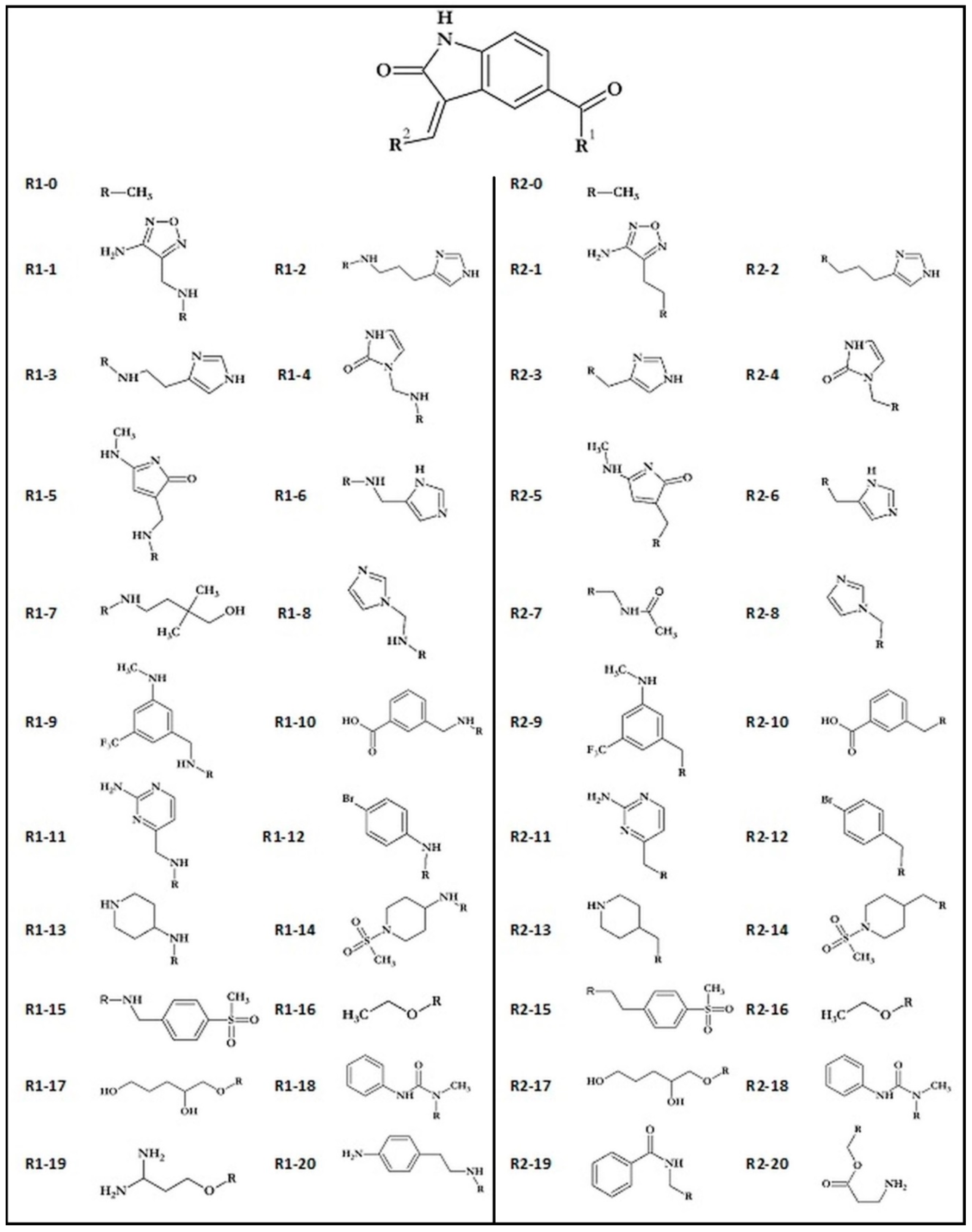
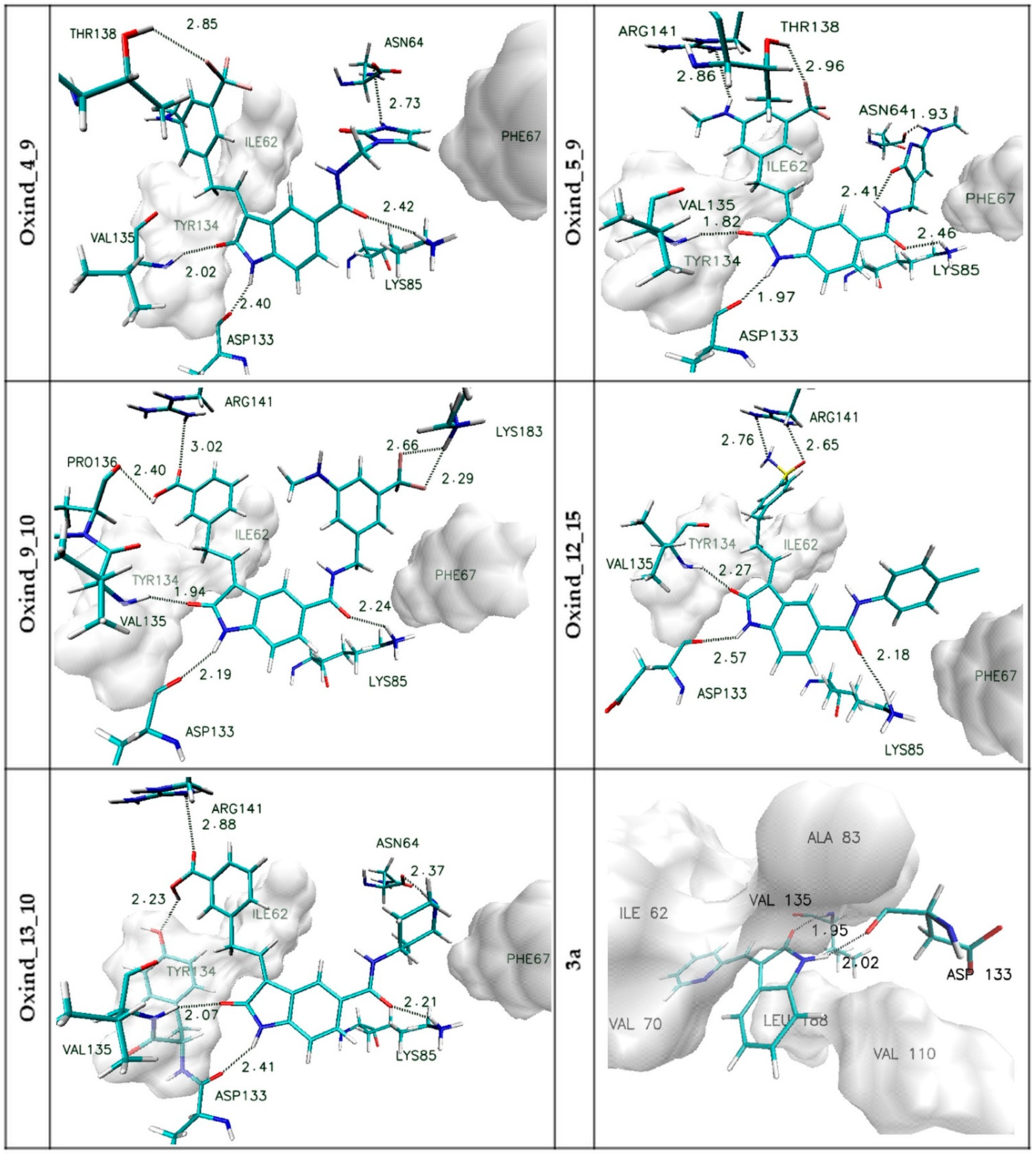
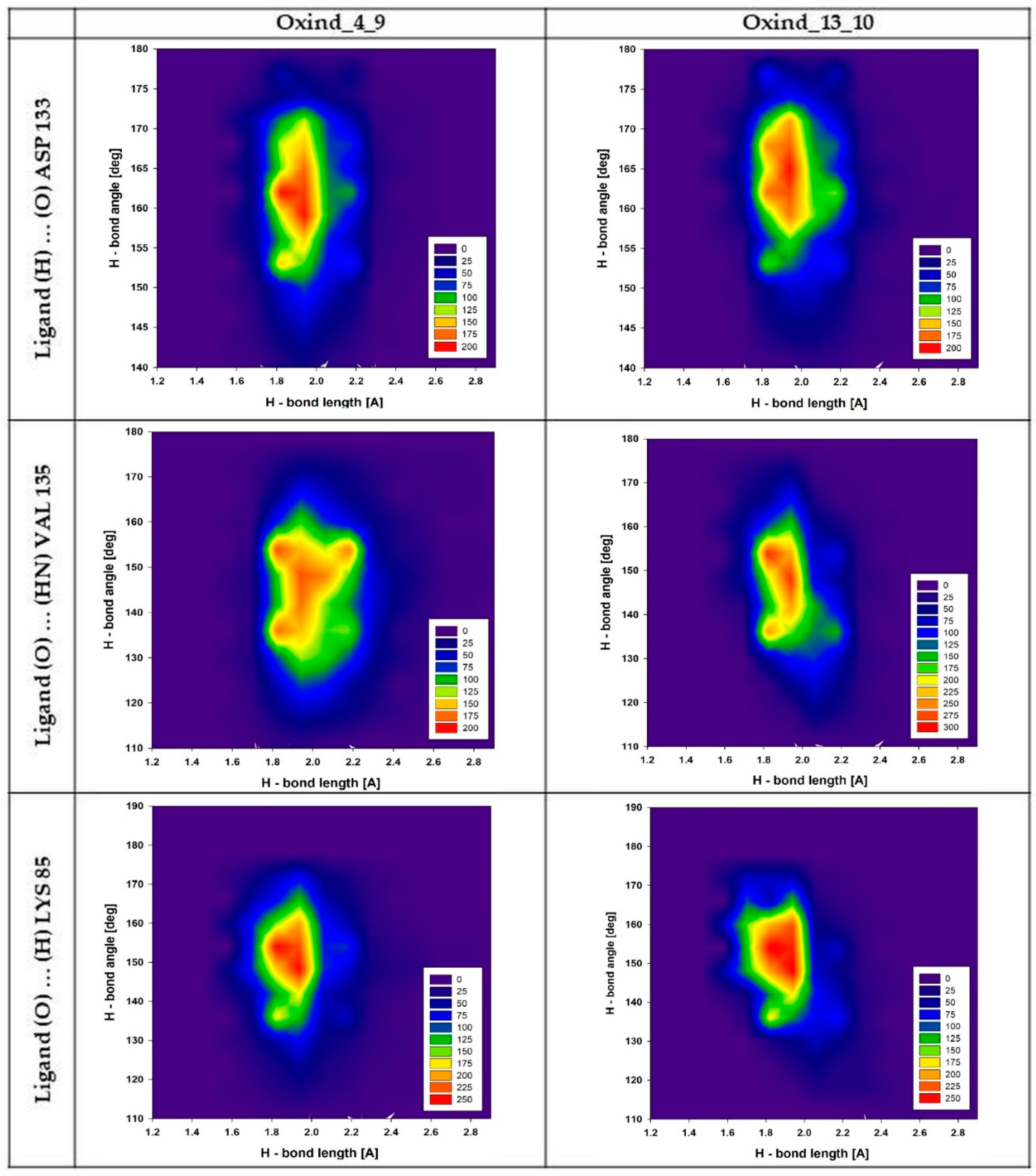

| Name | Binding Affinity (kcal/mol) | Increase in Binding Affinity (%) | Name | Binding Affinity (kcal/mol) | Increase in Binding Affinity (%) |
|---|---|---|---|---|---|
| Oxind_11_0 | −10.10 | 20.2 | Oxind_0_9 | −10.44 | 24.3 |
| Oxind_12_0 | −10.00 | 19.0 | Oxind_0_10 | −10.36 | 23.3 |
| Oxind_10_0 | −9.90 | 17.9 | Oxind_0_15 | −10.22 | 21.7 |
| Oxind_9_0 | −9.84 | 17.1 | Oxind_0_14 | −10.20 | 21.4 |
| Oxind_13_0 | −9.80 | 16.7 | Oxind_0_18 | −10.00 | 19.0 |
| Oxind_5_0 | −9.60 | 14.3 | Oxind_0_19 | −9.90 | 17.9 |
| Oxind_20_0 | −9.54 | 13.6 | Oxind_0_5 | −9.80 | 16.7 |
| Oxind_4_0 | −9.40 | 11.9 | Oxind_0_13 | −9.70 | 15.5 |
| Oxind_15_0 | −9.28 | 10.5 | Oxind_0_11 | −9.60 | 14.3 |
| Oxind_14_0 | −9.26 | 10.2 | Oxind_0_12 | −9.58 | 14.0 |
| Oxind_6_0 | −9.20 | 9.5 | Oxind_0_1 | −9.40 | 11.9 |
| Oxind_18_0 | −9.14 | 8.8 | Oxind_0_2 | −9.24 | 10.0 |
| Oxind_1_0 | −9.10 | 8.3 | Oxind_0_4 | −9.20 | 9.5 |
| Oxind_2_0 | −9.00 | 7.1 | Oxind_0_3 | −9.12 | 8.6 |
| Oxind_3_0 | −8.88 | 5.7 | Oxind_0_6 | −9.10 | 8.3 |
| Oxind_8_0 | −8.80 | 4.8 | Oxind_0_8 | −8.90 | 6.0 |
| Oxind_7_0 | −8.50 | 1.2 | Oxind_0_7 | −8.80 | 4.8 |
