Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance
Abstract
Simple Summary
Abstract
1. Introduction
2. Abiotic Stress
2.1. Transcriptomic Analyses of Abiotic Stress-Induced AS
2.1.1. Relationship between Stress-Induced AS and Stress-Tolerance
2.1.2. AS-Mediated Spatial Regulation of Stress-Responses
2.1.3. Stress-Induced Changes in AS Represent an Independent Layer of Gene Regulation
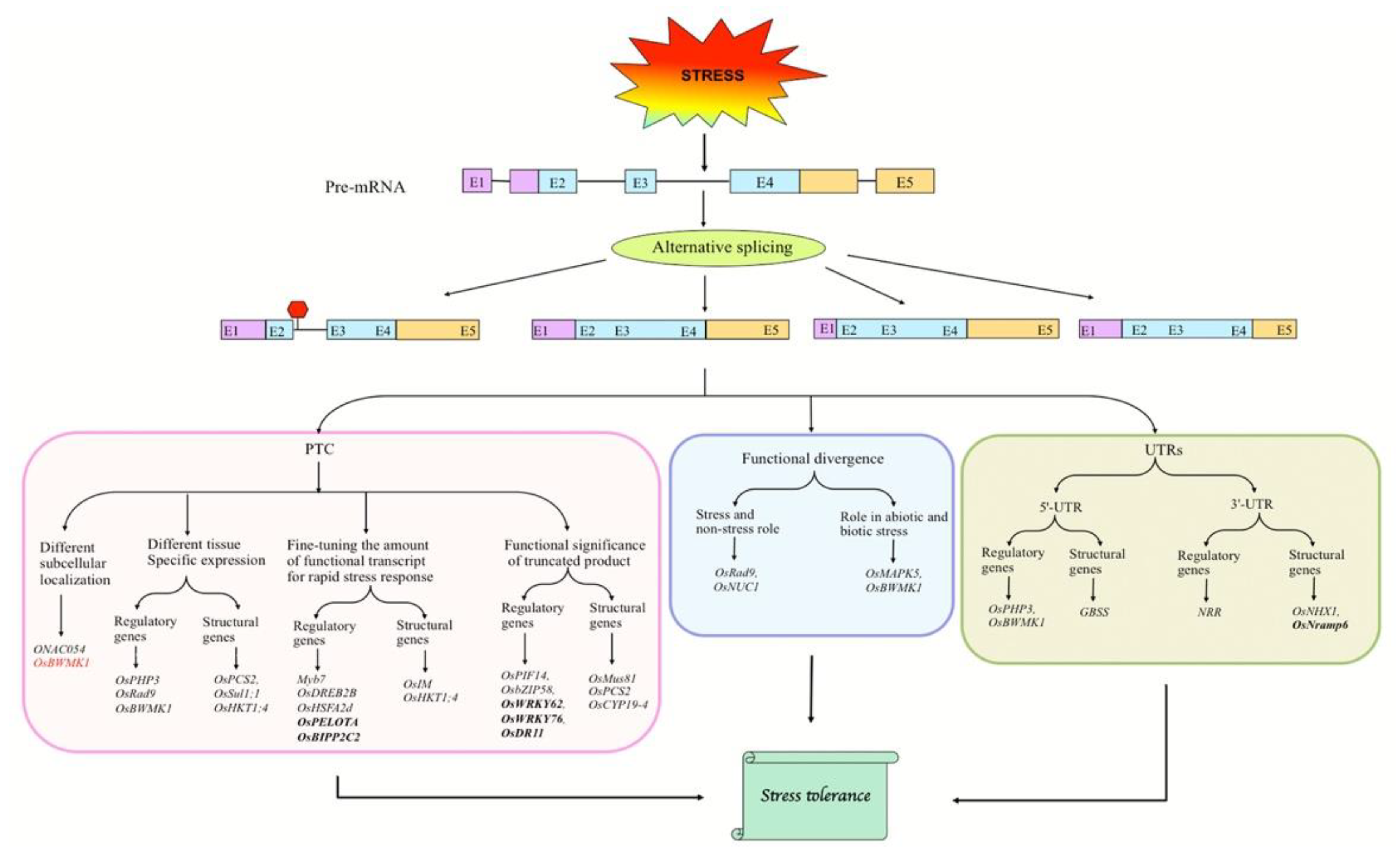
2.2. Abiotic Stress-Induced AS of Candidate Genes
2.2.1. Abiotic Stress-Responsive Genes
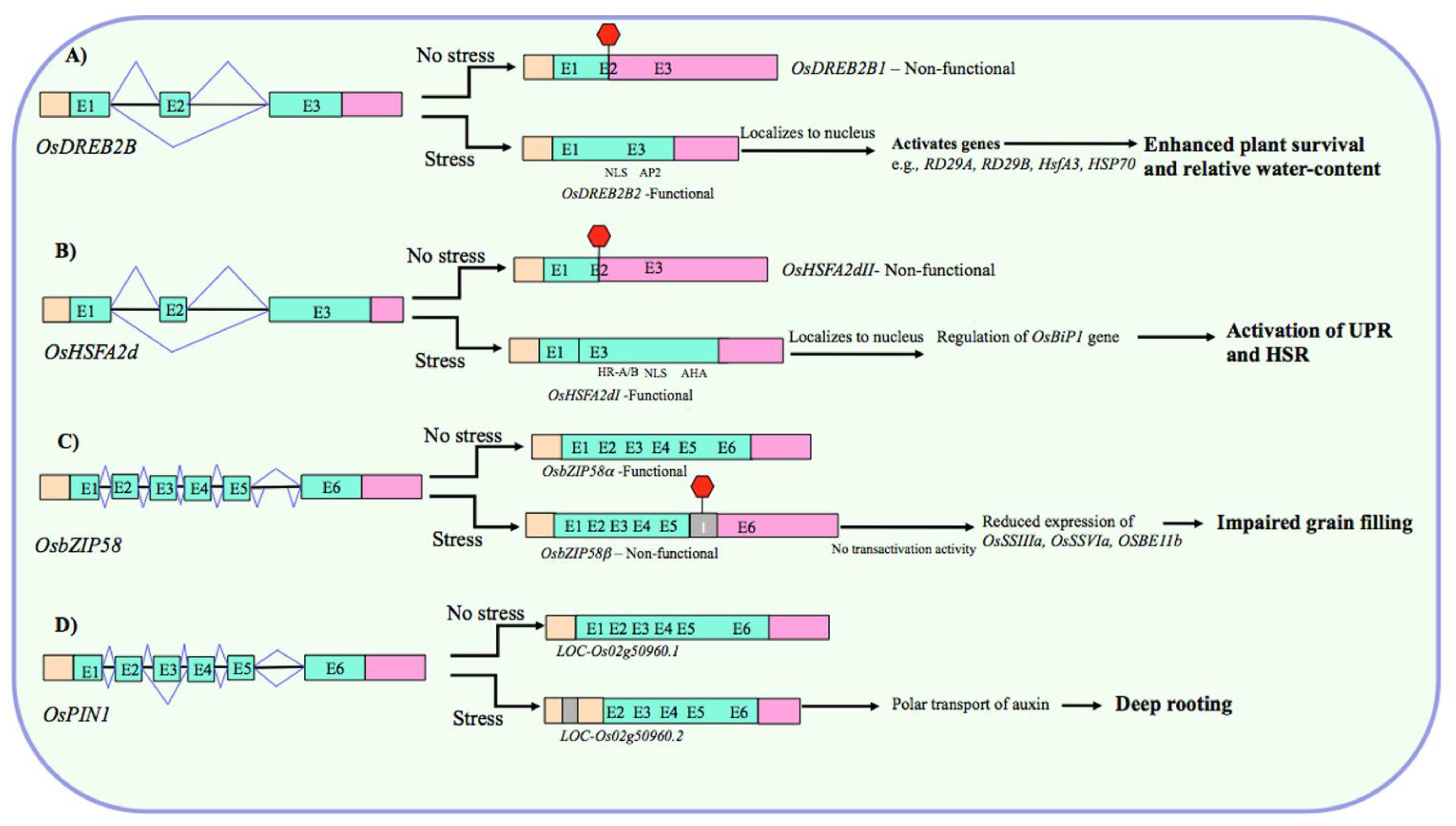
Fine-Tuning the Abundance of Functional Transcripts
Regulatory Role of IR in Stress Responses
Tissue- and/or Developmental Stage-Specific Expression of Transcript Variants
Stress and Non-Stress Roles of Alternative Transcripts
Regulation of Subcellular Localization of Transcript Variants
AS in UTRs of Pre-mRNAs
AS and ABA-Mediated Responses
Regulation of AS by Abiotic Stress-Responsive Genes
2.2.2. Circular RNAs
2.2.3. AS and Splicing Factors
3. Biotic Stress
3.1. AS in Biotic Stress-Responsive Genes
3.1.1. AS in Pre-mRNAs of Resistance (R) Genes
3.1.2. AS in Other Biotic Stress-Related Genes
Functional Significance of AS in Rice Immunity
Modulation of Subcellular Localization of Splice Variants
AS-Mediated Inverse Regulation of Rice Immunity and Abiotic Stress Tolerance
Impact of Sequence Variations in Biotic Stress-Related Genes on Splicing Events
Modulation of the Level of Functional Transcripts
Splicing Factors
Role of Noncoding RNA in AS for Rice Immunity
3.2. Transcriptomic Analysis of Biotic Stress-Induced AS in Rice
3.3. AS in Rice Pathogens
3.3.1. Role of AS in Fungal Virulence and Growth
3.3.2. Role of AS in Virulence of Insect Pests
3.3.3. Role of AS in Insect Growth, Development and Fecundity
3.3.4. Role of AS in Vectoring Rice Pathogens
3.3.5. Role of Splicing Factors in Growth, Development, and Virulence of Rice Pathogens
4. Future Outlook for Research on AS in Rice Stress Responses
- How do stresses regulate AS in rice? Recent studies in plants indicate that much of the AS occurs co-transcriptionally [171,172,173], and that chromatin architecture (epigenetic state—histone modifications and DNA methylation, and nucleosome occupancy) [25,26,27,174] plays a key role in modulating AS. Analysis of AS in chromatin-bound RNAs in rice in response to different stresses should reveal the extent of stress-regulated co-transcriptional AS. By performing AS analysis and epigenetic analyses simultaneously in response to stresses, it should be possible to identify stress-induced AS events that are regulated by specific epigenetic changes.
- The cross-talk between miRNA and AS pathways is poorly explored in rice abiotic stress responses. For example, it is not known how AS influences miRNA-mediated regulation of rice stress-responsive genes. The AS can have a major effect on miRNA-binding sites in the 3′-UTR of genes [41], and can also affect the process of miRNA excision from the primary-miRNA transcript to regulate the mature miRNA levels, and thus, the corresponding mRNAs in the cell [29].
- AS is a critical component of stress priming-induced memory, which promotes the tolerance of plants to subsequent lethal stress [46]. Except for a preliminary report [175], it is not known whether the stress priming alters the choice of alternative splice sites in pre-mRNA transcripts of rice genes and whether it induces splicing memory, and should be explored.
- The potential of CRISPR technology has been exploited in animals to modulate splicing for correcting the mutations associated with diseases [176]. However, in plants, especially rice, there is no report on improving any trait using this strategy. A recent study on Arabidopsis has developed an efficient base-editing tool for gene splicing [177]. This tool can be potentially used for elucidating the regulatory function of AS in plant responses to stress through generating the mutants for specific splice variants.
- Genetic variability can profoundly influence the AS and abundance of alternative transcripts. The functional impact of genetic variations in shaping AS events with stress-adaptive consequences has also been revealed in plants. For example, microsatellites are involved in modulating AS of miRNA genes under different stress conditions [178], and ‘splicing QTLs’ (sQTLs) potentially regulate AS of various stress-responsive genes [179]. However, the significance of microsatellites and sQTLs in regulating AS for rice stress tolerance has yet to be determined.
- The emergence of single-cell RNA-seq has revealed gene expression signatures, including AS, distinguishable at the cellular level, which is invisible in whole plant or organ RNA-seq. Isoform expression analysis at the single-cell level is crucial to gain deeper insights into AS biology. The specific stress responses emanating from AS in a particular cell-type of shoot or root tissues of rice have not been revealed yet and should be explored.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhandari, H.; Mishra, A.K. Impact of demographic transformation on future rice farming in Asia. Outlook Agric. 2018, 47, 125–132. [Google Scholar] [CrossRef]
- FAO. Crop Prospects and Food Situation-Quarterly Global Report No. 1; FAO: Rome, Italy, 2021. [Google Scholar] [CrossRef]
- Liu, W.; Wang, G.-L. Plant innate immunity in rice: A defense against pathogen infection. Natl. Sci. Rev. 2016, 3, 295–308. [Google Scholar] [CrossRef]
- Ganie, S.A.; Ahammed, G.J.; Wani, S.H. Vascular plant one zinc-finger (VOZ) transcription factors: Novel regulators of abiotic stress tolerance in rice (Oryza sativa L.). Genet. Resour. Crop. Evol. 2020, 67, 799–807. [Google Scholar] [CrossRef]
