Role of Pirin, an Oxidative Stress Sensor Protein, in Epithelial Carcinogenesis
Abstract
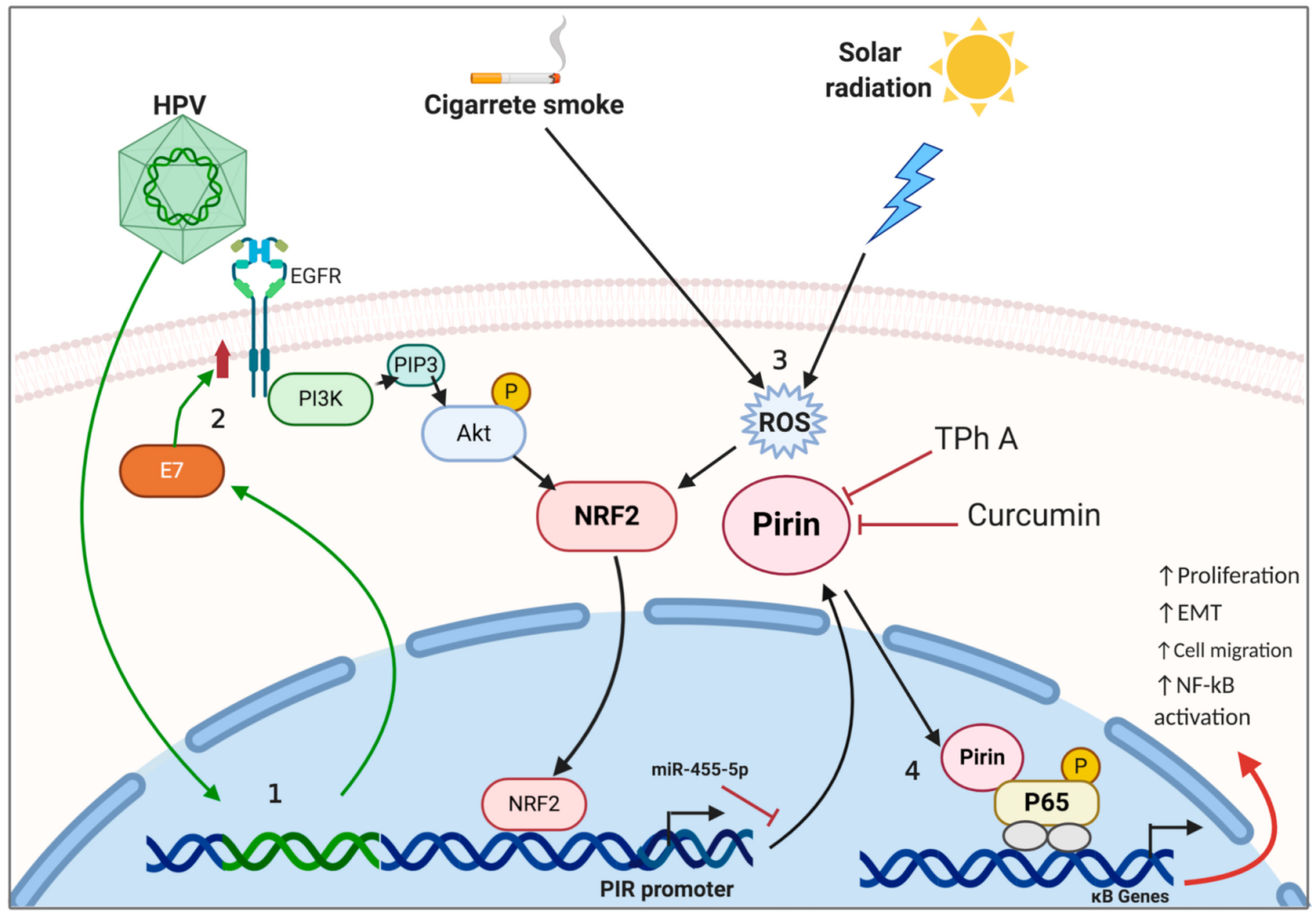
Simple Summary
Abstract
1. Introduction
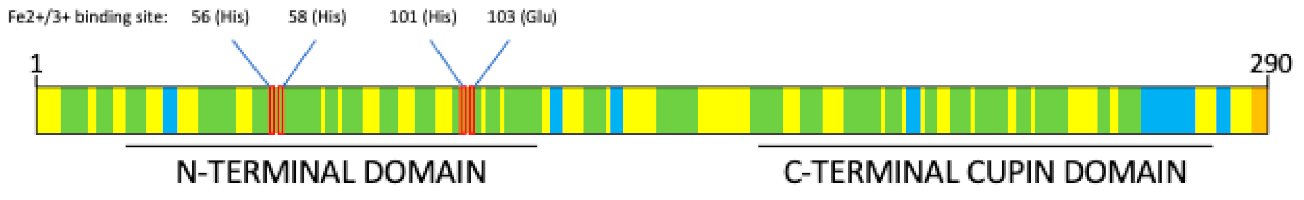
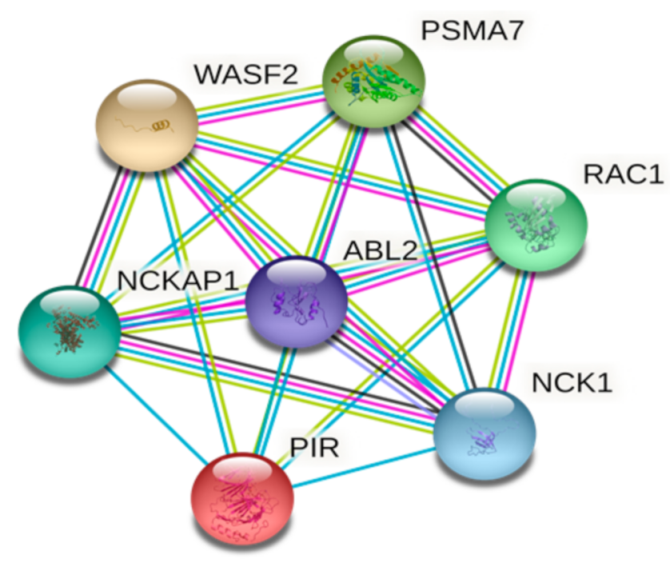
2. Pirin Structure and Biological Functions
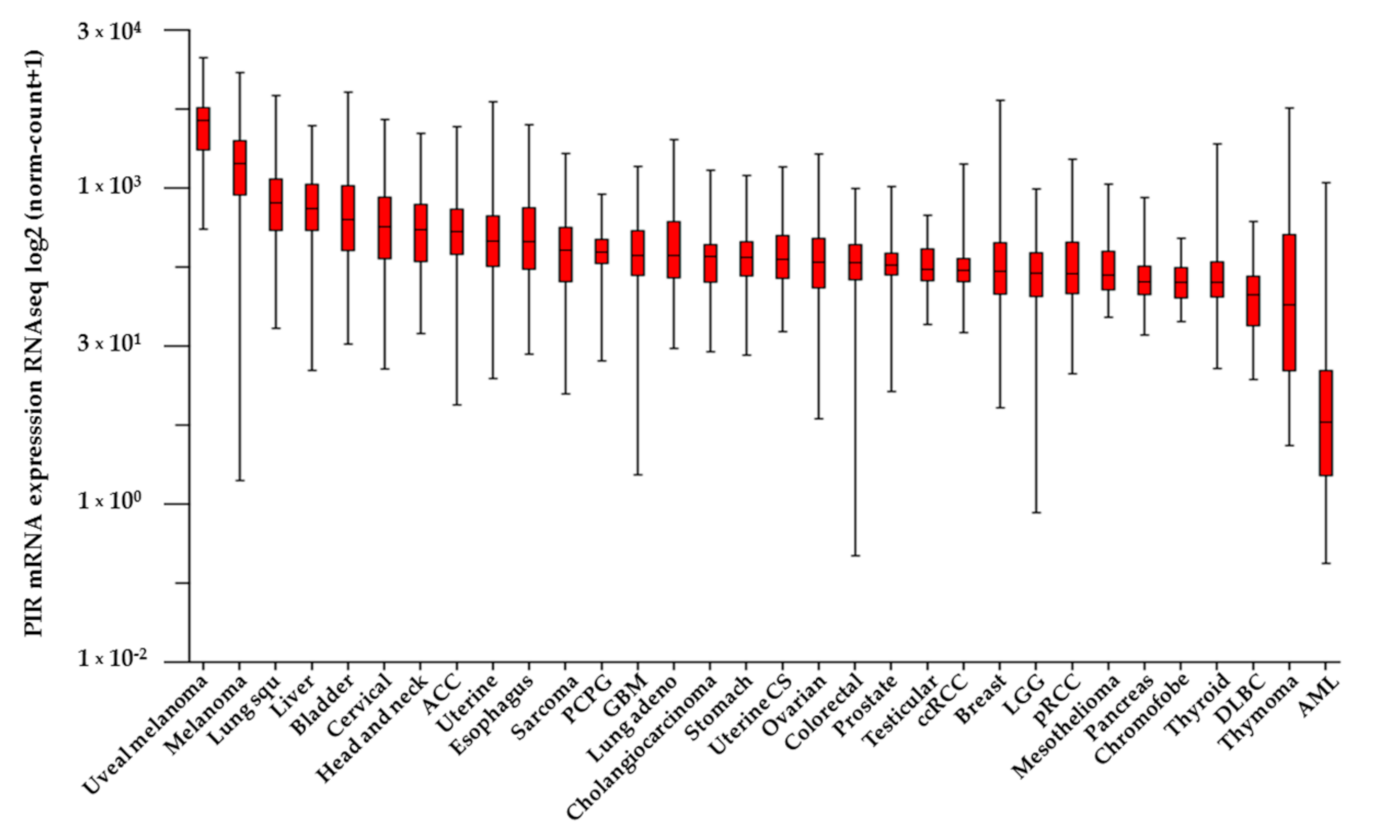
3. Role of Pirin in Cancer Development
3.1. Lung Cancer
3.2. Cervical Cancer
3.3. Skin Cancer
3.4. Breast Cancer
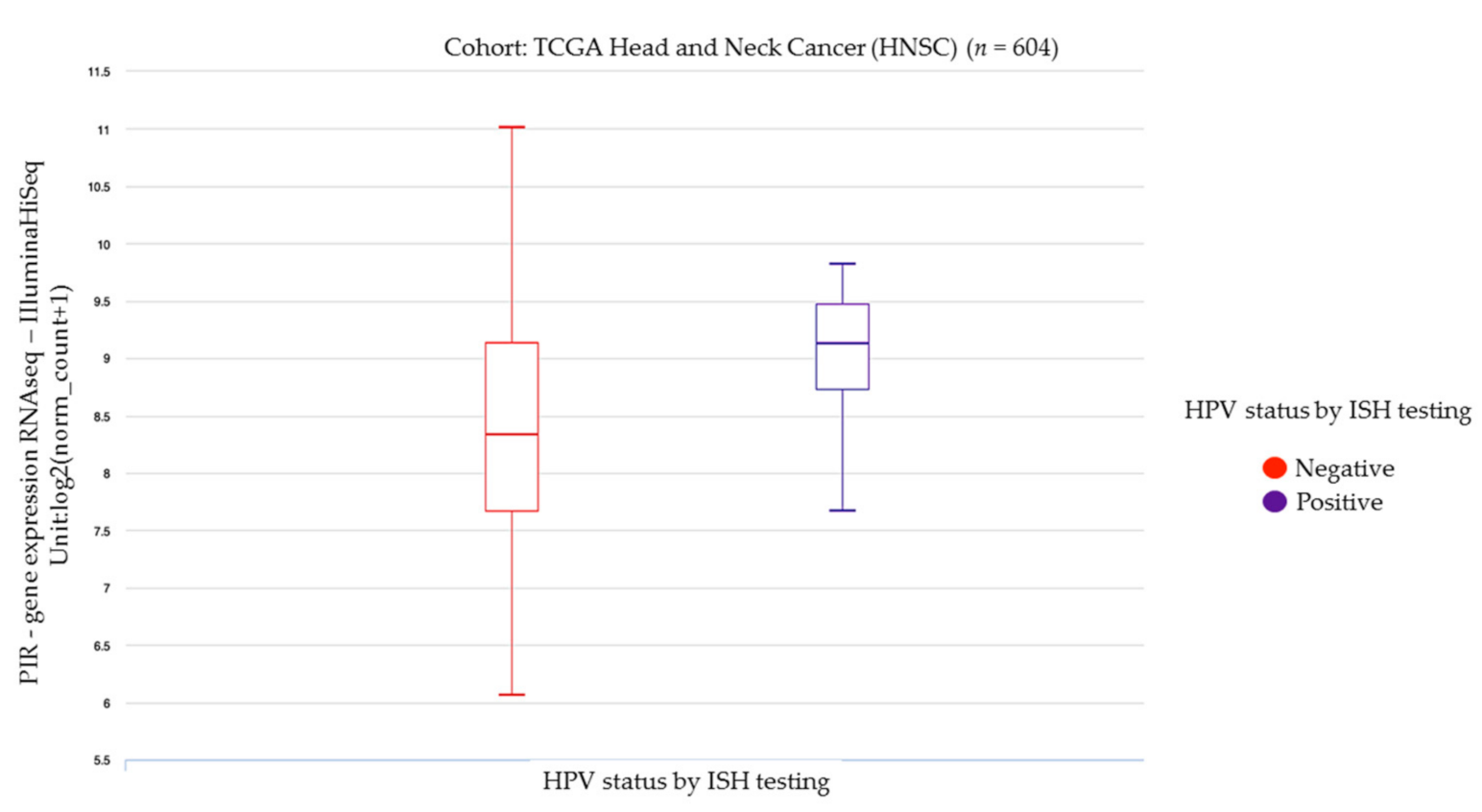
3.5. Head and Neck and Gastrointestinal Cancers
3.6. Non-Epithelial Cancers
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dunwell, J.M. Cupins: A new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol. Genet. Eng. Rev. 1998, 15, 1–32. [Google Scholar] [CrossRef]
- Woo, E.J.; Dunwell, J.M.; Goodenough, P.W.; Marvier, A.C.; Pickersgill, R.W. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 2000, 7, 1036–1040. [Google Scholar] [CrossRef]
- Dunwell, J.M.; Purvis, A.; Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry 2004, 65, 7–17. [Google Scholar] [CrossRef]
- Clissold, P.M.; Ponting, C. JmjC: Cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem. Sci. 2001, 26, 7–9. [Google Scholar] [CrossRef]
- Agarwal, G.; Rajavel, M.; Gopal, B.; Srinivasan, N. Structure-Based Phylogeny as a Diagnostic for Functional Characterization of Proteins with a Cupin Fold. PLoS ONE 2009, 4, e5736. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, J.M.; Culham, A.; Carter, C.E.; Sosa-Aguirre, C.R.; Goodenough, P.W. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 2001, 26, 740–746. [Google Scholar] [CrossRef]
- Dunwell, J.M.; Gane, P.J. Microbial Relatives of Seed Storage Proteins: Conservation of Motifs in a Functionally Diverse Superfamily of Enzymes. J. Mol. Evol. 1998, 46, 147–154. [Google Scholar] [CrossRef]
- Sarkar, B.; Kulharia, M.; Mantha, A.K. Understanding human thiol dioxygenase enzymes: Structure to function, and biology to pathology. Int. J. Exp. Pathol. 2017, 98, 52–66. [Google Scholar] [CrossRef]
