1. Introduction
Osteoclasts (OCs) are multi-nucleated cells, originating from the monocyte/macrophage lineage of hematopoietic stem cells, playing a fundamental role in skeletal development, bone remodeling, and fracture healing [
1].
Under stimulation by appropriate cytokines, OCs precursors undertake osteoclastogenesis, which is a very complex and sophisticated differentiation process, involving pre-OCs proliferation, their migration, cell–cell adhesion, and fusion, eventually leading to the formation of mature multinucleated OCs [
2]. The two essential cytokines are the Macrophage-Colony Stimulating Factor (M-CSF), stimulating migration and supporting pre-OCs proliferation, and the Receptor Activator of Nuclear factor κ-B Ligand (RANKL), which is crucial for the maturation of pre-OCs in multinucleated OCs. The events downstream the binding to their corresponding receptors, c-fms and RANK, respectively, have been widely described [
2]. The RANKL signaling pathway involves the recruitment of adapter proteins, i.e., TNF receptor associated factors (TRAFs), and leads to the activation of mitogen-activated protein kinases (MAPKs) and transcription factors, such as AP-1, NFATc2, and then NFATc1 [
2].
MAPKs are important molecules that transduce external signals into internal cellular responses. It has been demonstrated that p38, c-Jun N-terminal kinase (JNK), and extracellular signal-regulated kinase (ERK) transient phosphorylation promotes the expression of various OC marker genes including NFATc1 [
3,
4,
5]. More recently, the axis RANKL-RANK-TRAF6-p38 MAP kinase was related to the onset of osteoclastogenesis [
6] and RANKL-RANK-MEK-ERK-NFATc1 axis was related to migration-fusion in RAW 264.7 cells [
7]. NFATc1 has been shown to play a role as a master regulator of osteoclastogenesis, regulating the expression of numerous osteoclast-specific molecules involved in fusion, in differentiation, and maturation of multi-nucleated OCs, and in bone remodeling [
8]. Our previous results showed that the expression of osteoclast hallmarks depends on the induction of NFATc1 and ingenuity pathway analysis (IPA) correlated NFATc1 with the activation of ERK and p-38 MAPKs [
9].
In this study, we analyzed timing and behavior of OCs during their in vitro differentiation. We used the RAW 264.7 cells, which are a mouse macrophage cell line with the capacity to differentiate into OCs after RANKL stimulation. We investigated osteoclastogenesis at a morphological level, studying OCs phenotypes by electron microscopy and proteins localization, distribution by immunofluorescence analyses at a molecular level, and studying the mRNA expression of a panel of genes by qPCR analysis.
2. Materials and Methods
2.1. Cell Culture and Osteoclastogenesis In Vitro
The murine RAW 264.7 cell line was purchased from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Cells were grown in Dulbecco modified Eagle’s medium (DMEM, Gibco, NY, USA), with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, USA,), 100-U/mL penicillin, and 100-µg/mL streptomycin. To induce OC differentiation, cells were suspended in alpha-minimal essential medium (α-MEM Gibco, Grand Island, NY, USA) with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich, St. Louis, MO, USA), 100-U/mL penicillin, and 100-µg/mL streptomycin, with the addition of 50-ng/mL RANKL (Peprotech, Rocky Hill, CT, USA). RAW 264.7 differentiation toward multinucleated OCs has been followed in vitro over four days, considering as day 0, the day the cytokine RANKL was added, which was replaced every two days to maintain the signal.
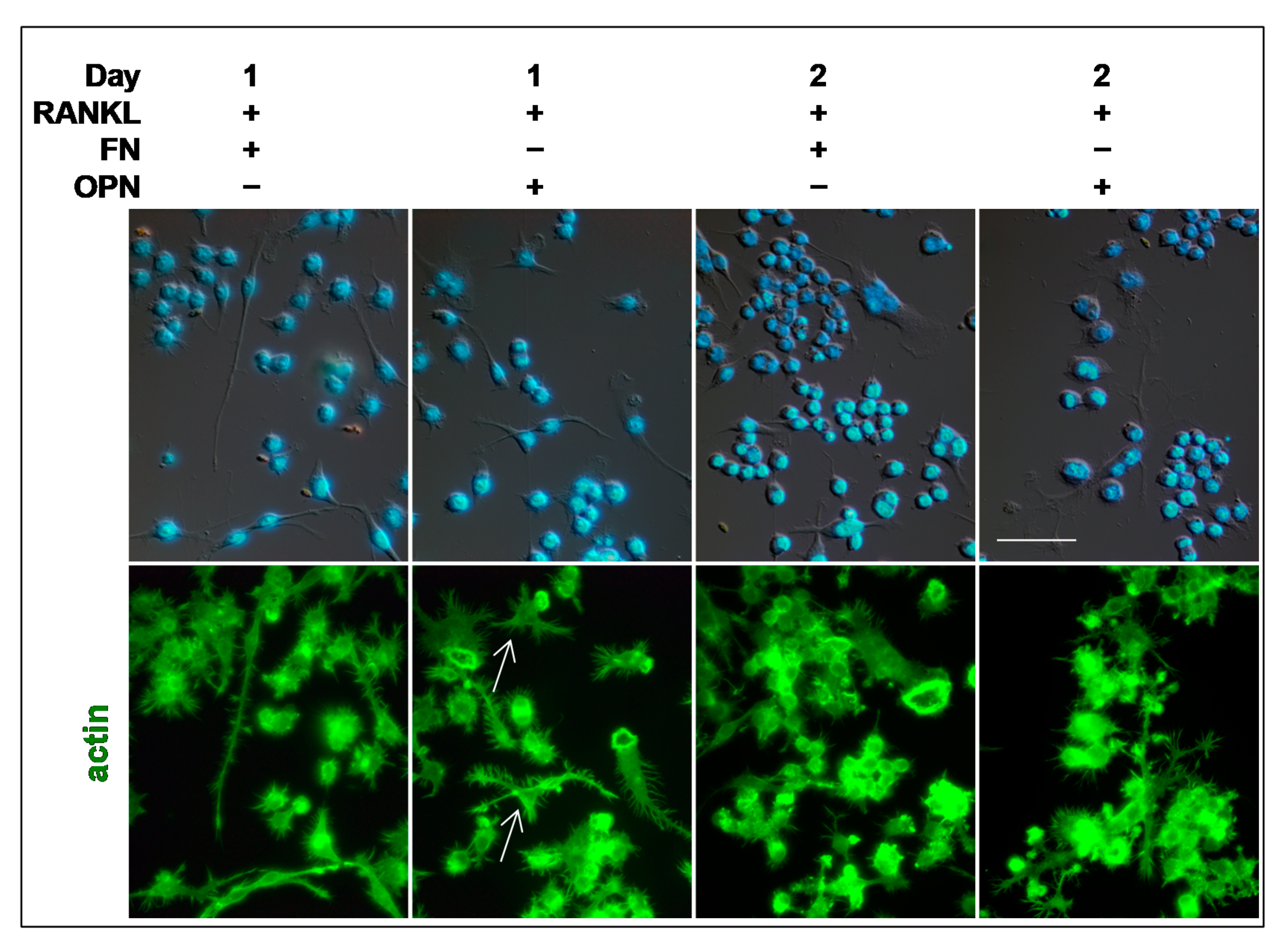
For the experiments of OC differentiation on extracellular matrix (ECM) proteins, 8-well chamber slides were pre-coated with 20 µg/mL of fibronectin (FN, Sigma-Aldrich, St. Louis, MO, USA) or recombinant osteopontin (OPN, Sigma-Aldrich, St. Louis, MO, USA) in phosphate buffered saline (PBS), overnight at 4 °C [
10]. To increase the amount of proteins physically adsorbed onto the wells, an additional incubation was performed for 1 h at 37 °C. The wells were then blocked using 1% bovine serum albumin (BSA) in PBS for 1 h at 37 °C in a CO