- Motion, G.B.; Amaro, T.M.M.M.; Kulagina, N.; Huitema, E. Nuclear processes associated with plant immunity and pathogen susceptibility. Brief. Funct. Genom. 2015, 14, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Haak, D.C.; Fukao, T.; Grene, R.; Hua, Z.; Ivanov, R.; Perrella, G.; Li, S. Multilevel regulation of abiotic stress responses in plants. Front. Plant Sci. 2017, 8, 1564. [Google Scholar] [CrossRef]
- Guerra, D.; Crosatti, C.; Khoshro, H.H.; Mastrangelo, A.M.; Mica, E.; Mazzucotelli, E. Post-transcriptional and post-translational regulations of drought and heat response in plants: A spider’s web of mechanisms. Front. Plant Sci. 2015, 6, 57. [Google Scholar] [CrossRef]
- Ganie, S.A. RNA chaperones: Potential candidates for engineering salt tolerance in rice. Crop. Sci. 2020, 60, 530–540. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Rigo, R.; Bazin, J.; Crespi, M.; Charon, C. Alternative splicing in the regulation of plant–microbe interactions. Plant Cell Physiol. 2019, 60, 1906–1916. [Google Scholar] [CrossRef]
- Dong, C.; He, F.; Berkowitz, O.; Liu, J.; Cao, P.; Tang, M.; Shi, H.; Wang, W.; Li, Q.; Shen, Z.; et al. Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 2018, 30, 2267–2285. [Google Scholar] [CrossRef] [PubMed]
- Jabre, I.; Reddy, A.S.; Kalyna, M.; Chaudhary, S.; Khokhar, W.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Does co-transcriptional regulation of alternative splicing mediate plant stress responses? Nucleic Acids Res. 2019, 47, 2716–2726. [Google Scholar] [CrossRef]
- Chaudhary, S.; Jabre, I.; Reddy, A.S.; Staiger, D.; Syed, N.H. Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 2019, 24, 496–506. [Google Scholar] [CrossRef]
- Zalabák, D.; Ikeda, Y. First Come, First Served: Sui Generis Features of the First Intron. Plants 2020, 9, 911. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, R.E.; Andreadis, A.; Nadal-Ginard, B. Alternative splicing: A ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 1987, 56, 467–495. [Google Scholar] [CrossRef]
- Reddy, A.S.N. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Ner-Gaon, H.; Halachmi, R.; Savaldi-Goldstein, S.; Rubin, E.; Ophir, R.; Fluhr, R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004, 39, 877–885. [Google Scholar] [CrossRef]
- Reddy, A.S.; Rogers, M.F.; Richardson, D.N.; Hamilton, M.; Ben-Hur, A. Deciphering the plant splicing code: Experimental and computational approaches for predicting alternative splicing and splicing regulatory elements. Front. Plant Sci. 2012, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Palusa, S.G.; Reddy, A.S.N. Differential recruitment of splice variants from SR pre-mRNAs to polysomes during development and in response to stresses. Plant Cell Physiol. 2015, 56, 421–427. [Google Scholar] [CrossRef]
- Reddy, A.S.N.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W.S. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of alternative pre-messenger rna splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Barta, A.; Kalyna, M.; Reddy, A.S.N. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.N.; Huang, J.; Syed, N.H.; Ben-Hur, A.; Dong, S.; Gu, L. Decoding co-/post-transcriptional complexities of plant transcriptomes and epitranscriptome using next-generation sequencing technologies. Biochem. Soc. Trans. 2020, 48, 2399–2414. [Google Scholar] [CrossRef] [PubMed]
- Ullah, F.; Hamilton, M.; Reddy, A.S.N.; Ben-Hur, A. Exploring the relationship between intron retention and chromatin accessibility in plants. BMC Genom. 2018, 19. [Google Scholar] [CrossRef]
- Wang, X.; Hu, L.; Wang, X.; Li, N.; Xu, C.; Gong, L.; Liu, B. DNA methylation affects gene alternative splicing in plants: An example from rice. Mol. Plant 2016, 9, 305–307. [Google Scholar] [CrossRef]
- Pajoro, A.; Severing, E.; Angenent, G.C.; Immink, R.G. Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Biol. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Meng, X.; Liu, Y.; Wang, X.; Wang, T.-J.; Zhang, A.; Li, N.; Qi, X.; Liu, B.; Xu, Z.-Y. The chromatin remodeler ZmCHB101 impacts alternative splicing contexts in response to osmotic stress. Plant Cell Rep. 2018, 38, 131–145. [Google Scholar] [CrossRef]
- Yan, K.; Liu, P.; Wu, C.-A.; Yang, G.-D.; Xu, R.; Guo, Q.-H.; Huang, J.-G.; Zheng, C.-C. Stress-Induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531. [Google Scholar] [CrossRef]
- Campo, S.; Peris-Peris, C.; Siré, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; San Segundo, B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef]
- Jia, F.; Rock, C.D. MIR846 and MIR842 comprise a cistronic MIRNA pair that is regulated by abscisic acid by alternative splicing in roots of Arabidopsis. Plant Mol. Biol. 2013, 81, 447–460. [Google Scholar] [CrossRef]
- Meng, Y.; Shao, C.; Ma, X.; Wang, H. Introns targeted by plant microRNAs: A possible novel mechanism of gene regulation. Rice 2013, 6. [Google Scholar] [CrossRef]
- Park, S.-Y.; Grabau, E. Bypassing miRNA-mediated gene regulation under drought stress: Alternative splicing affects CSD1 gene expression. Plant Mol. Biol. 2017, 95, 243–252. [Google Scholar] [CrossRef]
- Zhang, Y.; Rahmani, R.S.; Yang, X.; Chen, J.; Shi, T. Integrative expression network analysis of microRNA and gene isoforms in sacred lotus. BMC Genom. 2020, 21, 1–13. [Google Scholar] [CrossRef]
- Chen, L.; Tovar-Corona, J.M.; Urrutia, A.O. Alternative Splicing: A Potential Source of Functional Innovation in the Eukaryotic Genome. Int. J. Evol. Biol. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Brogna, S.; McLeod, T.; Petric, M. The meaning of NMD: Translate or perish. Trends Genet. 2016, 32, 395–407. [Google Scholar] [CrossRef]