- Schmidt, H.R.; Zheng, S.; Gurpinar, E.; Koehl, A.; Manglik, A.K.A.; Kruse, A.C. Crystal structure of the human σ1 receptor. Nature 2016, 532, 527–530. [Google Scholar] [CrossRef]
- Uekita, T.; Gotoh, I.; Kinoshita, T.; Itoh, Y.; Sato, H.; Shiomi, T.; Okada, Y.; Seiki, M. Membrane-type 1 Matrix Metalloproteinase Cytoplasmic Tail-binding Protein-1 Is a New Member of the Cupin Superfamily. A possible multifunctional protein acting as an invasion suppressor down-regulated in tumors. J. Biol. Chem. 2004, 279, 12734–12743. [Google Scholar] [CrossRef]
- Wendler, W.M.F.; Kremmer, E.; Förster, R.; Winnacker, E.-L. Identification of Pirin, a Novel Highly Conserved Nuclear Protein. J. Biol. Chem. 1997, 272, 8482–8489. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Bartlam, M.; Zeng, Q.; Miyatake, H.; Hisano, T.; Miki, K.; Wong, L.L.; Gao, G.F.; Rao, Z. Crystal Structure of Human Pirin. J. Biol. Chem. 2004, 279, 1491–1498. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.; Jia, Z. Structural and Biochemical Analysis Reveal Pirins to Possess Quercetinase Activity. J. Biol. Chem. 2005, 280, 28675–28682. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, L.; Lu, S. Evaluation of Antioxidant and Immunity Activities of Quercetin in Isoproterenol-Treated Rats. Molecules 2012, 17, 4281–4291. [Google Scholar] [CrossRef]
- Soo, P.-C.; Horng, Y.-T.; Lai, M.-J.; Wei, J.-R.; Hsieh, S.-C.; Chang, Y.-L.; Tsai, Y.-H.; Lai, H.-C. Pirin Regulates Pyruvate Catabolism by Interacting with the Pyruvate Dehydrogenase E1 Subunit and Modulating Pyruvate Dehydrogenase Activity. J. Bacteriol. 2006, 189, 109–118. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Dechend, R.; Hirano, F.; Lehmann, K.; Heissmeyer, V.; Ansieau, S.; Wulczyn, F.G.; Scheidereit, C.; Leutz, A. The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene 1999, 18, 3316–3323. [Google Scholar] [CrossRef]
- Maldonado, V.; Melendez-Zajgla, J. Role of Bcl-3 in solid tumors. Mol. Cancer 2011, 10, 1–7. [Google Scholar] [CrossRef]