2 incubator. Finally, the wells were washed with PBS and, to prevent denaturing of the proteins, left in PBS until cells were plated. RAW 264.7 cells were plated at 1 × 10
4 cells/well in 8-well chamber slides overnight.
2.2. Electron Microscopy Analysis
For electron microscopy experiments, cells (2.5 × 105/well/6 wells plate) were grown on coverslips, fixed in 2.5% glutaraldehyde in 0.1 M Sorenson’s buffer (pH 7.4) for 2 h with subsequent post-fixation in 1% OsO4 and embedded in Epon (Sigma-Aldrich, St. Louis, MO, USA). Serial ultrathin sections (90 nm) were cut with Leica Ultracut-E ultramicrotome (Leica Microsystems, Wetzlar, Germany), stained with 2% aqueous uranylacetate and lead citrate, and observed with JEM-1400 TEM microscope operated at 100 kV (JEOL, Tokyo, Japan).
2.3. Tartrate-Resistant Acid Phosphatase (TRAP) Staining
RAW 264.7 cells (2.5 × 105/well/plate 6 wells) were cultured for four days with RANKL (50 ng/mL) as previously described and then fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 10 min at room temperature. TRAP staining was carried out in accordance with the manufacturer’s instruction (Sigma Aldrich, St. Louis, MO, USA). Cells were observed under light microscopy (OLYMPUS CKX31, Olympus, Tokyo, Japan), and those containing more than three nuclei were considered OCs.
2.4. Immunofluorescence Analysis
For indirect immunofluorescence experiments, RAW 264.7 cells (2.5 × 10
5/well/plate 6 wells) were grown on coverslips for various time periods with or without RANKL (50 ng/mL). The analysis of the cell surface localization of RANK was performed according to Fiorino et al. [
11] with few modifications. Cells were washed with cold TBS/1 mM Ca
2+ and incubated with anti-RANK antibody (1:200, Santa Cruz, CA, USA) for 20 min at 4 °C. Then, cells were fixed with 4% paraformaldehyde (PFA, Sigma-Aldrich, St. Louis, MO, USA), blocked with 5% foetal calf serum, and incubated with secondary antibody Alexa Fluor-594 rabbit anti-mouse (1:500, Invitrogen Molecular Probes, Life Technologies, Carlsbad, CA, USA) for 1 h at room temperature in a dark, humidified chamber. For the analysis of cytoplasmic antigens, cells were fixed with 4% PFA in PBS for 15 min, permeabilized with 0.5% Triton X-100 (Sigma)/PBS for 10 min, and blocked with 1% BSA (Sigma)/0.1% Triton X-100/PBS for 30 min. Alexa Fluor-488 phalloidin (1:250, Invitrogen Molecular Probes, Carlsbad, CA, USA), WGA-FITC (1:400, Sigma-Aldrich, St. Louis, MO, USA), and antibodies anti α-tubulin (1:500, Sigma-Aldrich, St. Louis, MO, USA), RANK (1:20, Santa Cruz, CA, USA), TRAF6 (1:20, Santa Cruz, CA, USA), p-p38 (1:20, Cell Signaling Technology, Beverly, MA, USA), p-ERK (1:20, Cell Signaling Technology, Beverly, MA, USA), and NFATc1 (1:20, Santa Cruz, CA, USA) were incubated for 2 h at room temperature in a dark, humidified chamber and secondary antibodies Alexa Fluor-488 rabbit anti-mouse (1:200, Invitrogen Molecular Probes, Carlsbad, CA, USA) and Alexa Fluor-594 goat anti-rabbit (1:200, Invitrogen Molecular Probes, Carlsbad, CA, USA) were incubated for 1 h at room temperature. All the coverslips were mounted on glass slides with mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen Molecular Probes, Carlsbad, CA, USA) and observed under an Axioskop-2 plus microscope (Zeiss, Oberkochen, Germany), equipped for epifluorescence. Images were recorded using a digital camera system, then cropped, and grouped. The contrast and brightness were adjusted using Photoshop CS2 (Adobe Systems, San Jose, CA, USA). Samples labeled with p-ERK were observed under a Nikon Eclipse 80i microscope (Nikon, Kawasaki, Kanagaw, Japan) equipped for epifluorescence and recorded by a digital camera system (40×). The percentage of cells showing the nuclear localization of various proteins was calculated as the ratio of nuclei labeled with specific antibodies to the total number of nuclei labeled with DAPI (Molecular Probes, Eugene, OR, USA), counted using the ImageJ software (
https://imagej.nih.gov/ij/, 1997–2018).