- Ohtani, M.; Wachter, A. NMD-based gene regulation—A strategy for fitness enhancement in plants? Plant Cell Physiol. 2019, 60, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Gracz, J. Alternative splicing in plant stress response. BioTechnologia 2016, 1, 9–17. [Google Scholar] [CrossRef]
- Liu, J.; Sun, N.; Liu, M.; Liu, J.; Du, B.; Wang, X.; Qi, X. An autoregulatory loop controlling Arabidopsis HsfA2 expression: Role of heat shock-induced alternative splicing. Plant Physiol. 2013, 162, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, A.M.; Marone, D.; Laidò, G.; De Leonardis, A.M.; De Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012, 185–186, 40–49. [Google Scholar] [CrossRef]
- Srivastava, A.K.; Lu, Y.; Zinta, G.; Lang, Z.; Zhu, J.-K. UTR-dependent control of gene expression in plants. Trends Plant Sci. 2018, 23, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126. [Google Scholar] [CrossRef] [PubMed]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S.N. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, S.; Khokhar, W.; Jabre, I.; Reddy, A.S.; Byrne, L.J.; Wilson, C.M.; Syed, N.H. Alternative Splicing and Protein Diversity: Plants Versus Animals. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-X.; Zhang, K.-L.; Zhang, M.; Das, D.; Fang, Y.-M.; Dai, L.; Zhang, J.; Zhu, F.-Y. Alternative splicing and its regulatory role in woody plants. Tree Physiol. 2020, 40, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Serrano, N.; Gao, G.; Atia, M.; Mokhtar, M.; Woo, Y.H.; Bazin, J.; Veluchamy, A.; Benhamed, M.; Crespi, M.; et al. Thermopriming triggers Splicing memory in Arabidopsis. J. Exp. Bot. 2018, 69, 2659–2675. [Google Scholar] [CrossRef]
- Martin, L.B.; Fei, Z.; Giovannoni, J.J.; Rose, J.K. Catalyzing plant science research with RNA-seq. Front. Plant Sci. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Jain, M. Next-generation sequencing technologies for gene expression profiling in plants. Brief. Funct. Genom. 2011, 11, 63–70. [Google Scholar] [CrossRef]
- Takahagi, K.; Uehara-Yamaguchi, Y.; Yoshida, T.; Sakurai, T.; Shinozaki, K.; Mochida, K.; Saisho, D. Analysis of single nucleotide polymorphisms based on RNA sequencing data of diverse bio-geographical accessions in barley. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Liu, Q.; Zheng, L.; Cui, Y.; Shen, Z.; Zheng, L. RNA-seq analysis of rice roots reveals the involvement of post-transcriptional regulation in response to cadmium stress. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Lou, Q.; Xu, K.; Yan, M.; Xia, H.; Ma, X.; Yu, X.; Luo, L. Alternative splicing complexity contributes to genetic improvement of drought resistance in the rice maintainer HuHan2B. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Liu, H.; Sun, J.; Zheng, H.; Wang, J.; Yang, L.; Zhao, H.; Zou, D. Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef]
- Phule, A.S.; Barbadikar, K.M.; Maganti, S.M.; Seguttuvel, P.; Subrahmanyam, D.; Babu, M.B.; Kumar, P.A. RNA-seq reveals the involvement of key genes for aerobic adaptation in rice. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, G.M.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E.; et al. Gene expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zha, Z.; Cai, H.; Qin, D.; Jia, H.; Liu, C.; Qiu, D.; Zhang, Z.; Wan, Z.; Yang, Y.; et al. Dynamic transcriptome analysis of anther response to heat stress during anthesis in thermotolerant rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1155. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, B. Comparative alternative splicing analysis of two contrasting rice cultivars under drought stress and association of differential splicing genes with drought response QTLs. Euphytica 2018, 214. [Google Scholar] [CrossRef]
- Mansuri, R.M.; Shobbar, Z.S.; Jelodar, N.B.; Ghaffari, M.R.; Nematzadeh, G.A.; Asari, S. Dissecting molecular mechanisms underlying salt tolerance in rice: A comparative transcriptional profiling of the contrasting genotypes. Rice 2019, 12. [Google Scholar] [CrossRef]
- Fu, L.; Shen, Q.; Kuang, L.; Wu, D.; Zhang, G. Transcriptomic and alternative splicing analyses reveal mechanisms of the difference in salt tolerance between barley and rice. Environ. Exp. Bot. 2019, 166, 103810. [Google Scholar] [CrossRef]
- Shankar, R.; Bhattacharjee, A.; Jain, M. Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Junior, A.T.D.; Farias, D.D.; dos Santos, R.S.; do Amaral, M.N.; Arge, L.W.P.; Oliveira, D.D.C.; de Oliveira, A.C. The quest for more tolerant rice: How high concentrations of iron affect alternative splicing? Transcr. Open Access 2015, 3. [Google Scholar] [CrossRef]
- Chen, M.-X.; Zhu, F.-Y.; Wang, F.-Z.; Ye, N.-H.; Gao, B.; Chen, X.; Zhao, S.-S.; Fan, T.; Cao, Y.-Y.; Liu, T.-Y.; et al. Alternative splicing and translation play important roles in hypoxic germination in rice. J. Exp. Bot. 2019, 70, 817–833. [Google Scholar] [CrossRef] [PubMed]
- Sampangi-Ramaiah, M.H.; Ravishankar, K.V.; Nataraja, K.N.; Uma Shaanker, R. Endophytic fungus, Fusarium sp. reduces alternative splicing events in rice plants under salinity stress. Plant Physiol. Rep. 2019, 24, 487–495. [Google Scholar] [CrossRef]
- Li, Y.-F.; Zheng, Y.; Vemireddy, L.R.; Panda, S.K.; Jose, S.; Ranjan, A.; Panda, P.; Govindan, G.; Cui, J.; Wei, K.; et al. Comparative transcriptome and translatome analysis in contrasting rice genotypes reveals differential mRNA translation in salt-tolerant Pokkali under salt stress. BMC Genom. 2018, 19. [Google Scholar] [CrossRef]