- Li, Q.; Verma, I.M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002, 2, 725–734. [Google Scholar] [CrossRef]
- Sonenshein, G.E. Rel/NF-κB transcription factors and the control of apoptosis. Semin. Cancer Biol. 1997, 8, 113–119. [Google Scholar] [CrossRef]
- DiDonato, J.A.; Mercurio, F.; Karin, M. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012, 246, 379–400. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Rehmani, I.; Esaki, S.; Fu, R.; Chen, L.; De Serrano, V.; Liu, A. Pirin is an iron-dependent redox regulator of NF-B. Proc. Natl. Acad. Sci. USA 2013, 110, 9722–9727. [Google Scholar] [CrossRef] [PubMed]
- Barman, A.; Hamelberg, D. Fe(II)/Fe(III) Redox Process Can Significantly Modulate the Conformational Dynamics and Electrostatics of Pirin in NF-κB Regulation. ACS Omega 2016, 1, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Adeniran, C.; Hamelberg, D. Redox-Specific Allosteric Modulation of the Conformational Dynamics of κB DNA by Pirin in the NF-κB Supramolecular Complex. Biochemistry 2017, 56, 5002–5010. [Google Scholar] [CrossRef]
- Talà, A.; Damiano, F.; Gallo, G.; Pinatel, E.M.; Calcagnile, M.; Testini, M.; Fico, D.; Rizzo, D.; Sutera, A.; Renzone, G.; et al. Pirin: A novel redox-sensitive modulator of primary and secondary metabolism in Streptomyces. Metab. Eng. 2018, 48, 254–268. [Google Scholar] [CrossRef]
- Orzáez, D.; De Jong, A.J.; Woltering, E.J. A tomato homologue of the human protein PIRIN is induced during programmed cell death. Plant Mol. Biol. 2001, 46, 459–468. [Google Scholar] [CrossRef]
- Brzóska, K.; Stępkowski, T.M.; Kruszewski, M. Putative proto-oncogene Pir expression is significantly up-regulated in the spleen and kidney of cytosolic superoxide dismutase-deficient mice. Redox Rep. 2011, 16, 129–133. [Google Scholar] [CrossRef]
- Chen, Z.; Borek, D.; Padrick, S.B.; Gomez, T.S.; Metlagel, Z.; Ismail, A.M.; Umetani, J.; Billadeau, D.D.; Otwinowski, Z.; Rosen, M.K. Structure and control of the actin regulatory WAVE complex. Nature 2010, 468, 533–538. [Google Scholar] [CrossRef]
