Knockout of myoc Provides Evidence for the Role of Myocilin in Zebrafish Sex Determination Associated with Wnt Signalling Downregulation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. CRISPR/Cas 9 Gene Editing
2.3. Zebrafish DNA Extraction
2.4. Genotyping of CRISPR/Cas9-Induced Mutations by PAGE and Sanger Sequencing
2.5. Quantitative Reverse Transcription PCR (qRT-PCR)
2.6. Zebrafish Tissue Samples
2.7. Fluorescent Whole Mount Immunohistochemistry (FWIHC)
2.8. Fluorescence Immunohistochemistry
2.9. Hematoxilin and Eosin Staining
2.10. High Throughput RNA Sequencing
2.11. Statistics
3. Results
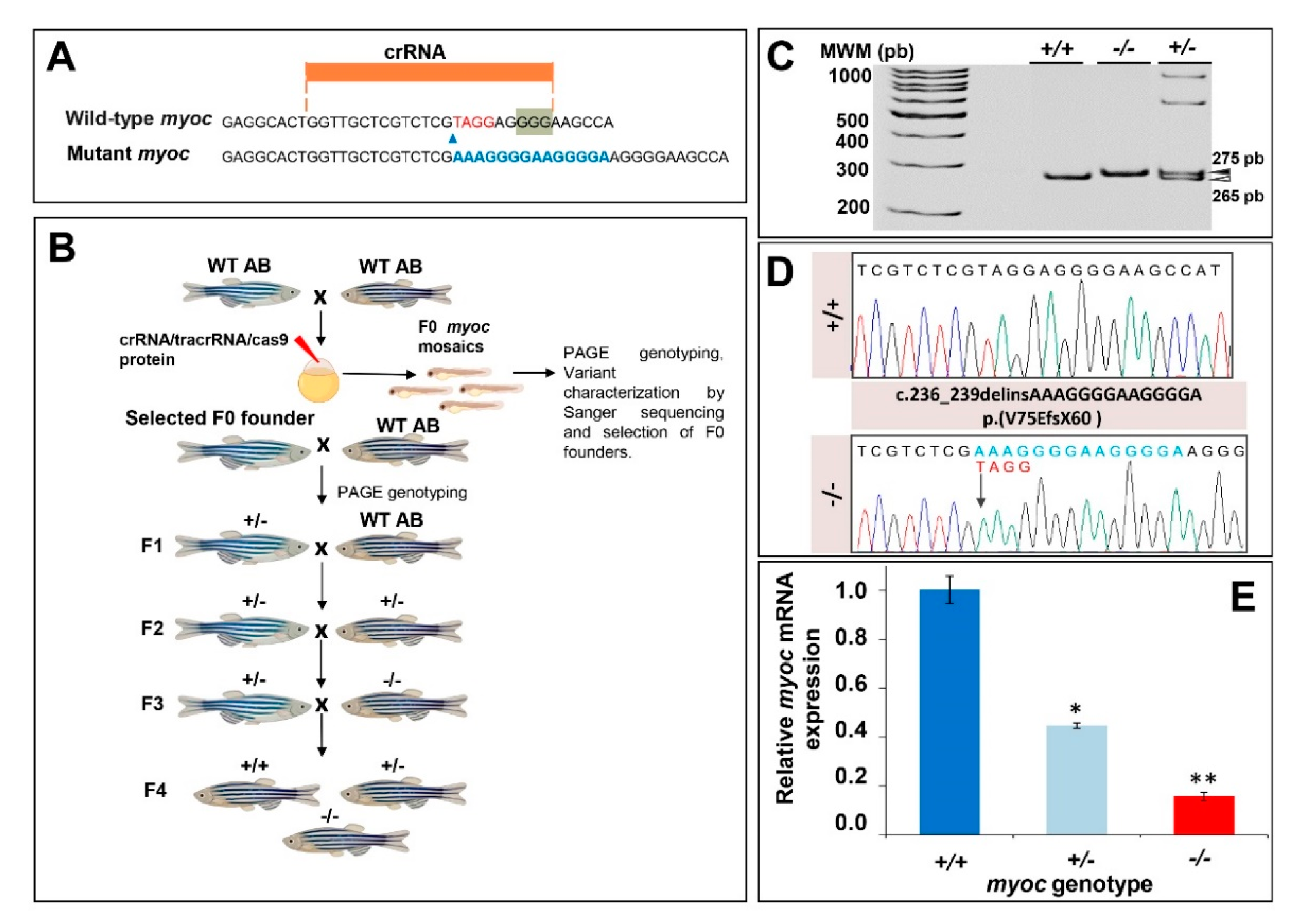
3.1. Generation of the KO Myoc Line in Zebrafish Using CRISPR/Cas9
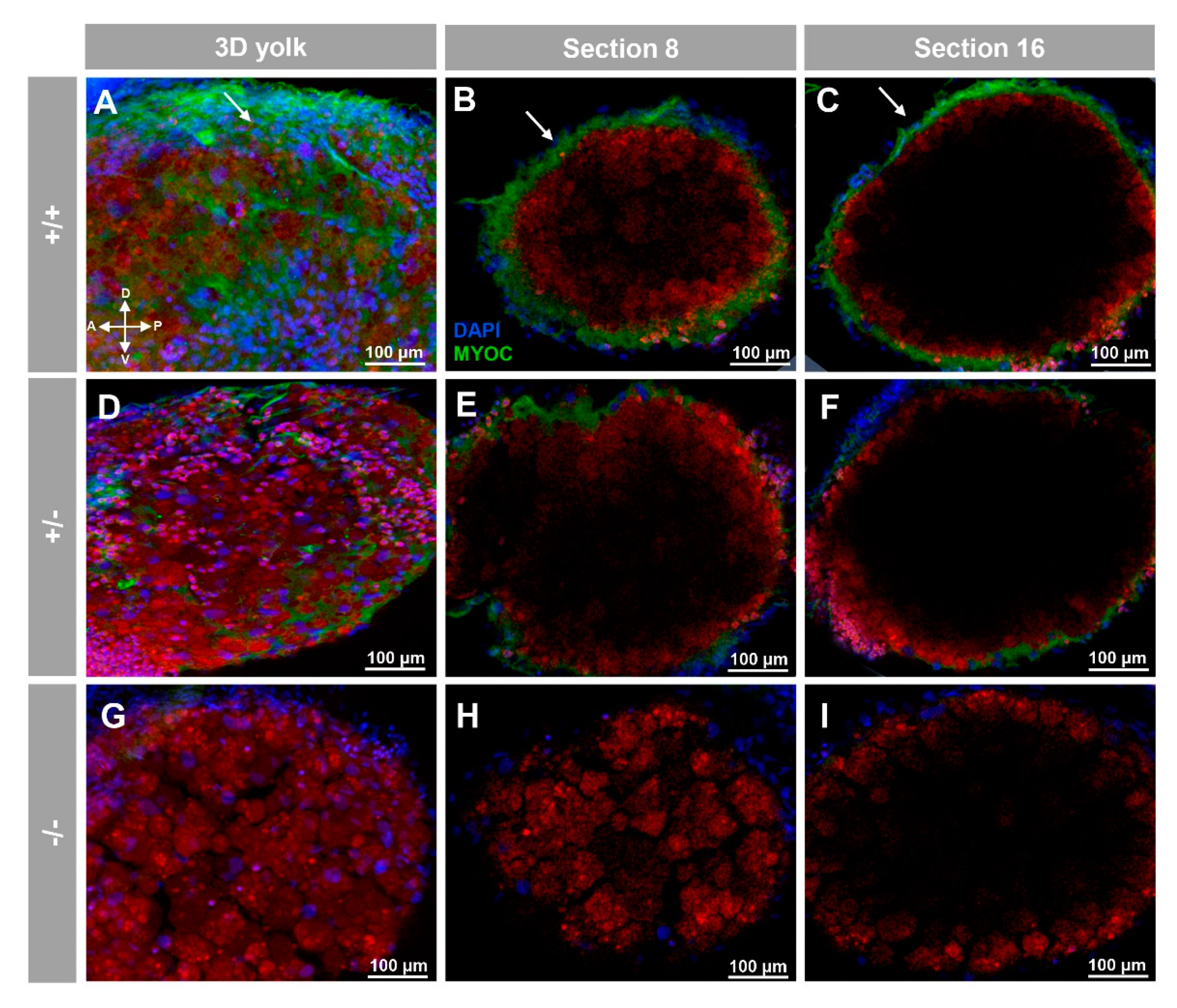
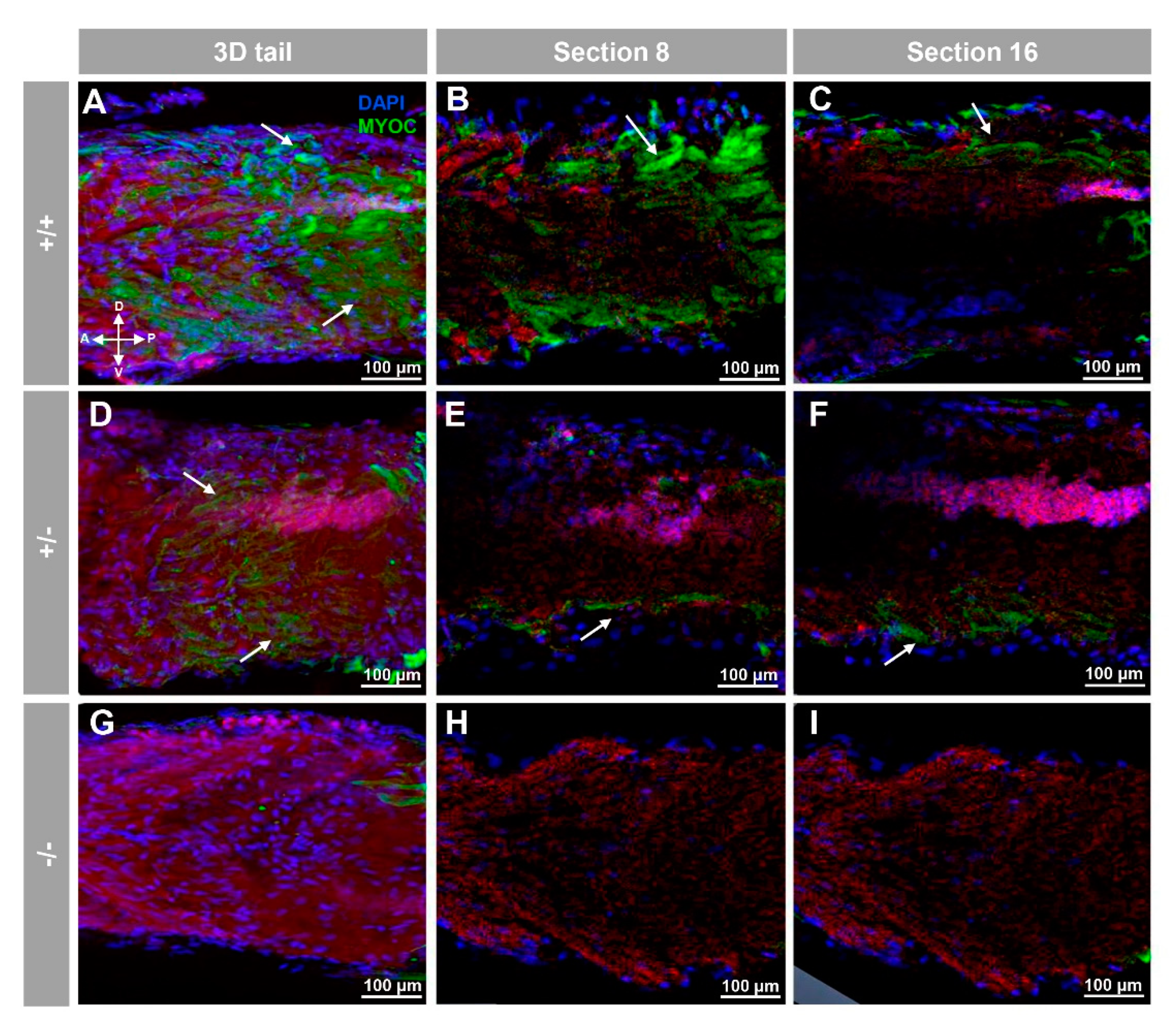
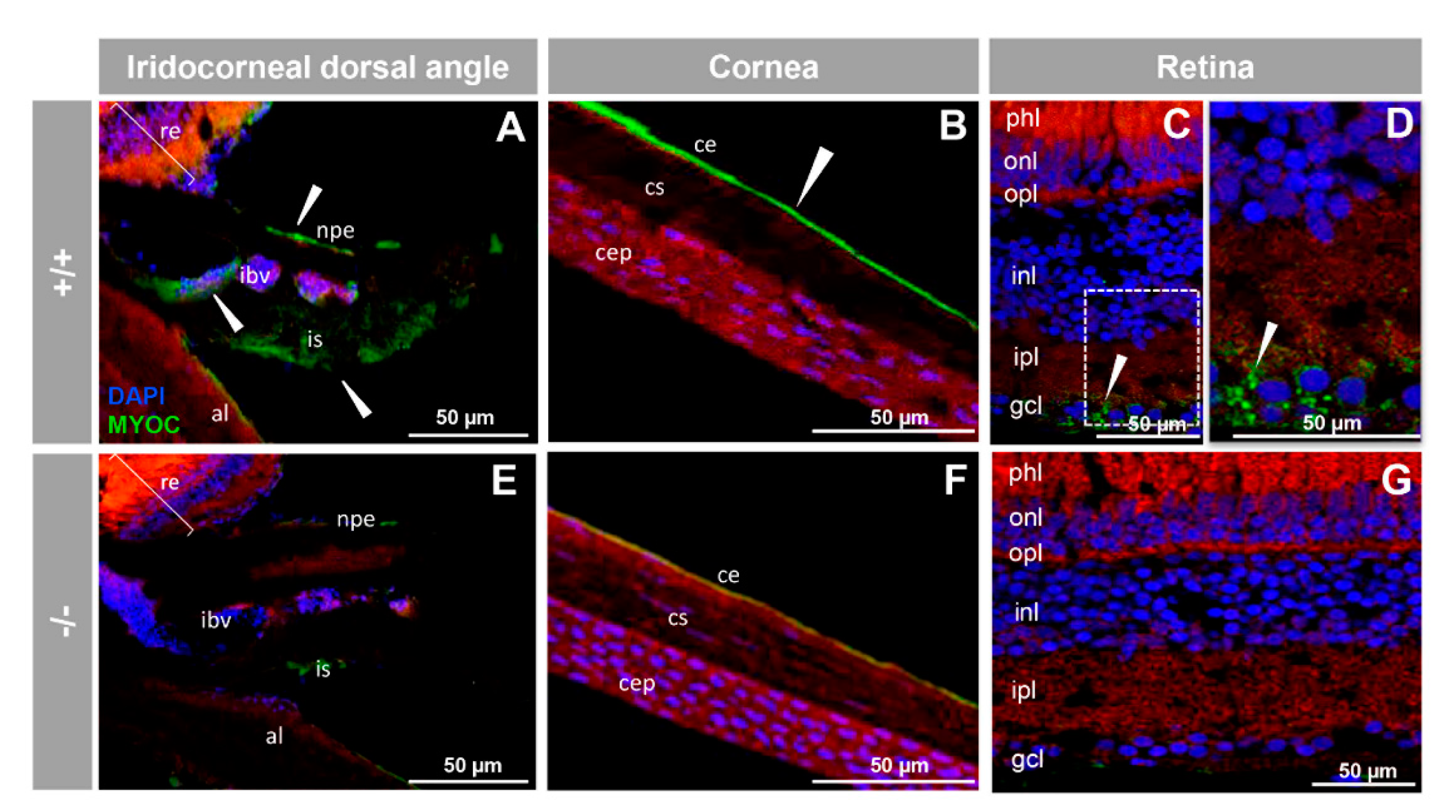
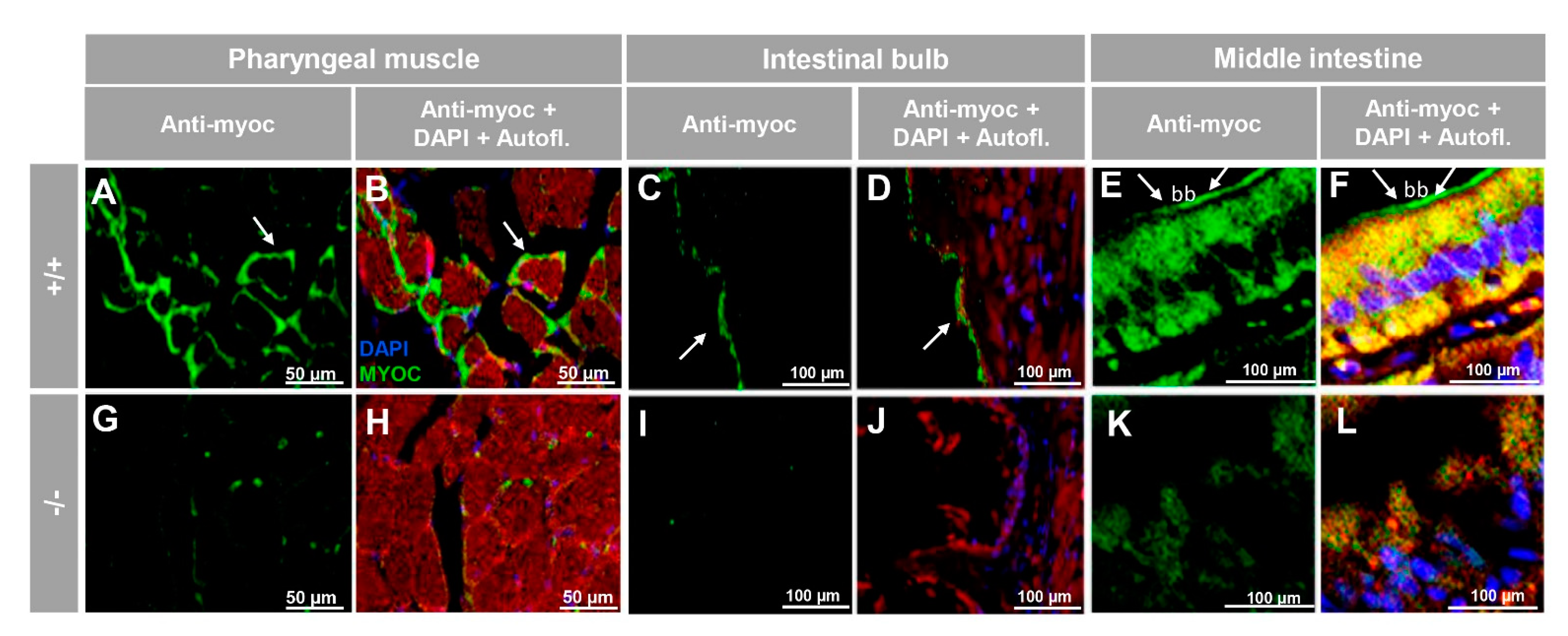
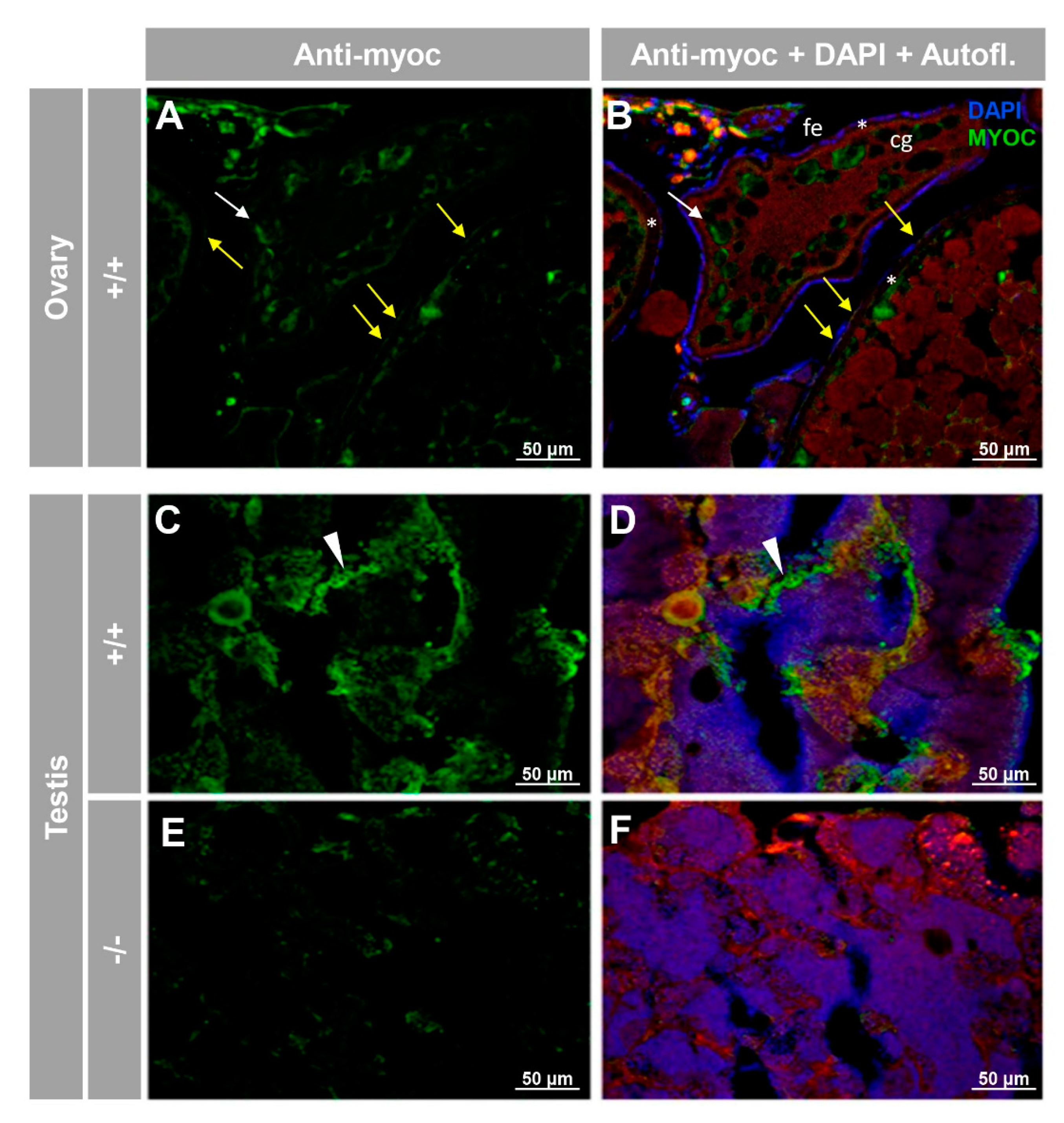
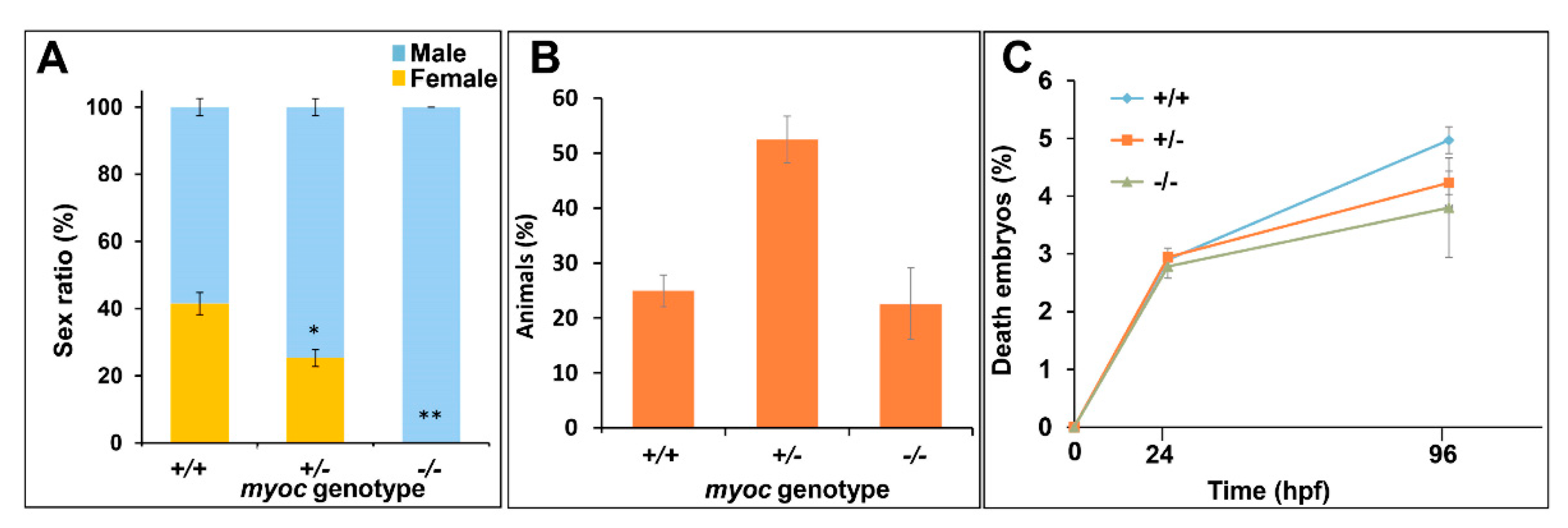
3.2. Expression of Myoc and Phenotypic Characterisation of the KO Zebrafish Line
3.3. Histology and Terminal dUTP Nick-End Labeling (TUNEL) of the Immature Gonad of the Myoc KO Zebrafish Line
3.4. Comparison of Transcriptomic Profiles of Adult Male −/− and +/+ Zebrafish
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stone, E.M.; Fingert, J.H.; Alward, W.L.M.; Nguyen, T.D.; Polansky, J.R.; Sunden, S.L.F.; Nishimura, D.; Clark, A.F.; Nystuen, A.; Nichols, B.E.; et al. Identification of a Gene That Causes Primary Open Angle Glaucoma. Science 1997, 275, 668–670. [Google Scholar] [CrossRef] [PubMed]
- Quigley, H.A. Ganglion cell death in glaucoma: Pathology recapitulates ontogeny. Aust. N. Z. J. Ophthalmol. 1995, 23, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Polansky, J.R.; Fauss, D.J.; Chen, P.; Chen, H.; Lütjen-Drecoll, E.; Johnson, D.; Kurtz, R.M.; Ma, Z.-D.; Bloom, E.; Nguyen, T.D. Cellular Pharmacology and molecular Biology of the Trabecular Meshwork Inducible glucocorticoid Response Gene Product. Ophthalmologica 1997, 211, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Kubota, R.; Noda, S.; Wang, Y.; Minoshima, S.; Asakawa, S.; Kudoh, J.; Mashima, Y.; Oguchi, Y.; Shimizu, N. A novel myosin-like protein (myocilin) expressed in the connecting cilium of the photoreceptor: Molecular cloning, tissue expression, and chromosomal mapping. Genomics 1997, 41, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Escribano, J.; Ortego, J.; Coca-Prados, M. Isolation and Characterization of Cell-Specific cDNA Clones from a Subtractive Library of the Ocular Ciliary Body of a Single Normal Human Donor: Transcription and Synthesis of Plasma Proteins. J. Biochem. 