- Ganie, S.A.; Ahammed, G.J. Dynamics of cell wall structure and related genomic resources for drought tolerance in rice. Plant Cell Rep. 2021, 40, 437–459. [Google Scholar] [CrossRef]
- Ganie, S.A.; Wani, S.H.; Henry, R.; Hensel, G. Improving rice salt tolerance by precision breeding in a new era. Curr. Opin. Plant Biol. 2021, 60, 101996. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, L.; Wang, Y.; Wang, W.; Zhao, X.; Zhang, S.; Zhang, J.; Hu, F.; Fu, B.; Li, Z. Differential transcriptome profiling of chilling stress response between shoots and rhizomes of Oryza longistaminata using RNA sequencing. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Deng, Q.; Bai, L.; Dai, L.; Chen, Y.; Fang, J.; Xie, J.; Luo, X. Identification of phosphorus stress related proteins in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.) using label-free quantitative proteomic analysis. Res. Square 2020. [Google Scholar] [CrossRef]
- Wei, H.; Lou, Q.; Xu, K.; Zhou, L.; Chen, S.; Chen, L.; Luo, L. Pattern of alternative splicing different associated with difference in rooting depth in rice. Plant Soil 2020, 449, 233–248. [Google Scholar] [CrossRef]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 2010, 283, 185–196. [Google Scholar] [CrossRef]
- Magaraggia, F.; Solinas, G.; Valle, G.; Giovinazzo, G.; Coraggio, I. Maturation and translation mechanisms involved in the expression of a myb gene of rice. Plant Mol. Biol. 1997, 35, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Zhou, Y.; Liu, Z.; Zhang, L.; Song, G.; Guo, Z.; Wang, W.; Qu, X.; Zhu, Y.; Yang, D. An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. 2015, 17, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Cotsaftis, O.; Plett, D.; Shirley, N.; Tester, M.; Hrmova, M. A Two-Staged model of Na+ exclusion in rice explained by 3D modeling of HKT transporters and alternative splicing. PLoS ONE 2012, 7, e39865. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Gong, J.-M.; Zhang, Z.-G.; Zhang, J.-S.; Chen, S.-Y. A new AOX homologous gene OsIM1 from rice (Oryza sativa L.) with an alternative splicing mechanism under salt stress. Theor. Appl. Genet. 2003, 107, 326–331. [Google Scholar] [CrossRef]
- Cordeiro, A.M.; Figueiredo, D.D.; Tepperman, J.; Borba, A.R.; Lourenço, T.; Abreu, I.A.; Ouwerkerk, P.B.F.; Quail, P.H.; Margarida Oliveira, M.; Saibo, N.J.M. Rice phytochrome-interacting factor protein OsPIF14 represses OsDREB1B gene expression through an extended N-box and interacts preferentially with the active form of phytochrome B. Biochim. Biophys. Acta Gene Regul. Mech. 2016, 1859, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, X.; Zhang, H.; Wang, L.; Zhu, Z.; Gao, J.; Li, C.; Zhu, Y. High temperature inhibits the accumulation of storage materials by inducing alternative splicing of OsbZIP58 during filling stage in rice. Plant Cell Environ. 2020, 43, 1879–1896. [Google Scholar] [CrossRef] [PubMed]
- Mimida, N.; Kitamoto, H.; Osakabe, K.; Nakashima, M.; Ito, Y.; Heyer, W.-D.; Toki, S.; Ichikawa, H. Two Alternatively Spliced Transcripts generated From OSMUS81, a Rice homolog of Yeast MUS81, are Up-Regulated by DNA-Damaging Treatments. Plant Cell Physiol. 2007, 48, 648–654. [Google Scholar] [CrossRef][Green Version]
- Lee, A.; Lee, S.; Jung, W.; Park, H.; Lim, B.; Kim, H.-S.; Ahn, J.; Cho, H. The OsCYP19-4 Gene Is Expressed as Multiple Alternatively Spliced Transcripts Encoding Isoforms with Distinct Cellular Localizations and PPIase Activities under Cold Stress. Int. J. Mol. Sci. 2016, 17, 1154. [Google Scholar] [CrossRef]
- Li, R.; Wang, W.; Li, F.; Wang, Q.; Wang, S.; Xu, Y.; Chen, F. Response of alternative splice isoforms of OsRad9 gene from Oryza sativa to environmental stress. Z. Naturforsch. C 2017, 72, 325–334. [Google Scholar] [CrossRef]
- Sripinyowanich, S.; Chamnanmanoontham, N.; Udomchalothorn, T.; Maneeprasopsuk, S.; Santawee, P.; Buaboocha, T.; Qu, L.-J.; Gu, H.; Chadchawan, S. Overexpression of a Partial fragment of the salt-responsive gene OsNUC1 enhances salt adaptation in transgenic Arabidopsis thaliana and rice (Oryza sativa L.) during salt stress. Plant Sci. 2013, 213, 67–78. [Google Scholar] [CrossRef]
- Koo, S.C.; Yoon, H.W.; Kim, C.Y.; Moon, B.C.; Cheong, Y.H.; Han, H.J.; Lee, S.M.; Kang, K.Y.; Kim, M.C.; Lee, S.Y.; et al. Alternative splicing of the OsBWMK1 gene generates three transcript variants showing differential subcellular localizations. Biochem. Biophys. Res. Commun. 2007, 360, 188–193. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Han, S.-H.; Kim, S.-H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.-C. Multilayered regulation of membrane-bound ONAC054 is essential for abscisic acid-induced leaf senescence in rice. Plant Cell 2020, 32, 630–649. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Tsai, Y.-C. Light-Regulated alternative splicing of Pseudo-histidine Phosphotransfer Protein 3 in Oryza sativa. J. Plant Growth Regul. 2019, 38, 1215–1227. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Lu, W.-C.; Ko, S.-S.; Sun, C.-M.; Hung, J.-C.; Chiou, T.-J. Upstream open reading frame and phosphate-regulated expression of rice OsNLA1 controls phosphate transport and reproduction. Plant Physiol. 2020, 182, 393–407. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Yan, Y.-S.; Wang, L.-N.; Yang, K.; Xiao, N.; Liu, Y.-F.; Fu, Y.-P.; Sun, Z.-X.; Fang, R.-X.; Chen, X.-Y. A novel rice gene, NRR responds to macronutrient deficiency and regulates root growth. Mol. Plant 2012, 5, 63–72. [Google Scholar] [CrossRef]
- Larkin, P.D.; Park, W.D. Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant Mol. Biol. 1999, 40, 719–727. [Google Scholar] [CrossRef]