2.5. RNA Extraction and cDNA Synthesis
RAW 264.7 cells were cultured in a 6-well plate (1 × 106 cells/well) overnight. Cells were treated or untreated (ctrl) with 50 ng/mL RANKL for 1, 2, 3, and 4 days. At each stimulation time, cells (treated and untreated) were detached from the wells and washed once with PBS. Total RNA was isolated using the “GenElute Mammalian Total RNA Miniprep Kit” (Sigma-Aldrich, St. Louis, MO, USA) and quantified by using the bio photometer (Eppendorf, Hamburg, Germany). Total RNA (3 μg) was converted to cDNA using the SuperScript Vilo (Invitrogen Molecular Probes, Carlsbad, CA, USA).
2.6. qPCR
Quantification of gene expression was performed using the StepOnePlus real-time PCR as described in the manufacturer’s manual (Applied Biosystems, Grand Island, NY, USA) with SYBR Green chemistry, and the Comparative Threshold Cycle Method [
12]. The qPCR was run as follows: 1× cycle, denaturing at 95 °C for 10 min for DNA polymerase activation, and 38 cycles, melting at 95 °C for 15 s and annealing/extension at 60 °C for 60 s. QPCR was then performed using primer by Qiagen (Germantown, MD, USA): QT001676692 (NFATc1), QT00166663 (TRAF6), QT00108815 (MMP9), QT00131313 (MITF1), QT00131012 (TRAP), QT01047032 (DC-STAMP), QT02589489 (CTSK2), QT00149415 (RelA), QT00197568 (RhoA), QT01167257 (OSCAR), QT009399 (RANK), QT01658692 (GAPDH), FOS F: 5′CACTCCAAGCGGAGACAGAT3′, and R: 5′TCGGTGGGCTGCCAAAATAA3′. The threshold cycle (CT) values were calculated against the housekeeping gene GAPDH, whose expression was not affected by the experimental conditions. The expression levels of the analyzed genes are presented as relative values of the cells treated with RANKL compared to the cells not treated with RANKL (ctrl). Each day has its appropriate control on the same day (assumed with a value of 1). At least three distinct biological samples were examined for each gene and treatment. Data are expressed as mean ± S.D.
2.7. Statistical Analysis