1995, 118, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Ortego, J.; Escribano, J.; Coca-Prados, M. Cloning and characterization of subtracted cDNAs from a human ciliary body library encoding TIGR, a protein involved in juvenile open angle glaucoma with homology to myosin and olfactomedin. FEBS Lett. 1997, 413, 349–353. [Google Scholar] [CrossRef]
- Huang, W.; Jaroszewski, J.; Ortego, J.; Escribano, J.; Coca-Prados, M. Expression of the TIGR gene in the iris, ciliary body, and trabecular meshwork of the human eye. Ophthalmic Genet. 2000, 21, 155–169. [Google Scholar] [CrossRef]
- Karali, A.; Russell, P.; Stefani, F.H.; Tamm, E.R. Localization of myocilin/trabecular meshwork--inducible glucocorticoid response protein in the human eye. Investig. Ophthalmol. Vis. Sci. 2000, 41, 729–740. [Google Scholar]
- Ezzat, M.-K.; Howell, K.G.; Bahler, C.K.; Beito, T.G.; Loewen, N.A.; Poeschla, E.M.; Fautsch, M.P. Characterization of monoclonal antibodies against the glaucoma-associated protein myocilin. Exp. Eye Res. 2008, 87, 376–384. [Google Scholar] [CrossRef]
- Russell, P.; Tamm, E.R.; Grehn, F.J.; Picht, G.; Johnson, M. The presence and properties of myocilin in the aqueous humor. Investig. Ophthalmol. Vis. Sci. 2001, 42, 983–986. [Google Scholar]
- Aroca-Aguilar, J.D.; Sánchez-Sánchez, F.; Ghosh, S.; Coca-Prados, M.; Escribano, J. Myocilin Mutations Causing Glaucoma Inhibit the Intracellular Endoproteolytic Cleavage of Myocilin between Amino Acids Arg226 and Ile227. J. Biol. Chem. 2005, 280, 21043–21051. [Google Scholar] [CrossRef] [PubMed]
- Fautsch, M.P.; Johnson, D.H. Characterization of myocilin-myocilin interactions. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2324–2331. [Google Scholar]
- Fautsch, M.P.; Vrabel, A.M.; Peterson, S.L.; Johnson, D.H. In vitro and in vivo characterization of disulfide bond use in myocilin complex formation. Mol. Vis. 2004, 10, 417–425. [Google Scholar] [PubMed]
- Aroca-Aguilar, J.-D.; Fernández-Navarro, A.; Ontañón, J.; Coca-Prados, M.; Escribano, J. Identification of myocilin as a blood plasma protein and analysis of its role in leukocyte adhesion to endothelial cell monolayers. PLoS ONE 2018, 13, e0209364. [Google Scholar] [CrossRef] [PubMed]