- Takenawa, T.; Miki, H. WASP and WAVE family proteins: Key molecules for rapid rearrangement of cortical actin filaments and cell movement. J. Cell Sci. 2001, 114, 1801–1809. [Google Scholar]
- Du, H.; Huang, X.; Wang, S.; Wu, Y.; Xu, W.; Li, M. PSMA7, a potential biomarker of diseases. Protein Pept. Lett. 2009, 16, 486–489. [Google Scholar] [CrossRef]
- Klooster, J.P.T.; Leeuwen, I.V.; Scheres, N.; Anthony, E.C.; Hordijk, P.L. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 2007, 26, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Watanabe, T.; Noritake, J.; Nakagawa, M.; Yamaga, M.; Kuroda, S.; Matsuura, Y.; Iwamatsu, A.; Perez, F.; Kaibuchi, K. Rac1 and Cdc42 Capture Microtubules through IQGAP1 and CLIP-170. Cell 2002, 109, 873–885. [Google Scholar] [CrossRef]
- Teng, Y.; Qin, H.; Bahassan, A.; Bendzunas, N.G.; Kennedy, E.J.; Cowell, J.K. The WASF3–NCKAP1–CYFIP1 Complex Is Essential for Breast Cancer Metastasis. Cancer Res. 2016, 76, 5133–5142. [Google Scholar] [CrossRef] [PubMed]
- Eden, S.; Rohatgi, R.; Podtelejnikov, A.V.; Mann, M.; Kirschner, M.W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 2002, 418, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Chaki, S.P.; Barhoumi, R.; Rivera, G.M. Nck adapter proteins promote podosome biogenesis facilitating extracellular matrix degradation and cancer invasion. Cancer Med. 2019, 8, 7385–7398. [Google Scholar] [CrossRef]
- Buvall, L.; Rashmi, P.; Lopez-Rivera, E.; Andreeva, S.; Weins, A.; Wallentin, H.; Greka, A.; Mundel, P. Proteasomal degradation of Nck1 but not Nck2 regulates RhoA activation and actin dynamics. Nat. Commun. 2013, 4, 1–12. [Google Scholar] [CrossRef]
- Carrillo, D.; Guerrero, N.; Muñoz, J.P.; Aedo-Agulera, V.; Tapia, J.C.; León, O.; Calaf, G.M.; Corvalán, A.; Boccardo, E.; Aguayo, F. Human papillomavirus E7 promotes EGFR/PI3K/Akt/NRF2 signaling pathway contributing to PIR/NF-kB activation in oral cancer cells. Cancers 2020, 12, 1904. [Google Scholar] [CrossRef]