Data are expressed as mean ± S.D. from at least three experiments and statistical analyses were performed by Student’s T test. p < 0.05 was considered to indicate a statistically significant difference.
4. Discussion
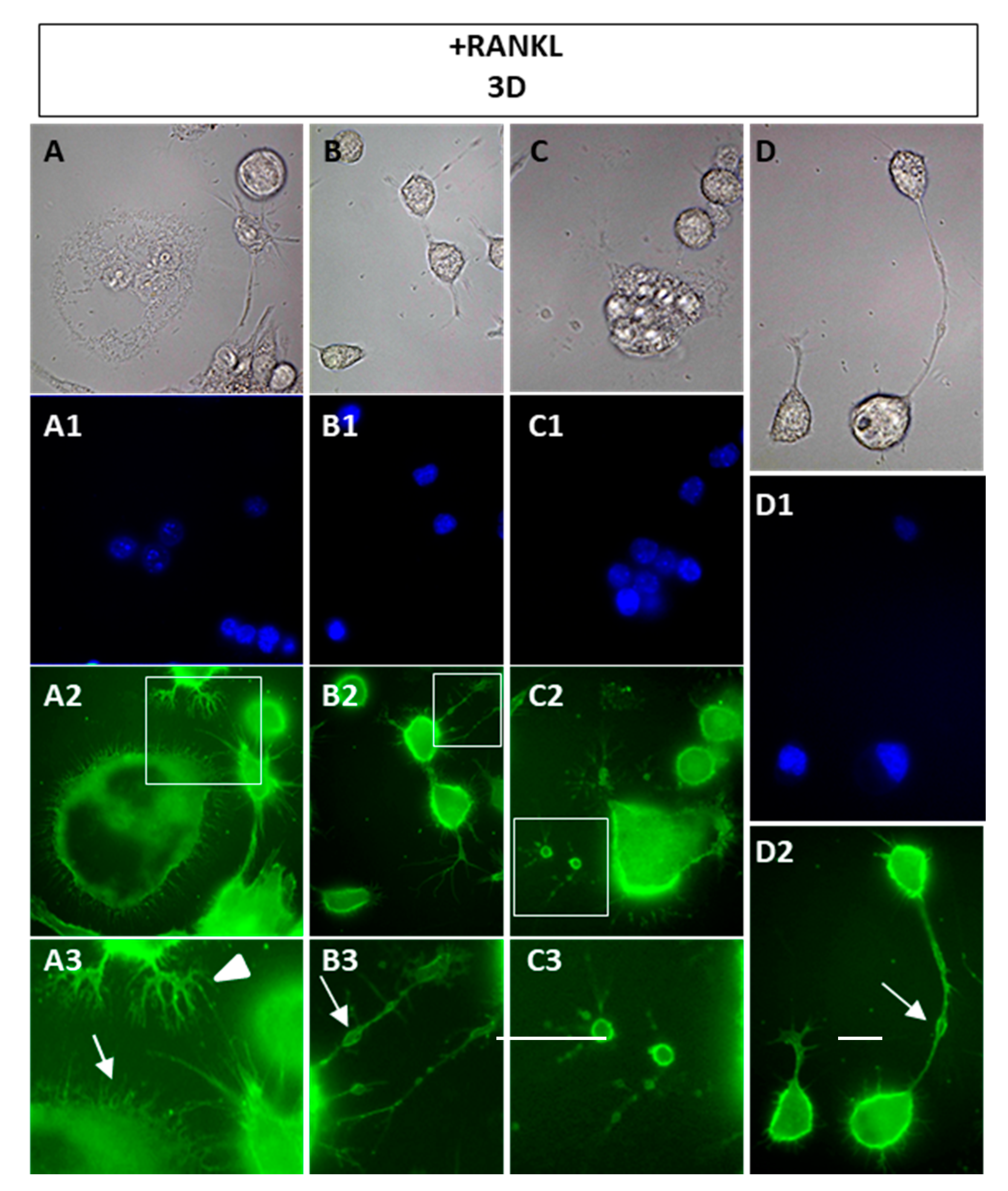
In the present study, we investigated the timing and behavior of OCs during their in vitro differentiation, evaluating morphological changes, cytoskeleton organization, and sub-cellular distribution of some proteins as well as the expression of a panel of OC-specific genes. Utilizing the RAW 264.7 cells, which come from a murine monocyte-macrophage cell line, we were able to study the sequential events leading to OC differentiation induced only by RANKL. Our results clarify several points in the OC differentiation process that have not been highlighted so far. First, we show that morphological changes and cytoskeleton organization are related to the timing of RANKL stimulation as well as to the substrate on which RANKL-stimulated cells adhere. Pre-OCs and mature OCs are very dynamic cells, which alternate among migration times, cell-cell contacts, fusion, and resorption, resulting in different morphologies and a series of cytoskeleton rearrangements [
15]. By our results, it appears evident that the first day after RANKL addition is characterized by pre-OCs search for a partner to fuse with, as shown by the presence of numerous cells with bipolar morphology. The filopodia emitted by the pre-OCs were of different sizes and shapes, depending on the substrate they adhered to, which might reflect the involvement of different adhesion molecules [
2]. Furthermore, the appearance of numerous actin rings during early times of OC differentiation (1 or 2 days of RANKL stimulation) on FN or OPN, but not on a glass substrate, confirms the role played by the ECM in modulating OCs morphology [
10].
The second interesting point are the vesicles observed along the filopodia of 3 days/RANKL