- Amin, U.S.; Biswas, S.; Elias, S.M.; Razzaque, S.; Haque, T.; Malo, R.; Seraj, Z.I. Enhanced salt tolerance conferred by the complete 2.3 kb cDNA of the rice vacuolar Na+/H+ antiporter gene compared to 1.9 kb coding region with 5′ UTR in transgenic lines of rice. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Costanzo, S.; Jia, Y. Alternatively spliced transcripts of Pi-ta blast resistance gene in Oryza sativa. Plant Sci. 2009, 177, 468–478. [Google Scholar] [CrossRef]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tian, D.; Gu, K.; Yang, X.; Wang, L.; Zeng, X.; Yin, Z. Induction of Xa10-like genes in rice cultivar Nipponbare confers disease resistance to rice bacterial blight. Mol. Plant Microbe Interact. 2017, 30, 466–477. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Liang, X.; Zhou, X.; Yang, F.; Liu, J.; He, S.Y.; Guo, Z. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in Pathogen defense. Plant Physiol. 2016, 171, 1427–1442. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Xiao, W.; Xia, F.; Liu, H.; Xiao, J.; Li, X.; Wang, S. Two different transcripts of A LAMMER Kinase Gene play Opposite roles in disease resistance. Plant Physiol. 2016, 172, 1959–1972. [Google Scholar] [CrossRef]
- Liu, D.; Shi, S.; Hao, Z.; Xiong, W.; Luo, M. OsbZIP81, A Homologue of Arabidopsis VIP1, May Positively Regulate JA Levels by Directly Targetting the Genes in JA Signaling and Metabolism Pathway in Rice. Int. J. Mol. Sci. 2019, 20, 2360. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, H.; Li, G.; Yang, Y.; Zheng, Z.; Song, F. Ectopic expression of a rice protein phosphatase 2C gene OsBIPP2C2 in tobacco improves disease resistance. Plant Cell Rep. 2009, 28, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-B.; Feng, B.-H.; Wang, H.-M.; Xu, X.; Shi, Y.-F.; He, Y.; Chen, Z.; Sathe, A.P.; Shi, L.; Wu, J.-L. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 2018, 60, 160–172. [Google Scholar] [CrossRef]
- Feng, W.; Hongbin, W.; Bing, L.; Jinfa, W. Cloning and characterization of a novel splicing isoform of the iron-superoxide dismutase gene in rice (Oryza sativa L.). Plant Cell Rep. 2006, 24, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Favory, J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010, 33, 88–103. [Google Scholar] [CrossRef]
- Zhao, B.; Tang, Y.; Zhang, B.; Wu, P.; Li, M.; Xu, X.; Wu, G.; Jiang, H.; Chen, Y. The Temperature-Dependent Retention of Introns in GPI8 Transcripts Contributes to a Drooping and Fragile Shoot Phenotype in Rice. Int. J. Mol. Sci. 2020, 21, 299. [Google Scholar] [CrossRef]
- Das, N.; Bhattacharya, S.; Bhattacharyya, S.; Maiti, M.K. Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Mol. Biol. 2017, 94, 167–183. [Google Scholar] [CrossRef] [PubMed]
- Sellamuthu, G.; Jegadeeson, V.; Sajeevan, R.S.; Rajakani, R.; Parthasarathy, P.; Raju, K.; Shabala, L.; Chen, Z.-H.; Zhou, M.; Sowdhamini, R.; et al. Distinct evolutionary origins of intron retention splicing events in NHX1 antiporter transcripts relate to sequence specific distinctions in Oryza species. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Koo, S.C.; Choi, M.S.; Chun, H.J.; Park, H.C.; Kang, C.H.; Shim, S.I.; Chung, J.I.; Cheong, Y.H.; Lee, S.Y.; Yun, D.-J.; et al. Identification and characterization of alternative promoters of the rice MAP kinase gene OsBWMK1. Mol. Cells 2009, 27, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Dubey, R.S.; Trivedi, P.K. Comprehensive analysis of regulatory elements of the promoters of rice sulfate transporter gene family and functional characterization of OsSul1;1 promoter under different metal stress. Plant Signal. Behav. 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Khurana, N.; Chauhan, H.; Khurana, P. Expression analysis of A Heat-inducible, Myo-inositol-1-phosphate synthase (MIPS) gene from wheat and the alternatively spliced variants of rice and Arabidopsis. Plant Cell Rep. 2011, 31, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Asif, M.H.; Chakrabarty, D.; Tripathi, R.D.; Trivedi, P.K. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct. Integr. Genom. 2011, 11, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bian, M.; Yang, Z.; Lin, C.; Shi, W. Preliminary functional analysis of the Isoforms of OsHsfA2a (Oryza sativa L.) generated by alternative splicing. Plant Mol. Biol. Rep. 2013, 31, 38–46. [Google Scholar] [CrossRef]
- Fang, Z. Differential expression pattern of splice variants of amino acid transporter genes from rice grown under various nitrogen conditions and during development. Int. J. Agric. Biol. 2017, 19, 1246–1258. [Google Scholar] [CrossRef]
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Almadanim, M.C.; Gonçalves, N.M.; Rosa, M.T.G.; Alexandre, B.M.; Cordeiro, A.M.; Rodrigues, M.; Saibo, N.J.M.; Soares, C.M.; Romão, C.V.; Oliveira, M.M.; et al. The rice cold-responsive calcium-dependent protein kinase OsCPK17 is regulated by alternative splicing and post-translational modifications. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 231–246. [Google Scholar] [CrossRef]
- Xiong, L.; Yang, Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid–inducible mitogen-activated protein kinase. Plant Cell 2003, 15, 745–759. [Google Scholar] [CrossRef]
- Zeng, C.; Hamada, M. RNA-Seq analysis reveals localization-associated alternative splicing across 13 cell lines. Genes 2020, 11, 820. [Google Scholar] [CrossRef]
- Lu, S.-J.; Yang, Z.-T.; Sun, L.; Sun, L.; Song, Z.-T.; Liu, J.-X. Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol. Plant 2012, 5, 504–514. [Google Scholar] [CrossRef]