- Miyazaki, I.; Simizu, S.; Okumura, H.; Takagi, S.; Osada, H. A small-molecule inhibitor shows that pirin regulates migration of melanoma cells. Nat. Chem. Biol. 2010, 6, 667–673. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Yoshikawa, R.; Yanagi, H.; Hashimoto-Tamaoki, T.; Morinaga, T.; Nakano, Y.; Noda, M.; Fujiwara, Y.; Okamura, H.; Yamamura, T. Gene expression in response to anti-tumour intervention by polysaccharide-K (PSK) in colorectal carcinoma cells. Oncol. Rep. 2004, 12, 1287–1293. [Google Scholar] [CrossRef]
- Licciulli, S.; Cambiaghi, V.; Scafetta, G.; Gruszka, A.M.; Alcalay, M. Pirin downregulation is a feature of AML and leads to impairment of terminal myeloid differentiation. Leukemia 2009, 24, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Jungk, C.; Mock, A.; Exner, J.; Geisenberger, C.; Warta, R.; Capper, D.; Abdollahi, A.; Friauf, S.; Lahrmann, B.; Grabe, N.; et al. Spatial transcriptome analysis reveals Notch pathway-associated prognostic markers in IDH1 wild-type glioblastoma involving the subventricular zone. BMC Med. 2016, 14, 170. [Google Scholar] [CrossRef] [PubMed]
- Lisabeth, E.M.; Kahl, D.; Gopallawa, I.; Haynes, S.E.; Misek, S.A.; Campbell, P.L.; Dexheimer, T.S.; Khanna, D.; Fox, D.A.; Jin, X.; et al. Identification of Pirin as a Molecular Target of the CCG-1423/CCG-203971 Series of Antifibrotic and Antimetastatic Compounds. ACS Pharmacol. Transl. Sci. 2019, 2, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Newcomb, P.A.; Carbone, P.P. The health consequences of smoking: Cancer. Med. Clin. N. Am. 1992, 76, 305–331. [Google Scholar] [CrossRef]
- Spira, A.E.; Beane, J.; Shah, V.; Liu, G.; Schembri, F.; Yang, X.; Palma, J.; Brody, J.S. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc. Natl. Acad. Sci. USA 2004, 101, 10143–10148. [Google Scholar] [CrossRef]
- Gelbman, B.D.; Heguy, A.; O’Connor, T.P.; Zabner, J.; Crystal, R.G. Upregulation of pirin expression by chronic cigarette smoking is associated with bronchial epithelial cell apoptosis. Respir. Res. 2007, 8, 10. [Google Scholar] [CrossRef]