+ cells, or sometimes disconnected by cells, shown by the WGA-FITC labeling. To our knowledge, this type of structure has never been described in the OCs and it is difficult to explain its presence and possible function during osteoclastogenesis. However, these vesicles resemble “migrasomes,” i.e., cellular organelles recently described, which form on retraction fibers of migrating cells as large vesicle-like structures [
16]. The migrasome role has not yet been demonstrated, but it is proposed to provide spatio-temporal chemical information for cell-cell communication during cell migration [
17]. Recently, Hu et al. [
18] showed the release of a large number of cholesterol-rich particles by macrophages during their locomotion. The particles were fragments of plasma membrane, released during projection and retraction of filopodia and lamellipodia, which remained anchored to the substrate [
18]. In this case, the particles were suggested to contribute to cholesterol transport.
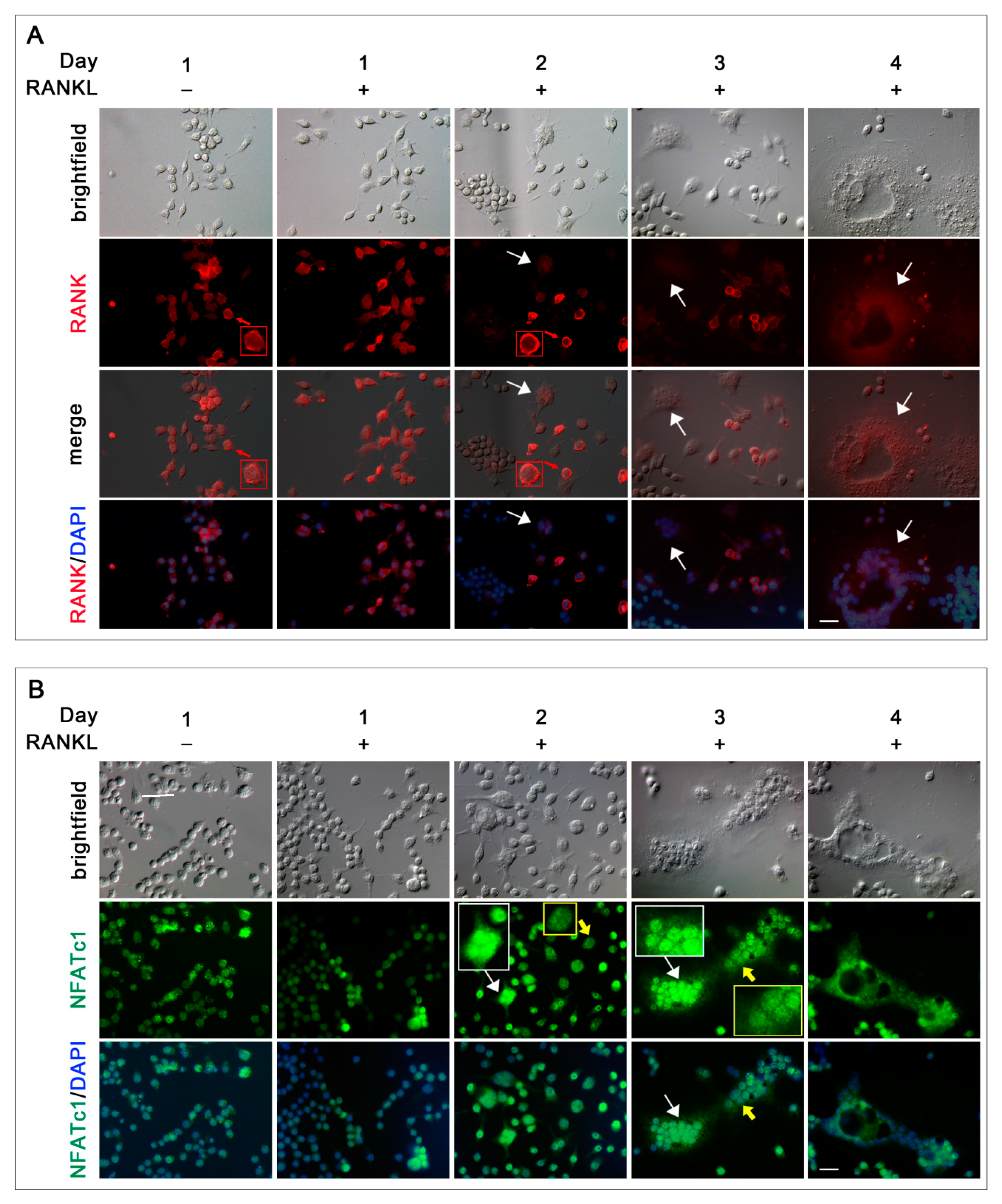
The sub-cellular distribution of molecules involved in the RANKL-RANK pathway related to the OC differentiation timing has not been previously described. Studying RANK distribution during RANKL stimulation, its surface localization in almost all unstimulated cells as well as in mononuclear RANKL stimulated cells was expected, suggesting that they likely corresponded to OC-precursors, which are a pool of cells ready to undergo OC differentiation since they are able to rapidly respond to RANKL [
19]. However, it is not clear the reason why, during RANKL treatment timing, the number of mononuclear cells expressing RANK on the surface decreased. The finding that a small percentage of RAW 264.7 cells are RANK
+ pre-OCs is related to the observation that a little amount of cells undergo fusion after RANKL stimulation (
Movie S1). Levaot et al. [
20] assessed that OCs fusion began by a small subset of RANKL-stimulated pre-OCs (about 2.4%, but always below 10%), termed “fusion founders,” which fuse with other founders or with non-stimulated progenitors, termed “fusion followers.” The authors described an initial pre-fusion phase, during which a founder and a follower cell pair together, developing cytoplasmic communication. However, both of them maintain their own morphology for a while before the fusion event becomes apparent [