| Oxind_17_0 | −8.30 | −1.2 | Oxind_0_16 | −8.60 | 2.4 |
| Oxind_19_0 | −8.16 | −2.9 | Oxind_0_17 | −8.30 | −1.2 |
| Oxind_16_0 | −8.00 | −4.8 | Oxind_0_20 | −8.24 | −1.9 |
| Name | LogP | Toxicity | Binding Affinity (kcal/mol) | Increase in Binding Affinity (%) | Inhibition Constant (nM) | IC/IC Ref | |
|---|---|---|---|---|---|---|---|
| 1. | Oxind_13_10 | 2.38 | 0.28 | −11.60 | 38.10 | 3.14 | 221.6 |
| 2. | Oxind_12_15 | 3.07 | 0.38 | −11.50 | 36.90 | 3.72 | 187.2 |
| 3. | Oxind_9_10 | 5.12 | 0.39 | −11.46 | 36.43 | 3.98 | 175.0 |
| 4. | Oxind_4_9 | 3.58 | 0.32 | −11.40 | 35.71 | 4.40 | 158.1 |
| 5. | Oxind_5_9 | 2.68 | 0.39 | −11.22 | 33.57 | 5.97 | 116.7 |
| 6. | Oxind_9_15 | 3.64 | 0.19 | −11.20 | 33.33 | 6.17 | 112.8 |
| 7. | Oxind_4_10 | 2.04 | 0.07 | −11.14 | 32.62 | 6.83 | 102.0 |
| 8. | Oxind_15_9 | 2.58 | 0.25 | −11.12 | 32.38 | 7.06 | 98.6 |
| 9. | Oxind_12_14 | 2.99 | 0.1 | −11.10 | 32.14 | 7.30 | 95.3 |
| 10. | Oxind_10_14 | 1.82 | 0.04 | −11.02 | 31.19 | 8.36 | 83.3 |
| 11. | Oxind_4_18 | 1.67 | 0.29 | −10.96 | 30.48 | 9.25 | 75.2 |
| 12. | Oxind_20_15 | 2.18 | 0.19 | −10.90 | 29.76 | 10.24 | 68.0 |
| 13. | Oxind_10_13 | 3.38 | 0.28 | −10.90 | 29.76 | 10.24 | 68.0 |