- Hardy, K.M.; Hoffman, E.A.; Gonzalez, P.; McKay, B.S.; Stamer, W.D. Extracellular Trafficking of Myocilin in Human Trabecular Meshwork Cells. J. Biol. Chem. 2005, 280, 28917–28926. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.A.; Perkumas, K.M.; Highstrom, L.M.; Stamer, W.D. Regulation of Myocilin-Associated Exosome Release from Human Trabecular Meshwork Cells. Investig. Ophthalmol. Vis. Sci. 2009, 50, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Gobeil, S.; Letartre, L.; Raymond, V. Functional analysis of the glaucoma-causing TIGR/myocilin protein: Integrity of amino-terminal coiled-coil regions and olfactomedin homology domain is essential for extracellular adhesion and secretion. Exp. Eye Res. 2006, 82, 1017–1029. [Google Scholar] [CrossRef]
- Stamer, W.D.; Perkumas, K.M.; Hoffman, E.A.; Roberts, B.C.; Epstein, D.L.; McKay, B.S. Coiled–coil targeting of myocilin to intracellular membranes. Exp. Eye Res. 2006, 83, 1386–1395. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, F.; Martínez-Redondo, F.; Aroca-Aguilar, J.D.; Coca-Prados, M.; Escribano, J. Characterization of the Intracellular Proteolytic Cleavage of Myocilin and Identification of Calpain II as a Myocilin-processing Protease. J. Biol. Chem. 2007, 282, 27810–27824. [Google Scholar] [CrossRef]
- Anholt, R.R.H. Olfactomedin proteins: Central players in development and disease. Front. Cell Dev. Biol. 2014, 2, 6. [Google Scholar] [CrossRef]
- Donegan, R.K.; Hill, S.E.; Freeman, D.M.; Nguyen, E.; Orwig, S.D.; Turnage, K.C.; Lieberman, R.L. Structural basis for misfolding in myocilin-associated glaucoma. Hum. Mol. Genet. 2015, 24, 2111–2124. [Google Scholar] [CrossRef] [PubMed]
- Hill, S.E.; Nguyen, E.; Donegan, R.K.; Patterson-Orazem, A.C.; Hazel, A.; Gumbart, J.C.; Lieberman, R.L. Structure and Misfolding of the Flexible Tripartite Coiled-Coil Domain of Glaucoma-Associated Myocilin. Structure 2017, 25, 1697–1707.e5. [Google Scholar] [CrossRef] [PubMed]
- Aroca-Aguilar, J.-D.; Martínez-Redondo, F.; Sánchez-Sánchez, F.; Coca-Prados, M.; Escribano, J. Functional Role of Proteolytic Processing of Recombinant Myocilin in Self-Aggregation. Investig. Ophthalmology Vis. Sci. 2010, 51, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Aroca-Aguilar, J.-D.; Sánchez-Sánchez, F.; Ghosh, S.; Fernández-Navarro, A.; Coca-Prados, M.; Escribano, J. Interaction of Recombinant Myocilin with the Matricellular Protein SPARC: Functional Implications. Investig. Ophthalmol. Vis. Sci. 2011, 52, 179–189. [Google Scholar] [CrossRef]
- Furutani, Y.; Manabe, R.-I.; Tsutsui, K.; Yamada, T.; Sugimoto, N.; Fukuda, S.; Kawai, J.; Sugiura, N.; Kimata, K.; Hayashizaki, Y.; et al. Identification and characterization of photomedins: Novel olfactomedin-domain-containing proteins with chondroitin sulphate-E-binding activity. Biochem. J. 2005, 389, 675–684. [Google Scholar] [CrossRef]