- Mercer, B.A.; Lemaitre, V.; Powell, C.A.; D’Armiento, J. The Epithelial Cell in Lung Health and Emphysema Pathogenesis. Curr. Respir. Med. Rev. 2006, 2, 101–142. [Google Scholar] [CrossRef]
- Park, E.-J.; Park, Y.-J.; Lee, S.J.; Lee, K.; Yoon, C. Whole cigarette smoke condensates induce ferroptosis in human bronchial epithelial cells. Toxicol. Lett. 2019, 303, 55–66. [Google Scholar] [CrossRef]
- Hübner, R.-H.; Schwartz, J.D.; De, B.P.; Ferris, B.; Omberg, L.; Mezey, J.G.; Hackett, N.R.; Crystal, R.G. Coordinate Control of Expression of Nrf2-Modulated Genes in the Human Small Airway Epithelium Is Highly Responsive to Cigarette Smoking. Mol. Med. 2009, 15, 203–219. [Google Scholar] [CrossRef]
- Itoha, K.; Chibabc, T.; Takahashia, S.; Ishiia, T.; Igarashia, K.; Katoha, Y.; Oyaked, T.; Hayashid, N.; Satohe, K.; Hatayamae, I.; et al. An Nrf2/Small Maf Heterodimer Mediates the Induction of Phase II Detoxifying Enzyme Genes through Antioxidant Response Elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Ohta, T.; Iijima, K.; Miyamoto, M.; Nakahara, I.; Tanaka, H.; Ohtsuji, M.; Suzuki, T.; Kobayashi, A.; Yokota, J.; Sakiyama, T.; et al. Loss of Keap1 Function Activates Nrf2 and Provides Advantages for Lung Cancer Cell Growth. Cancer Res. 2008, 68, 1303–1309. [Google Scholar] [CrossRef] [PubMed]
- Chorley, B.N.; Campbell, M.R.; Wang, X.; Karaca, M.; Sambandan, D.; Bangura, F.; Xue, P.; Pi, J.; Kleeberger, S.R.; Bell, D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: Influence on retinoid X receptor alpha. Nucleic Acids Res. 2012, 40, 7416–7429. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.M.; Rocha, C.R.R.; Kinker, G.S.; Pelegrini, A.L.; Menck, C.F.M. The balance between NRF2/GSH antioxidant mediated pathway and DNA repair modulates cisplatin resistance in lung cancer cells. Sci. Rep. 2019, 9, 17639. [Google Scholar] [CrossRef] [PubMed]