| 14. | Oxind_10_18 | 3.43 | 0.03 | −10.88 | 29.52 | 10.59 | 65.7 |
| 15. | Oxind_12_18 | 3.48 | 0.18 | −10.88 | 29.52 | 10.59 | 65.7 |
| Oxind_4_9 | Oxind_5_9 | Oxind_9_10 | Oxind_12_15 | Oxind_13_10 | 3a | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSK3β | LIG | GSK3β | LIG | GSK3β | LIG | GSK3β | LIG | GSK3β | LIG | GSK3β | LIG | |
| RMSD | 2.47 | 1.24 | 2.42 | 1.62 | 2.28 | 0.77 | 2.61 | 1.97 | 2.25 | 0.50 | 2.37 | 1.14 |
| SD | 0.17 | 0.14 | 0.18 | 0.25 | 0.21 | 0.21 | 0.19 | 0.54 | 0.17 | 0.15 | 0.22 | 0.13 |
| Hydrogen Bond | Population% | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| ∑ | 1.6 Å | 1.8 Å | 2.0 Å | 2.2 Å | 2.4 Å | 2.6 Å | 2.8 Å | 3.0 Å | |
| Oxind_4_9 | |||||||||
| Ligand (H3) … (O) ASP 133 | 100 | 1.8 | 39.4 | 44.2 | 12.4 | 2.0 | 0.2 | 0.1 | 0,0 |
| Ligand(O2) … (HN) VAL 135 | 99.9 | 0.3 | 19.9 | 48.0 | 22.9 | 6.4 | 1.9 | 0.5 | 0.1 |
| Ligand (O3) … (H) LYS 85 | 99.2 | 2.7 | 43.7 | 37.1 | 10.7 | 3.4 | 1.0 | 0.4 | 0.2 |
| Ligand (O1) … (H) LYS 85 | 50.0 | 0.0 | 0.3 | 1.2 | 3.5 | 6.8 | 10.2 | 14.0 | 14.0 |
| Ligand (H4) … (O) ASN 64 | 7.4 | 0.1 | 1.7 | 2.4 | 1.0 | 0.4 | 0.2 | 0.5 | 1.1 |
| Ligand (H4) … (O) GLY 68 | 90.6 | 0.8 | 26.0 | 36.2 | 16.2 | 6.0 | 2.9 | 1.4 | 0.9 |
| Oxind_5_9 | |||||||||