- Maertens, B.; Hopkins, D.; Franzke, C.-W.; Keene, D.R.; Bruckner-Tuderman, L.; Greenspan, D.S.; Koch, M. Cleavage and Oligomerization of Gliomedin, a Transmembrane Collagen Required for Node of Ranvier Formation. J. Biol. Chem. 2007, 282, 10647–10659. [Google Scholar] [CrossRef]
- Volynski, K.E.; Silva, J.P.; Lelianova, V.G.; Atiqur Rahman, M.; Hopkins, C.; Ushkaryov, Y.A. Latrophilin fragments behave as independent proteins that associate and signal on binding of LTX(N4C). EMBO J. 2004, 23, 4423–4433. [Google Scholar] [CrossRef]
- Aroca-Aguilar, J.-D.; Martínez-Redondo, F.; Martin-Gil, A.; Pintor, J.; Coca-Prados, M.; Escribano, J. Bicarbonate-Dependent Secretion and Proteolytic Processing of Recombinant Myocilin. PLoS ONE 2013, 8, e54385. [Google Scholar] [CrossRef]
- Kwon, H.-S.; Lee, H.-S.; Ji, Y.; Rubin, J.S.; Tomarev, S.I. Myocilin Is a Modulator of Wnt Signaling. Mol. Cell. Biol. 2009, 29, 2139–2154. [Google Scholar] [CrossRef]
- Shen, X.; Ying, H.; Yue, B.Y.J.T. Wnt Activation by Wild Type and Mutant Myocilin in Cultured Human Trabecular Meshwork Cells. PLoS ONE 2012, 7, e44902. [Google Scholar] [CrossRef]
- Wentz-Hunter, K.; Kubota, R.; Shen, X.; Yue, B.Y. Extracellular myocilin affects activity of human trabecular meshwork cells. J. Cell. Physiol. 2004, 200, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Wentz-Hunter, K.; Shen, X.; Okazaki, K.; Tanihara, H.; Yue, B.Y. Overexpression of myocilin in cultured human trabecular meshwork cells. Exp. Cell Res. 2004, 297, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.A.; High, S.K.; McCluskey, B.M.; Amores, A.; Yan, Y.-L.; Titus, T.A.; Anderson, J.L.; Batzel, P.; Carvan, M.J.; Schartl, M.; et al. Wild Sex in Zebrafish: Loss of the Natural Sex Determinant in Domesticated Strains. Genetics 2014, 198, 1291–1308. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; Luzio, A.; Coimbra, A.M. Zebrafish sex differentiation and gonad development: A review on the impact of environmental factors. Aquat. Toxicol. 2017, 191, 141–163. [Google Scholar] [CrossRef] [PubMed]
- Leerberg, D.M.; Sano, K.; Draper, B.W. Fibroblast growth factor signaling is required for early somatic gonad development in zebrafish. PLoS Genet. 2017, 13, e1006993. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Marí, A.; Yan, Y.-L.; BreMiller, R.A.; Wilson, C.; Cañestro, C.; Postlethwait, J.H. Characterization and expression pattern of zebrafish anti-Müllerian hormone (amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns 2005, 5, 655–667. [Google Scholar] [CrossRef]
- Wang, X.G.; Bartfai, R.; Sleptsova-Freidrich, I.; Orban, L. The timing and extent of ‘juvenile ovary’ phase are highly variable during zebrafish testis differentiation. J. Fish Biol. 2007, 70, 33–44. [Google Scholar] [CrossRef]