20] (see
Figure 1B,D and
Movie S1 in this study). Thus, an emerging concept concerns the heterogeneity between fusion partners, which show differences in both biological and molecular characteristics. It seems that fusion preferably occurs between partners of dissimilar motility and differentiation stages, i.e., a more mature and immobile OC prefers to fuse with a less mature and mobile pre-OC, which could be a strategy to control fusion in vivo [
21].
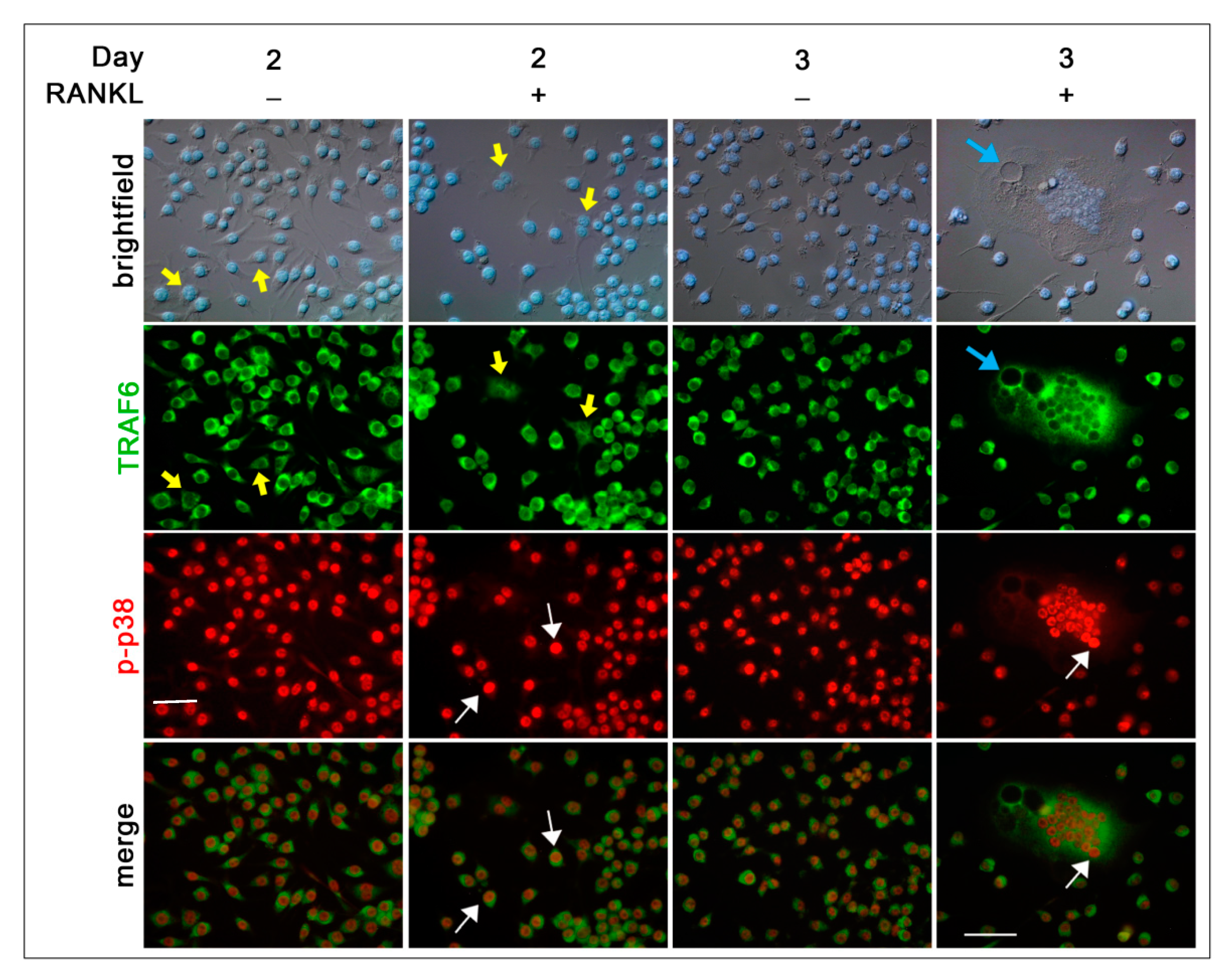
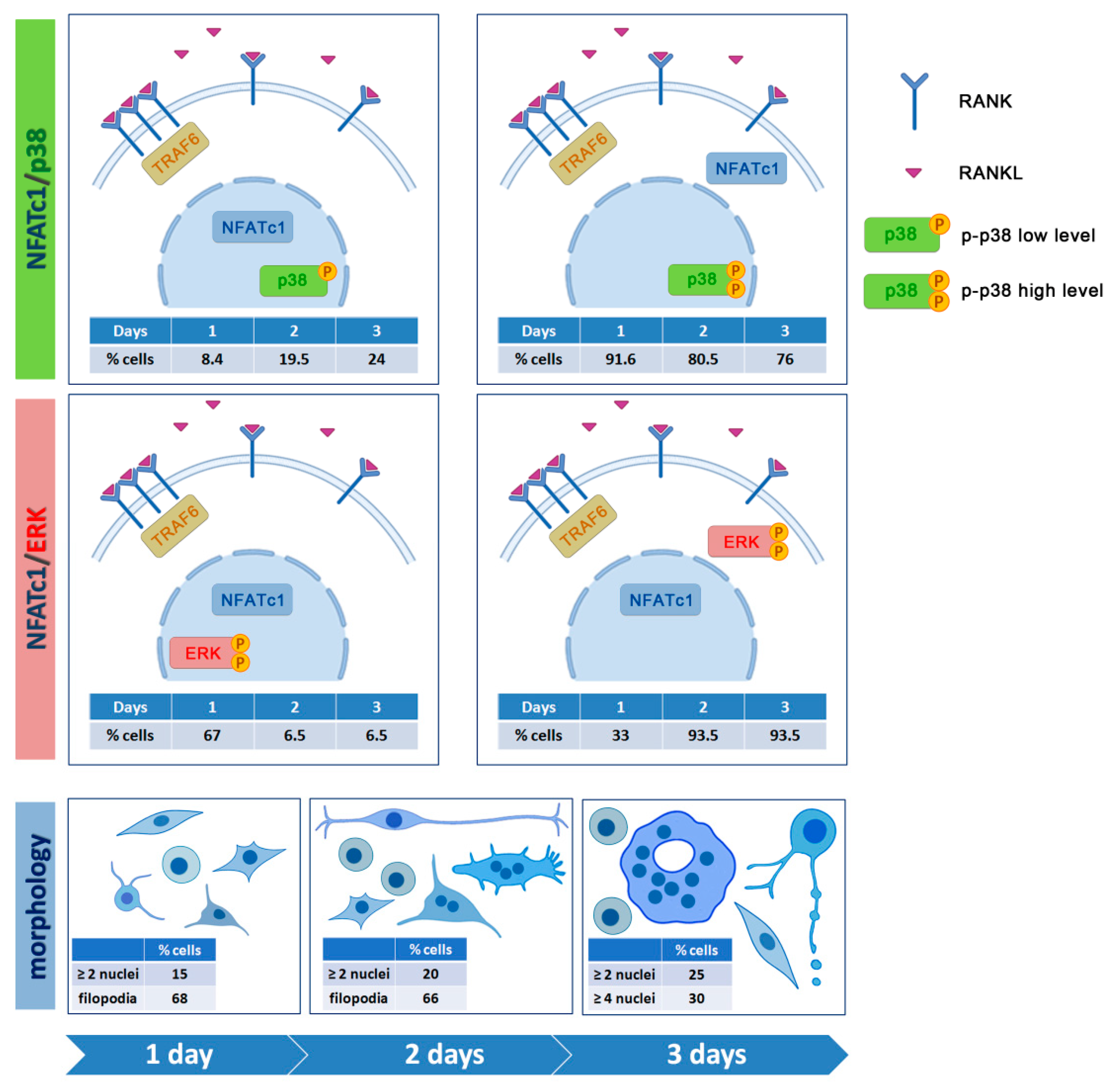
To study the intracellular signaling pathways involved in pre-OC differentiation, we focused on the 1, 2, and 3 days after RANKL addition, as, during these days, multinucleated OCs appear and increase in number and size, leading to a heterogeneous population of cells in different phases of OC differentiation, i.e., mononucleated and small and large multi-nucleated cells (see schematic representation of morphologies in
Figure 10). The binding of RANKL to its RANK receptor on the surface of the pre-OCs leads to a series of signaling cascades, involving, among others, the p38 and ERK MAPK signaling pathways [
22]. Since RANK lacks an intrinsic enzymatic activity, the intracellular transduction of the RANKL signal is performed by recruiting adapter molecules, such as TRAF6, which has been shown to activate p38 and JNK MAPK during OC differentiation [
6]. The cytoplasmic distribution of TRAF6 in RAW 264.7 cells at different phases of OC differentiation is a validation of its crucial role in transducing RANKL–RANK signaling. The nuclear co-localization of the stronger TRAF6 (cytoplasm) and p-p38 (nucleus) signals in RAW 264.7 cells is in agreement with the requirement of TRAF6 recruitment to the cytoplasmic tail of RANK for the RANKL-dependent p38 activation in bone marrow-derived mononuclear pre-OCs [
6].