| Ligand (H4) … (O) ASP 133 | 100.0 | 1.6 | 48.1 | 40.2 | 8.6 | 1.1 | 0.3 | 0.1 | 0.0 |
| Ligand(O2) … (HN) VAL 135 | 99.8 | 0.2 | 21.5 | 44.2 | 23.4 | 7.2 | 2.1 | 0.9 | 0.1 |
| Ligand (O3) … (H) LYS 85 | 95.1 | 2.4 | 41.5 | 34.5 | 10.6 | 3.2 | 1.4 | 0.8 | 0.6 |
| Ligand (H1) … (O) PRO 136 | 33.8 | 0.2 | 6.2 | 12.2 | 6.4 | 4.1 | 1.7 | 1.5 | 1.5 |
| Ligand (H2) … (O) ASN 64 | 25.0 | 0.1 | 2.4 | 5.6 | 4.9 | 3.3 | 2.9 | 2.4 | 3.4 |
| Oxind_9_10 | |||||||||
| Ligand (H3) … (O) ASP 133 | 100.0 | 1.4 | 34.1 | 46.0 | 14.4 | 3.4 | 0.6 | 0.0 | 0.1 |
| Ligand(O2) … (HN) VAL 135 | 100.0 | 0.3 | 26.6 | 48.7 | 19.3 | 4.3 | 0.7 | 0.0 | 0.1 |
| Ligand (O2) … (H) LYS 85 | 99.3 | 4.2 | 49.7 | 34.7 | 7.3 | 2.4 | 0.6 | 0.2 | 0.2 |
| Ligand (Fx) … (H) LYS 181 | 45.4 | 0.0 | 0.0 | 0.9 | 3.3 | 6.4 | 11.4 | 11.4 | 12.1 |
| Ligand (H3) … (O) PRO 136 | 79.9 | 1.6 | 10.6 | 18.6 | 16.2 | 13.5 | 8.5 | 6.3 | 4.6 |
| Oxind_12_15 | |||||||||
| Ligand (H3) … (O) ASP 133 | 95.6 | 0.1 | 21.7 | 41.9 | 20.4 | 7.4 | 2.7 | 0.8 | 0.7 |
| Ligand(O3) … (HN) VAL 135 | 98.3 | 0.8 | 25.3 | 42.9 | 21.4 | 5.1 | 1.7 | 0.5 | 0.8 |
| Ligand (O4) … (H) LYS 85 | 43.9 | 0.3 | 13.0 | 15.9 | 7.8 | 3.1 | 1.9 | 1.3 | 0.7 |