- Sreenivasan, R.; Jiang, J.; Wang, X.; Bártfai, R.; Kwan, H.Y.; Christoffels, A.; Orbán, L. Gonad Differentiation in Zebrafish Is Regulated by the Canonical Wnt Signaling Pathway. Biol. Reprod. 2014, 90, 45. [Google Scholar] [CrossRef]
- Monte, W. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 5th ed.; University of Oregon Press: Eugene, OR, USA, 2013. [Google Scholar]
- Meeker, N.D.; Hutchinson, S.A.; Ho, L.; Trede, N.S. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques 2007, 43, 610, 612, 614. [Google Scholar] [CrossRef]
- Morales-Cámara, S.; Alexandre-Moreno, S.; Bonet-Fernández, J.-M.; Atienzar-Aroca, R.; Aroca-Aguilar, J.-D.; Ferre-Fernández, J.-J.; Mendez, C.D.; Morales, L.; Fernández-Sánchez, L.; Cuenca, N.; et al. Role of GUCA1C in Primary Congenital Glaucoma and in the Retina: Functional Evaluation in Zebrafish. Genes 2020, 11, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ferre-Fernández, J.-J.; Aroca-Aguilar, J.-D.; Medina-Trillo, C.; Bonet-Fernández, J.-M.; Méndez-Hernández, C.D.; Morales-Fernández, L.; Corton, M.; Cabañero-Valera, M.-J.; Gut, M.; Tonda, R.; et al. Whole-Exome Sequencing of Congenital Glaucoma Patients Reveals Hypermorphic Variants in GPATCH3, a New Gene Involved in Ocular and Craniofacial Development. Sci. Rep. 2017, 7, 46175. [Google Scholar] [CrossRef] [PubMed]
- Bonet-Fernández, J.-M.; Aroca-Aguilar, J.-D.; Corton, M.; Ramírez, A.-I.; Alexandre-Moreno, S.; García-Antón, M.-T.; Salazar, J.-J.; Ferre-Fernández, J.-J.; Atienzar-Aroca, R.; Villaverde, C.; et al. CPAMD8 loss-of-function underlies non-dominant congenital glaucoma with variable anterior segment dysgenesis and abnormal extracellular matrix. Hum. Genet. 2020, 139, 1209–1231. [Google Scholar] [CrossRef] [PubMed]
- Ye, D.; Zhu, L.; Zhang, Q.; Xiong, F.; Wang, H.; Wang, X.; He, M.; Zhu, Z.; Sun, Y. Abundance of Early Embryonic Primordial Germ Cells Promotes Zebrafish Female Differentiation as Revealed by Lifetime Labeling of Germline. Mar. Biotechnol. 2019, 21, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Brogna, S.; Wen, J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef]
- Parichy, D.M.; Elizondo, M.R.; Mills, M.G.; Gordon, T.N.; Engeszer, R.E. Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Dev. Dyn. 2009, 238, 2975–3015. [Google Scholar] [CrossRef]
- Uchida, D.; Yamashita, M.; Kitano, T.; Iguchi, T. Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J. Exp. Biol. 2002, 205, 711–718. [Google Scholar]