The nuclear localization of the active form of p38 (p-p38) in almost all RAW 264.7 cells regardless of the culture conditions was surprising. Usually, p38 MAPK is localized in the cytoplasm of resting cells and translocates to the nucleus after extracellular stimulation [
23]. The ability of p38 to shuttle to the nucleus and back after extracellular stimulation is well known, even though it does not contain nuclear localization or nuclear export signals [
24]. Nevertheless, in a few cases, p38 has been detected in the nucleus of resting cells, thus, suggesting its dual ability to phosphorylate substrates in the cytoplasm as well as in the nucleus [
24]. NFATc1 is one of the targets of p38 MAPK. However, contrasting data have been reported concerning the relationship between them. Among various reports, Matsumoto and colleagues [
3] demonstrated that p-p38 directly phosphorylates NFATc1 in murine bone marrow cells, re-locating it from the cytoplasm into the nuclei, while p-p38 promotes, albeit moderately, NFATc1 nuclear expulsion in T cells [
25]. Our observation that the nuclear localization of NFATc1 was always associated with a less intense signal of nuclear p-p38, both in mononucleated and multinucleated OCs, suggests that they are mutually exclusive in nuclei after RANKL stimulation, supporting the idea that p-p-38 might contribute to NFATc1 nuclear export in RAW 264.7 cells. The relationship between p-p38 and NFATc1 in RAW 264.7 cells deserves further study (see schematic representation in
Figure 10).
We recently deepened the relationship between p-ERK and NFATc1, showing that the long-lasting ERK activity depends on RANKL-dependent NFATc1 induction [
7]. A significant decrease of ERK phosphorylation was observed after 24 h NFATc1 impairment (siRNA silencing) and, conversely, a decreased expression of NFATc1 when ERK phosphorylation was affected by FR180204 inhibitor treatment for 24 h [
7]. The present results on p-ERK localization are in agreement with the previous ones, i.e., the major number of p-ERK
+ cells was observed at 1 day after RANKL addition, while this amount severely decreased after 2 and 3 days. Furthermore, the major number of cells showing p-ERK/NFATc1 co-localization was observed on day 1, suggesting a strict relationship between them as a consequence of RANKL stimulation (see schematic representation in
Figure 10).
During OC differentiation, NFATc1 translocates to the nucleus and activates both OC-specific genes and its own transcription [
26]. In this study, we noticed a selective distribution of the NFATc1 protein in certain nuclei within individual large multi-nucleated OCs, shown by an unequal labeling intensity of the various NFATc1+ nuclei or even by its complete absence in others. This observation suggests that only a selection of nuclei within multinucleated OCs are transcriptionally active, even with different levels of activity, as already shown by Youn et al. [
27] in RAW 264.7 cells.
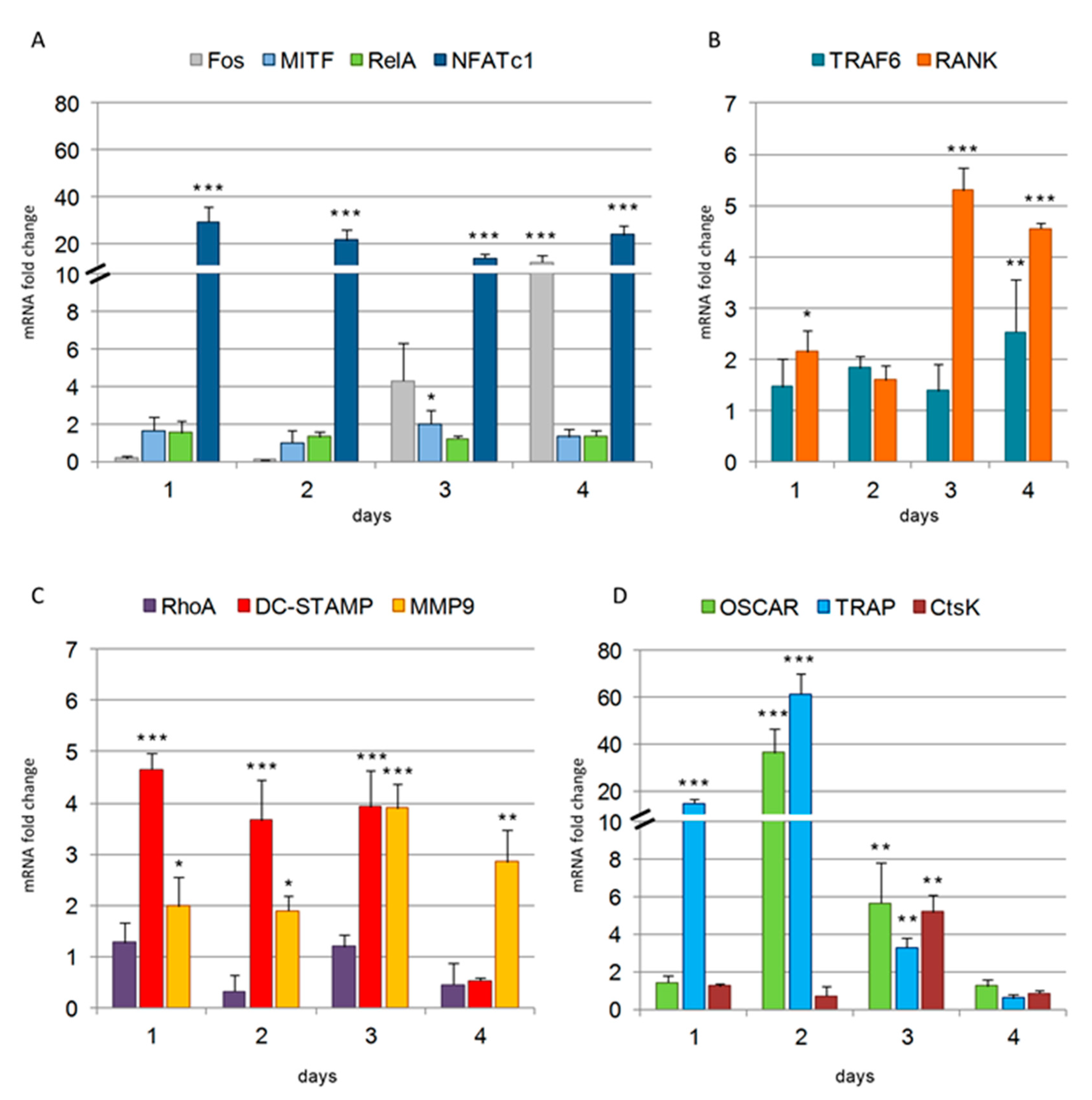
The remarkable upregulation of
NFATc1 mRNA observed in RAW 264.7 cells since the 1st day after RANKL stimulation is a hallmark aspect of osteoclastogenesis. By our previous qPCR and bioinformatic analyses, we found that NFATc1 significantly influences the expression of 31 genes, which are differently involved in osteoclastogenesis [
9]. A variety of data in literature support the idea that the initial induction of NFATc1 protein depends upon the cooperation of NFATc2 and NF-κB (in particular, components p50 and p65, named