| Ligand (N1) … (H) ARG 141 | 51.0 | 0.0 | 2.9 | 13.0 | 11.8 | 7.8 | 6.4 | 4.1 | 5.1 |
| Oxind_13_10 | |||||||||
| Ligand (H3) … (O) ASP 133 | 100.0 | 0.6 | 29.2 | 48.6 | 18.1 | 2.6 | 0.8 | 0.2 | 0.0 |
| Ligand(O1) … (HN) VAL 135 | 100.0 | 0.6 | 34.2 | 49.5 | 13.3 | 2.2 | 0.2 | 0.0 | 0.0 |
| Ligand (O2) … (H) LYS 85 | 93.9 | 3.8 | 46.5 | 31.6 | 8.1 | 2.9 | 0.5 | 0.2 | 0.3 |
| Ligand (H2) … (O) PRO 136 | 80.3 | 6.3 | 24.4 | 21.9 | 11.5 | 6.4 | 4.0 | 2.9 | 2.9 |
| Ligand (H2) … (O) TYR 134 | 50.1 | 0.0 | 0.7 | 1.7 | 3.5 | 5.6 | 7.6 | 13.7 | 17.4 |
| 3a | |||||||||
| Ligand (H3) … (O) ASP 133 | 100 | 2.5 | 55.6 | 36.0 | 5.1 | 0.9 | 0.0 | 0.0 | 0.0 |
| Ligand(O1) … (HN) VAL 135 | 96.5 | 0.5 | 18.2 | 35.9 | 22.1 | 10.3 | 4.1 | 2.7 | 2.7 |
| Oxind_4_9 | Oxind_5_9 | Oxind_9_10 | Oxind_12_15 | Oxind_13_10 | 3a | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔE | SD | ΔE | SD | ΔE | SD | ΔE | SD | ΔE | SD | ΔE | SD | |
| EVDWAALS | −48.31 | 2.63 | −45.52 | 3.21 | −50.63 | 3.79 | −46.56 | 6.62 | −49.80 | 2.98 | −47.91 | 2.76 |
| EEL | −51.29 | 3.88 | −49.62 | 4.95 | −49.80 | 5.20 | −47.52 | 7.38 | −50.85 | 3.66 | −49.84 | 3.67 |
| EPB | 65.76 | 4.94 | 67.84 | 6.89 | 70.68 | 5.97 | 68.54 | 9.50 | 66.19 | 4.38 | 65.21 | 4.15 |