- Lau, E.S.-W.; Zhang, Z.; Qin, M.; Ge, W. Knockout of Zebrafish Ovarian Aromatase Gene (cyp19a1a) by TALEN and CRISPR/Cas9 Leads to All-male Offspring Due to Failed Ovarian Differentiation. Sci. Rep. 2016, 6, 37357. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.D.; Chen, P.; Huang, W.D.; Chen, H.; Johnson, D.; Polansky, J.R. Gene Structure and Properties of TIGR, an Olfactomedin-related Glycoprotein Cloned from Glucocorticoid-induced Trabecular Meshwork Cells. J. Biol. Chem. 1998, 273, 6341–6350. [Google Scholar] [CrossRef] [PubMed]
- Fingert, J.H.; Ying, L.; Swiderski, R.E.; Nystuen, A.M.; Arbour, N.C.; Alward, W.L.; Sheffield, V.C.; Stone, E.M. Characterization and comparison of the human and mouse GLC1A glaucoma genes. Genome Res. 1998, 8, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yan, M.; Liu, Y.; Wang, R.; Li, C.; Deng, C.; Singh, A.; Coleman, W.G.; Rodgers, G.P. Olfactomedin 4 down-regulates innate immunity against Helicobacter pylori infection. Proc. Natl. Acad. Sci. USA 2010, 107, 11056–11061. [Google Scholar] [CrossRef]
- Kim, B.S.; Savinova, O.V.; Reedy, M.V.; Martin, J.; Lun, Y.; Gan, L.; Smith, R.S.; Tomarev, S.I.; John, S.W.M.; Johnson, R.L. Targeted Disruption of the Myocilin Gene (Myoc) Suggests that Human Glaucoma-Causing Mutations Are Gain of Function. Mol. Cell. Biol. 2001, 21, 7707–7713. [Google Scholar] [CrossRef]
- Lam, D.S.; Leung, Y.F.; Chua, J.K.; Baum, L.; Fan, D.S.; Choy, K.W.; Pang, C.P. Truncations in the TIGR gene in individuals with and without primary open-angle glaucoma. Investig. Ophthalmol. Vis. Sci. 2000, 41, 1386–1391. [Google Scholar]
- Bradley, K.M.; Breyer, J.P.; Melville, D.B.; Broman, K.W.; Knapik, E.W.; Smith, J.R. An SNP-Based Linkage Map for Zebrafish Reveals Sex Determination Loci. G3 2011, 1, 3–9. [Google Scholar] [CrossRef]
- Anderson, J.L.; Marí, A.R.; Braasch, I.; Amores, Á.; Hohenlohe, P.; Batzel, P.; Postlethwait, J.H. Multiple Sex-Associated Regions and a Putative Sex Chromosome in Zebrafish Revealed by RAD Mapping and Population Genomics. PLoS ONE 2012, 7, e40701. [Google Scholar] [CrossRef]
- Liew, W.C.; Bartfai, R.; Lim, Z.; Sreenivasan, R.; Siegfried, K.R.; Orbán, L. Polygenic Sex Determination System in Zebrafish. PLoS ONE 2012, 7, e34397. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Santos, E.M.; Workman, V.L.; Paull, G.C.; Filby, A.L.; Van Look, K.J.W.; Kille, P.; Tyler, C.R. Molecular basis of sex and reproductive status in breeding zebrafish. Physiol. Genom. 2007, 30, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.; Ha, N.-T.; Simianer, H.; Falker-Gieske, C.; Brenig, B.; Franke, A.; Hörstgen-Schwark, G.; Tetens, J.; Herzog, S.; Sharifi, A.R. Genetic mechanism underlying sexual plasticity and its association with colour patterning in zebrafish (Danio rerio). BMC Genom. 2019, 20, 341. [Google Scholar] [CrossRef] [PubMed]