- Hammad, A.; Namani, A.; Elshaer, M.; Wang, X.J.; Tang, X. “NRF2 addiction” in lung cancer cells and its impact on cancer therapy. Cancer Lett. 2019, 467, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, P.G.; Ellwanger, A.; Solon, M.; Cambier, C.J.; Pinkerton, K.E.; Koth, L.L. Alveolar Macrophage Recruitment and Activation by Chronic Second Hand Smoke Exposure in Mice. COPD 2009, 6, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Tien, C.-P.; Chen, C.-H.; Lin, W.-Y.; Liu, C.-S.; Liu, K.-J.; Hsiao, M.; Chang, Y.-C.; Hung, S.-C. Ambient particulate matter attenuates Sirtuin1 and augments SREBP1-PIR axis to induce human pulmonary fibroblast inflammation: Molecular mechanism of microenvironment associated with COPD. Aging 2019, 11, 4654–4671. [Google Scholar] [CrossRef]
- Hamra, G.B.; Guha, N.; Cohen, A.; Laden, F.; Raaschou-Nielsen, O.; Samet, J.M.; Vineis, P.; Forastiere, F.; Saldiva, P.; Yorifuji, T.; et al. Outdoor Particulate Matter Exposure and Lung Cancer: A Systematic Review and Meta-Analysis. Environ. Health Perspect. 2014, 122, 906–911. [Google Scholar] [CrossRef]
- Yokoi, S.; Yasui, K.; Iizasa, T.; Imoto, I.; Fujisawa, T.; Inazawa, J. TERC identified as a probable target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers. Clin. Cancer Res. 2003, 9, 4705–4713. [Google Scholar]
- Wu, X.; Ruan, L.; Yang, Y.; Mei, Q. Identification of crucial regulatory relationships between long non-coding RNAs and protein-coding genes in lung squamous cell carcinoma. Mol. Cell. Probes 2016, 30, 146–152. [Google Scholar] [CrossRef]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; De Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef]
- Iarc, W.G. Human papillomaviruses. IARC Monogr. Eval. Carcinog. Risks Hum. 2007, 90, 1–636. [Google Scholar]
- Carrillo, D.; Muñoz, J.P.; Huerta, H.; Leal, G.; Corvalan, A.H.; León, O.; Calaf, G.M.; Urzúa, U.; Boccardo, E.; Tapia, J.C.; et al. Upregulation of PIR gene expression induced by human papillomavirus E6 and E7 in epithelial oral and cervical cells. Open Biol. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Komai, K.; Niwa, Y.; Sasazawa, Y.; Simizu, S. Pirin regulates epithelial to mesenchymal transition independently of Bcl3-Slug signaling. FEBS Lett. 2015, 589, 738–743. [Google Scholar] [CrossRef]