| ECAVITY | −3.97 | 1.15 | −4.29 | 1.66 | −3.95 | 1.96 | −3.58 | 1.99 | −4.17 | 1.59 | −3.85 | 1.35 |
| ΔH | −37.80 | 6.91 | −31.59 | 9.22 | −33.71 | 8.99 | −29.13 | 13.87 | −38.63 | 6.63 | −36.39 | 6.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czeleń, P.; Szefler, B. The Oxindole Derivatives, New Promising GSK-3β Inhibitors as One of the Potential Treatments for Alzheimer’s Disease—A Molecular Dynamics Approach. Biology 2021, 10, 332. https://doi.org/10.3390/biology10040332
Czeleń P, Szefler B. The Oxindole Derivatives, New Promising GSK-3β Inhibitors as One of the Potential Treatments for Alzheimer’s Disease—A Molecular Dynamics Approach. Biology. 2021; 10(4):332. https://doi.org/10.3390/biology10040332
Chicago/Turabian StyleCzeleń, Przemysław, and Beata Szefler. 2021. "The Oxindole Derivatives, New Promising GSK-3β Inhibitors as One of the Potential Treatments for Alzheimer’s Disease—A Molecular Dynamics Approach" Biology 10, no. 4: 332. https://doi.org/10.3390/biology10040332
APA StyleCzeleń, P., & Szefler, B. (2021). The Oxindole Derivatives, New Promising GSK-3β Inhibitors as One of the Potential Treatments for Alzheimer’s Disease—A Molecular Dynamics Approach. Biology, 10(4), 332. https://doi.org/10.3390/biology10040332