- Webster, K.A.; Schach, U.; Ordaz, A.; Steinfeld, J.S.; Draper, B.W.; Siegfried, K.R. Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 2017, 422, 33–46. [Google Scholar] [CrossRef]
- Yan, Y.-L.; Batzel, P.; Titus, T.; Sydes, J.; Desvignes, T.; BreMiller, R.; Draper, B.W.; Postlethwait, J.H. A Hormone That Lost Its Receptor: Anti-Müllerian Hormone (AMH) in Zebrafish Gonad Development and Sex Determination. Genetics 2019, 213, 529–553. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, Y.; Saito, K.; Shinya, M.; Kawasaki, T.; Sakai, N. Evaluation of Sycp3, Plzf and Cyclin B3 expression and suitability as spermatogonia and spermatocyte markers in zebrafish. Gene Expr. Patterns 2011, 11, 309–315. [Google Scholar] [CrossRef]
- Miller, W.L. Molecular Biology of Steroid Hormone Synthesis. Endocr. Rev. 1988, 9, 295–318. [Google Scholar] [CrossRef]
- Clark, B.J.; Wells, J.; King, S.R.; Stocco, D.M. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR). J. Biol. Chem. 1994, 269, 28314–28322. [Google Scholar] [CrossRef]
- Zhang, Q.; Ye, D.; Wang, H.; Wang, Y.; Hu, W.; Sun, Y. Zebrafish cyp11c1 Knockout Reveals the Roles of 11-ketotestosterone and Cortisol in Sexual Development and Reproduction. Endocrinology 2020, 161, 161. [Google Scholar] [CrossRef]
- Borg, B. Androgens in teleost fishes. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 1994, 109, 219–245. [Google Scholar] [CrossRef]
- Xing, Y.-Y.; Cheng, X.-N.; Li, Y.-L.; Zhang, C.; Saquet, A.; Liu, Y.-Y.; Shao, M.; Shi, D.-L. Mutational analysis of dishevelled genes in zebrafish reveals distinct functions in embryonic patterning and gastrulation cell movements. PLoS Genet. 2018, 14, e1007551. [Google Scholar] [CrossRef]
- Cadigan, K.M.; Waterman, M.L. TCF/LEFs and Wnt Signaling in the Nucleus. Cold Spring Harb. Perspect. Biol. 2012, 4, a007906. [Google Scholar] [CrossRef] [PubMed]
- Tago, K.; Nakamura, T.; Nishita, M.; Hyodo, J.; Nagai, S.; Murata, Y.; Adachi, S.; Ohwada, S.; Morishita, Y.; Shibuya, H.; et al. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev. 2000, 14, 1741–1749. [Google Scholar] [PubMed]
- Krupnik, V.E.; Sharp, J.D.; Jiang, C.; Robison, K.; Chickering, T.W.; Amaravadi, L.; Brown, D.E.; Guyot, D.; Mays, G.; Leiby, K.; et al. Functional and structural diversity of the human Dickkopf gene family. Gene 1999, 238, 301–313. [Google Scholar] [CrossRef]