- Kornblihtt, A.R. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 2005, 17, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-Y.; Huang, T.-K.; Chiou, T.-J. NITROGEN limitation adaptation, a target of MicroRNA827, Mediates degradation of Plasma Membrane–Localized Phosphate transporters to Maintain Phosphate homeostasis in Arabidopsis. Plant Cell 2013, 25, 4061–4074. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Sotta, N.; Yamazumi, Y.; Yamashita, Y.; Miwa, K.; Murota, K.; Chiba, Y.; Hirai, M.Y.; Akiyama, T.; Onouchi, H.; et al. The Minimum Open Reading Frame, AUG-Stop, Induces Boron-Dependent Ribosome Stalling and mRNA Degradation. Plant Cell 2016, 28, 2830–2849. [Google Scholar] [CrossRef]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. Characterization of alternative splicing products of bZIP transcription factors OsABI5. Biochem. Biophys. Res. Commun. 2007, 360, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.T.; Almeida, D.M.; Pires, I.S.; da Rosa Farias, D.; Martins, A.G.; da Maia, L.C.; de Oliveira, A.C.; Saibo, N.J.; Oliveira, M.M.; Abreu, I.A. Insights into the transcriptional And Post-transcriptional regulation of the rice SUMOylation machinery and into the role of two rice SUMO proteases. BMC Plant Biol. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Xie, K.; Yao, J.; Qi, Z.; Xiong, L. A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc. Natl. Acad. Sci. USA 2009, 106, 6410–6415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Deng, H.; Xiao, F.; Liu, Y. Alterations of Alternative Splicing Patterns of Ser/Arg-Rich (SR) Genes in Response to Hormones and Stresses Treatments in Different Ecotypes of Rice (Oryza sativa). J. Integr. Agric. 2013, 12, 737–748. [Google Scholar] [CrossRef]
- Kababji, A.M. Targeted Mutagenesis and Functional Analysis of CWC25 Splicing Factor in Rice via CRISPR/Cas9. Ph.D. Thesis, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia, 2019. [Google Scholar]
- Park, H.J.; You, Y.N.; Lee, A.; Jung, H.; Jo, S.H.; Oh, N.; Kim, H.S.; Lee, H.J.; Kim, J.K.; Kim, Y.S.; et al. OsFKBP20-1b interacts with the splicing factor OsSR45 and participates in the environmental stress response at the post-transcriptional level in rice. Plant J. 2020, 102, 992–1007. [Google Scholar] [CrossRef]
- Zhang, P.; Li, S.; Chen, M. Characterization and function of circular RNAs in plants. Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef]
- Ye, C.Y.; Chen, L.; Liu, C.; Zhu, Q.H.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol. 2015, 208, 88–95. [Google Scholar] [CrossRef]
- Wang, K.; Wang, C.; Guo, B.; Song, K.; Shi, C.; Jiang, X.; Wang, K.; Tan, Y.; Wang, L.; Wang, L.; et al. CropCircDB: A comprehensive circular RNA resource for crops in response to abiotic stress. Database 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Zhang, H.; Liu, S.; Wang, Y.; Gao, Y.; Xi, F.; Zhao, L.; Liu, B.; Reddy, A.S.; et al. The interplay between microRNA and alternative splicing of linear and circular RNAs in eleven plant species. Bioinformatics 2019, 35, 3119–3126. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Bindereif, A. Circular RNAs: Coding or noncoding? Cell Res. 2017, 27, 724–725. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Xi, F.; Wang, H.; Han, X.; Wei, W.; Zhang, H.; Zhang, Q.; Zheng, Y.; Zhu, Q.; et al. Profiling of circular RNA N6 -methyladenosine in moso bamboo (Phyllostachys edulis) using nanopore-based direct RNA sequencing. J. Integr. Plant Biol. 2020, 62, 1823–1838. [Google Scholar] [CrossRef]
- Richardson, D.N.; Rogers, M.F.; Labadorf, A.; Ben-Hur, A.; Guo, H.; Paterson, A.H.; Reddy, A.S. Comparative Analysis of Serine/Arginine-Rich Proteins across 27 Eukaryotes: Insights into Sub-Family Classification and Extent of Alternative Splicing. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-A.; Huang, C.-K.; Huang, W.-S.; Huang, T.-S.; Liu, H.-Y.; Chen, Y.-F. DEAD-Box RNA Helicase 42 plays a critical role in pre-mRNA splicing under cold stress. Plant Physiol. 2020, 182, 255–271. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Piatek, A.; Li, L.; Reddy, A.S.N.; Mahfouz, M.M. Multiplex CRISPR mutagenesis of the Serine/arginine-rich (SR) gene family in rice. Genes 2019, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M. Disease resistance mechanisms in plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef]
- Yang, S.; Tang, F.; Zhu, H. Alternative splicing in plant immunity. Int. J. Mol. Sci. 2014, 15, 10424–10445. [Google Scholar] [CrossRef]
- Jiang, G.; Yin, D.; Shi, Y.; Zhou, Z.; Li, C.; Liu, P.; Jia, Y.; Wang, Y.; Liu, Z.; Yu, M.; et al. OsNPR3.3-dependent salicylic acid signaling is involved in recessive gene xa5-mediated immunity to rice bacterial blight. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef]
- Peng, Y.; Bartley, L.E.; Chen, X.; Dardick, C.; Chern, M.; Ruan, R.; Canlas, P.E.; Ronald, P.C. OsWRKY62 is a Negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 2008, 1, 446–458. [Google Scholar] [CrossRef]
- Xiao, X.; Tang, Z.; Li, X.; Hong, Y.; Li, B.; Xiao, W.; Gao, Z.; Lin, D.; Li, C.; Luo, L.; et al. Overexpressing OsMAPK12-1 inhibits plant growth and enhances resistance to bacterial disease in rice. Funct. Plant Biol. 2017, 44, 694. [Google Scholar] [CrossRef]
- Liu, Q.; Ning, Y.; Zhang, Y.; Yu, N.; Zhao, C.; Zhan, X.; Wu, W.; Chen, D.; Wei, X.; Wang, G.-L.; et al. OsCUL3a negatively regulates cell death and immunity by degrading OsNPR1 in rice. Plant Cell 2017, 29, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, H.; Chern, M.; Yin, J.; Chen, Y.; Wang, J.; Zhu, X.; Chen, Z.; Yuan, C.; Zhao, W.; et al. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2018, 115, 3174–3179. [Google Scholar]
- Al-Bader, N.; Meier, A.; Geniza, M.; Gongora, Y.S.; Oard, J.; Jaiswal, P. Loss of premature stop codon in the wall-associated kinase 91 (OsWAK91) gene confers sheath blight disease resistance in rice. bioRxiv 2019. [Google Scholar] [CrossRef]