RelA), both recruited to the NFATc1 promoter at the very early phase of OC differentiation, i.e., 1 h after RANKL stimulation. Both NFATc2 and NF-κB are present in the cytoplasm of unstimulated cells, to be readily available to enter the nucleus and activate target genes soon after RANKL stimulation.
NFATc1 and the osteoclast hallmarks were not transcribed after 1 h of RANKL-induction whereas they are expressed after 24 h [
7]. Consistent with these data are the levels of
RelA mRNA measured in our cellular system, which are present in resting cells as well as in the first day and during all OC differentiation (
Figure 9). On the contrary, from day 1 onwards after RANKL stimulation, c-Fos and NFATc1 itself are recruited to the NFATc1 promoter, as shown by ChIP analysis, and this occupancy persists toward terminal differentiation of OCs [
28] in agreement with our results (
Figure 9). MITF has been suggested to function as the most distal factor in osteoclastogenesis, being an NFATc1 signal modulator [
29]. In particular, it has been shown that MITF can amplify the NFATc1-dependent expression of many downstream OC-specific genes, including
CtsK,
OSCAR, and
TRAP [
29] in agreement with our results showing
MITF expression significantly increased at 3 days after RANKL-induction.
Cell migration is driven by the activity of the cytoskeleton, which undergoes rapid changes in its organization to accomplish cycles of movement and attachment during pre- and OCs migration. Our results showing the constant and significant increase in the expression of
MMP9 induced by RANKL from the first day after its addition are coherent with the known migrating activity of pre- and mature OCs under normal conditions. DC-STAMP is one of fusion-mediating molecules directly regulated by NFATc1, since the gene possesses a binding site for this TF in its promoter region [
30]. DC-STAMP is a transmembrane molecule, which is found on the surface of most of the unstimulated pre-OCs, while it moves from the cell surface to become mainly cytoplasmic after RANKL stimulation, suggesting an event of internalization when cells begin to differentiate toward OCs [
30]. Given that the expression of
DC-STAMP mRNA is upregulated from the 1st day of RANKL stimulation, we can hypothesize that RANKL-induced DC-STAMP expression is necessary to restore it on the surface of differentiating OCs as a response to its previous internalization, which is in agreement with data reported in literature [
30]. The mRNA expression levels of
DC-STAMP and
NFATc1 peaked at the same time points after RANKL stimulation and NFATc1-knockdown significantly inhibited
DC-STAMP expression [
9]. Several results suggest that there is a mutual regulation between DC-STAMP and NFATc1 at both gene and protein expression levels [
30]. The decreased expression of
NFATc1 has been recently described in DC-STAMP−/− cells [
31].
OSCAR is an osteoclast-specific immuno-receptor, which acts as a co-stimulatory signal required for RANKL-mediated activation of NFATc1 and, in turn, it is a direct target of NFATc1, implying the presence of a positive feedback circuit OSCAR-NFATc1-OSCAR [
8]. Recently, OSCAR has been shown to bind to specific motifs within fibrillar collagens, suggesting that ECM plays an active role in the OSCAR-mediated regulation of osteoclastogenesis [
32]. It is generally acknowledged that the ECM molecules significantly affect cell behavior and influence the activities of several intracellular signaling pathways [
33].