- Kossack, M.E.; High, S.K.; Hopton, R.E.; Yan, Y.-L.; Postlethwait, J.H.; Draper, B.W. Female Sex Development and Reproductive Duct Formation Depend on Wnt4a in Zebrafish. Genetics 2019, 211, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Parakh, T.N.; Hernandez, J.A.; Grammer, J.C.; Weck, J.; Hunzicker-Dunn, M.; Zeleznik, A.J.; Nilson, J.H. Follicle-stimulating hormone/cAMP regulation of aromatase gene expression requires beta-catenin. Proc. Natl. Acad. Sci. USA 2006, 103, 12435–12440. [Google Scholar] [CrossRef] [PubMed]
- Nicol, B.; Guiguen, Y. Expression profiling of Wnt signaling genes during gonadal differentiation and gametogenesis in rainbow trout. Sex. Dev. 2011, 5, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Amberg, J.J.; Goforth, R.R.; Sepúlveda, M.S. Antagonists to the Wnt Cascade Exhibit Sex-Specific Expression in Gonads of Sexually Mature Shovelnose Sturgeon. Sex. Dev. 2013, 7, 308–315. [Google Scholar] [CrossRef]
- Vainio, S.; Heikkilä, M.; Kispert, A.; Chin, N.; McMahon, A.P. Female development in mammals is regulated by Wnt-4 signalling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atienzar-Aroca, R.; Aroca-Aguilar, J.-D.; Alexandre-Moreno, S.; Ferre-Fernández, J.-J.; Bonet-Fernández, J.-M.; Cabañero-Varela, M.-J.; Escribano, J. Knockout of myoc Provides Evidence for the Role of Myocilin in Zebrafish Sex Determination Associated with Wnt Signalling Downregulation. Biology 2021, 10, 98. https://doi.org/10.3390/biology10020098
Atienzar-Aroca R, Aroca-Aguilar J-D, Alexandre-Moreno S, Ferre-Fernández J-J, Bonet-Fernández J-M, Cabañero-Varela M-J, Escribano J. Knockout of myoc Provides Evidence for the Role of Myocilin in Zebrafish Sex Determination Associated with Wnt Signalling Downregulation. Biology. 2021; 10(2):98. https://doi.org/10.3390/biology10020098
Chicago/Turabian StyleAtienzar-Aroca, Raquel, José-Daniel Aroca-Aguilar, Susana Alexandre-Moreno, Jesús-José Ferre-Fernández, Juan-Manuel Bonet-Fernández, María-José Cabañero-Varela, and Julio Escribano. 2021. "Knockout of myoc Provides Evidence for the Role of Myocilin in Zebrafish Sex Determination Associated with Wnt Signalling Downregulation" Biology 10, no. 2: 98. https://doi.org/10.3390/biology10020098
APA StyleAtienzar-Aroca, R., Aroca-Aguilar, J.-D., Alexandre-Moreno, S., Ferre-Fernández, J.-J., Bonet-Fernández, J.-M., Cabañero-Varela, M.-J., & Escribano, J. (2021). Knockout of myoc Provides Evidence for the Role of Myocilin in Zebrafish Sex Determination Associated with Wnt Signalling Downregulation. Biology, 10(2), 98. https://doi.org/10.3390/biology10020098