- Chen, X.; Hao, L.; Pan, J.; Zheng, X.; Jiang, G.; Jin, Y.; Gu, Z.; Qian, Q.; Zhai, W.; Ma, B. SPL5, a cell death and defense-related gene, encodes a Putative Splicing Factor 3b subunit 3 (SF3b3) in rice. Mol. Breed. 2012, 30, 939–949. [Google Scholar] [CrossRef]
- Chen, X.; Fu, S.; Zhang, P.; Gu, Z.; Liu, J.; Qian, Q.; Ma, B. Proteomic analysis of a disease-resistance-enhanced lesion mimic mutant spotted leaf 5 in rice. Rice 2013, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Zhou, X.; Jiang, B.; Gu, Z.; Zhang, P.; Qian, Q.; Chen, X.; Ma, B. Transcriptome profiling of the spl5 mutant reveals that SPL5 has a negative role in the biosynthesis of serotonin for rice disease resistance. Rice 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, P.; Segundo, B.S. MicroRNAs in rice innate immunity. Rice 2016, 9. [Google Scholar] [CrossRef]
- Li, Y.; Feng, H. Cooperation of alternative splicing and microRNA targeting in the gene regulation network of Arabidopsis thaliana. Preprints 2020. [Google Scholar] [CrossRef]
- Peris-Peris, C.; Serra-Cardona, A.; Sánchez-Sanuy, F.; Campo, S.; Ariño, J.; San Segundo, B. Two NRAMP6 isoforms function as iron and manganese transporters and contribute to disease resistance in rice. Mol. Plant Microbe Interact. 2017, 30, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Quan, W.; Li, G.-B.; Hu, X.-H.; Wang, Q.; Wang, H.; Li, X.-P.; Luo, X.; Feng, Q.; Hu, Z.-J.; et al. circRNAs are involved in the rice-Magnaporthe oryzae interaction. Plant Physiol. 2020, 182, 272–286. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Gheysen, G.; Denil, S.; Lindsey, K.; Topping, J.F.; Nahar, K.; Haegeman, A.; De Vos, W.H.; Trooskens, G.; Van Criekinge, W.; et al. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J. Exp. Bot. 2013, 64, 3885–3898. [Google Scholar] [CrossRef] [PubMed]
- Bagnaresi, P.; Biselli, C.; Orrù, L.; Urso, S.; Crispino, L.; Abbruscato, P.; Piffanelli, P.; Lupotto, E.; Cattivelli, L.; Valè, G. Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS ONE 2012, 7, e51609. [Google Scholar] [CrossRef] [PubMed]
- Venu, R.C.; Sheshu Madhav, M.; Sreerekha, M.V.; Nobuta, K.; Zhang, Y.; Carswell, P.; Boehm, M.J.; Meyers, B.C.; Korth, K.L.; Wang, G.-L. Deep and comparative transcriptome analysis of rice plants infested by the Beet Armyworm (Spodoptera exigua) and Water Weevil (Lissorhoptrus oryzophilus). Rice 2010, 3, 22–35. [Google Scholar] [CrossRef]
- Jung, K.-H.; Bartley, L.E.; Cao, P.; Canlas, P.E.; Ronald, P.C. Analysis of Alternatively Spliced Rice Transcripts Using microarray data. Rice 2008, 2, 44–55. [Google Scholar] [CrossRef]
- Muhammad, S.; Tan, J.; Deng, P.; Li, T.; He, H.; Bian, J.; Wu, L. Pesticide application has little influence on coding and non-coding gene expressions in rice. BMC Genom. 2019, 20. [Google Scholar] [CrossRef]
- van der Does, H.C.; Rep, M. Adaptation to the host environment by plant-pathogenic fungi. Annu. Rev. Phytopathol. 2017, 55, 427–450. [Google Scholar] [CrossRef]
- Liao, Z.-X.; Ni, Z.; Wei, X.-L.; Chen, L.; Li, J.-Y.; Yu, Y.-H.; Jiang, W.; Jiang, B.-L.; He, Y.-Q.; Huang, S. Dual RNA-seq of Xanthomonas oryzae pv. oryzicola infecting rice reveals novel insights into bacterial-plant interaction. PLoS ONE 2019, 14. [Google Scholar] [CrossRef]
- Zhang, J.; Guan, W.; Huang, C.; Hu, Y.; Chen, Y.; Guo, J.; Zhou, C.; Chen, R.; Du, B.; Zhu, L.; et al. Combining next-generation sequencing and single-molecule sequencing to explore brown plant hopper responses to contrasting genotypes of japonica rice. BMC Genom. 2019, 20. [Google Scholar] [CrossRef]
- Xia, Y.; Fei, B.; He, J.; Zhou, M.; Zhang, D.; Pan, L.; Li, S.; Liang, Y.; Wang, L.; Zhu, J.; et al. Transcriptome analysis reveals the host selection fitness mechanisms of the Rhizoctonia solani AG1IA pathogen. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Ebbole, D.J.; Jin, Y.; Thon, M.; Pan, H.; Bhattarai, E.; Thomas, T.; Dean, R. Gene discovery and gene expression in the rice blast fungus, Magnaporthe grisea: Analysis of expressed sequence tags. Mol. Plant Microbe Interact. 2004, 17, 1337–1347. [Google Scholar] [CrossRef]
- Li, Z.; Wu, L.; Wu, H.; Zhang, X.; Mei, J.; Zhou, X.; Wang, G.L.; Liu, W. Arginine methylation is required for remodelling pre-mRNA splicing and induction of autophagy in rice blast fungus. New Phytol. 2019, 225, 413–429. [Google Scholar] [CrossRef]
- Franceschetti, M.; Bueno, E.; Wilson, R.A.; Tucker, S.L.; Gómez-Mena, C.; Calder, G.; Sesma, A. Fungal virulence and development is regulated by alternative pre-mRNA 3′end processing in Magnaporthe oryzae. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, Z.; Xing, J.; Yang, Q.; Chen, X.-L. Genome-wide identification and characterization of circular RNAs in the rice blast fungus Magnaporthe oryzae. Sci. Rep. 2018, 8, 1–18. [Google Scholar] [CrossRef]
- Wan, P.-J.; Zhou, R.-N.; Nanda, S.; He, J.-C.; Yuan, S.-Y.; Wang, W.-X.; Lai, F.-X.; Fu, Q. Phenotypic and transcriptomic responses of two Nilaparvata lugens populations to the Mudgo rice containing Bph1. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Rao, W.; Zheng, X.; Liu, B.; Guo, Q.; Guo, J.; Wu, Y.; Shangguan, X.; Wang, H.; Wu, D.; Wang, Z.; et al. Secretome Analysis and In Planta Expression of Salivary Proteins Identify Candidate Effectors from the Brown Planthopper Nilaparvata lugens. Mol. Plant Microbe Interact. 2019, 32, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chen, L.; Li, D.; Kang, K.; Liu, K.; Yue, L.; Zhang, W. Two alternative splicing variants of a sugar gustatory receptor modulate fecundity through different signalling pathways in the brown planthopper, Nilaparvata lugens. J. Insect Physiol. 2019, 119, 103966. [Google Scholar] [CrossRef]
- Fu, X.; Li, T.; Chen, J.; Dong, Y.; Qiu, J.; Kang, K.; Zhang, W. Functional screen for microRNAs of Nilaparvata lugens reveals that targeting of glutamine synthase by miR-4868b regulates fecundity. J. Insect Physiol. 2015, 83, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.Y.; Merzendorfer, H.; Zhang, W.; Zhang, J.; Muthukrishnan, S. Biosynthesis, turnover, and functions of chitin in insects. Annu. Rev. Entomol. 2016, 61, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, H.; Zhou, C.; Yang, W.-J.; Jin, D.-C.; Long, G.-Y. Molecular cloning, expression, and functional analysis of the chitin synthase 1 gene and its two alternative splicing variants in the white-backed Planthopper, Sogatella furcifera (Hemiptera: Delphacidae). Sci. Rep. 2019, 9, 1–4. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, H.-W.; Huang, H.-J.; Xue, J.; Wu, W.-J.; Bao, Y.-Y.; Xu, H.-J.; Zhu, Z.-R.; Cheng, J.-A.; Zhang, C.-X. Chitin synthase 1 gene and its two alternative splicing variants from two sap-sucking insects, Nilaparvata lugens and Laodelphax striatellus (Hemiptera: Delphacidae). Insect Biochem. Mol. Biol. 2012, 42, 637–646. [Google Scholar] [CrossRef]
- Xiao, H.; Yuan, Z.; Guo, D.; Hou, B.; Yin, C.; Zhang, W.; Li, F. Genome-wide identification of long noncoding RNA genes and their potential association with fecundity and virulence in rice Brown planthopper, Nilaparvata lugens. BMC Genom. 2015, 16. [Google Scholar] [CrossRef]
- Ng, T.H.; Chiang, Y.-A.; Yeh, Y.-C.; Wang, H.-C. Review of dscam-mediated immunity in shrimp and other arthropods. Dev. Comp. Immunol. 2014, 46, 129–138. [Google Scholar] [CrossRef]
- Zhang, F.; Li, Q.; Chen, X.; Huo, Y.; Guo, H.; Song, Z.; Cui, F.; Zhang, L.; Fang, R. Roles of the Laodelphax striatellusDown Syndrome cell adhesion molecule in rice stripe virus infection of its insect vector. Insect Mol. Biol. 2016, 25, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.Y.; Ye, W.Y.; Xiao, H.M.; Li, M.Z.; Cao, Z.H.; Ye, X.H.; Zhao, X.X.; Kang, H.E.; Fei, L.I. LncRNAs are potentially involved in the immune interaction between small brown planthopper and rice stripe virus. J. Integr. Agric. 2019, 18, 2814–2822. [Google Scholar] [CrossRef]
- Gao, X.; Yin, C.; Liu, X.; Peng, J.; Chen, D.; He, D.; Shi, W.; Zhao, W.; Yang, J.; Peng, Y.-L. A glycine-rich protein MoGrp1 functions as a novel Splicing factor to regulate fungal virulence and growth in Magnaporthe oryzae. Phytopathol. Res. 2019, 1. [Google Scholar] [CrossRef]
- Zhuo, J.-C.; Zhang, H.-H.; Xie, Y.-C.; Li, H.-J.; Hu, Q.-L.; Zhang, C.-X. Identification of a female determinant gene for the sexual determination of a hemipteran insect, the brown planthopper. bioRxiv 2019. [Google Scholar] [CrossRef]
- Jia, J.; Long, Y.; Zhang, H.; Li, Z.; Liu, Z.; Zhao, Y.; Lu, D.; Jin, X.; Deng, X.; Xia, R.; et al. Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants. Nat. Plants 2020, 6, 780–788. [Google Scholar] [CrossRef]
- Li, S.; Wang, Y.; Zhao, Y.; Zhao, X.; Chen, X.; Gong, Z. Global co-transcriptional splicing in Arabidopsis and the correlation with splicing regulation in mature RNAs. Mol. Plant 2020, 13, 266–277. [Google Scholar] [CrossRef]
- Zhu, D.; Mao, F.; Tian, Y.; Lin, X.; Gu, L.; Gu, H.; Qu, L.J.; Wu, Y.; Wu, Z. The features and regulation of co-transcriptional splicing in Arabidopsis. Mol. Plant 2020, 13, 278–294. [Google Scholar] [CrossRef]
- Jabre, I.; Chaudhary, S.; Guo, W.; Kalyna, M.; Reddy, A.S.; Chen, W.; Zhang, R.; Wilson, C.; Syed, N.H. Differential nucleosome occupancy modulates alternative splicing in Arabidopsis thaliana. New Phytol. 2020, 229, 1937–1945. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, P.; Jin, G.; Gui, D.; Liu, L.; Zhang, C. Temporal regulation of alternative splicing events in rice memory under drought stress. Plant Divers. 2020. [Google Scholar] [CrossRef]
- Yuan, J.; Ma, Y.; Huang, T.; Chen, Y.; Peng, Y.; Li, B.; Chang, X. Genetic Modulation of RNA Splicing with a CRISPR-Guided Cytidine Deaminase. Mol. Cell 2018, 72, 380–394. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Zhang, H.; Lin, Q.; Fan, R.; Gao, C. Manipulating mRNA splicing by base editing in plants. Sci. China Life Sci. 2018, 61, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Joy, N.; MaimoonathBeevi, Y.P.; Soniya, E.V. A deeper view into the significance of simple sequence repeats in pre-miRNAs provides clues for its possible roles in determining the function of microRNAs. BMC Genet. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Khokhar, W.; Hassan, M.A.; Reddy, A.S.; Chaudhary, S.; Jabre, I.; Byrne, L.J.; Syed, N.H. Genome-Wide identification of Splicing quantitative Trait Loci (sQTLs) in Diverse Ecotypes of Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1160. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganie, S.A.; Reddy, A.S.N. Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance. Biology 2021, 10, 309. https://doi.org/10.3390/biology10040309
Ganie SA, Reddy ASN. Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance. Biology. 2021; 10(4):309. https://doi.org/10.3390/biology10040309
Chicago/Turabian StyleGanie, Showkat Ahmad, and Anireddy S. N. Reddy. 2021. "Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance" Biology 10, no. 4: 309. https://doi.org/10.3390/biology10040309
APA StyleGanie, S. A., & Reddy, A. S. N. (2021). Stress-Induced Changes in Alternative Splicing Landscape in Rice: Functional Significance of Splice Isoforms in Stress Tolerance. Biology, 10(4), 309. https://doi.org/10.3390/biology10040309