- Tabrizi, R.; Vakili, S.; Akbari, M.; Mirhosseini, N.; Lankarani, K.B.; Rahimi, M.; Mobini, M.; Jafarnejad, S.; Vahedpoor, Z.; Asemi, Z. The effects of curcumin-containing supplements on biomarkers of inflammation and oxidative stress: A systematic review and meta-analysis of randomized controlled trials. Phytother. Res. 2019, 33, 253–262. [Google Scholar] [CrossRef]
- Calaf, G.M.; Urzúa, U.; Termini, L.; Aguayo, F. Oxidative stress in female cancers. Oncotarget 2018, 9, 23824–23842. [Google Scholar] [CrossRef]
- Thacker, P.C.; Karunagaran, D. Curcumin and Emodin Down-Regulate TGF-β Signaling Pathway in Human Cervical Cancer Cells. PLoS ONE 2015, 10, e0120045. [Google Scholar] [CrossRef]
- Gallardo, M.; Calaf, G.M. Curcumin inhibits invasive capabilities through epithelial mesenchymal transition in breast cancer cell lines. Int. J. Oncol. 2016, 49, 1019–1027. [Google Scholar] [CrossRef]
- Gallardo, M.; Kemmerling, U.; Aguayo, F.; Bleak, T.C.; Calaf, G.M. Curcumin rescues breast cells from epithelial-mesenchymal transition and invasion induced by anti-miR-34a. Int. J. Oncol. 2019, 56, 480–493. [Google Scholar] [CrossRef]
- Aedo-Aguilera, V.; Carrillo-Beltrán, D.; Calaf, G.M.; Muñoz, J.P.; Guerrero, N.; Osorio, J.C.; Tapia, J.C.; León, O.; Contreras, H.R.; Aguayo, F. Curcumin decreases epithelial-mesenchymal transition by a Pirin-dependent mechanism in cervical cancer cells. Oncol. Rep. 2019, 42, 2139–2148. [Google Scholar] [CrossRef]
- Brzóska, K.; Stępkowski, T.M.; Kruszewski, M. Basal PIR expression in HeLa cells is driven by NRF2 via evolutionary conserved antioxidant response element. Mol. Cell. Biochem. 2014, 389, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Rogers, H.W.; Weinstock, M.A.; Feldman, S.R.; Coldiron, B.M. Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the US Population, 2012. JAMA Dermatol. 2015, 151, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Licciulli, S.; Luise, C.; Zanardi, A.; Giorgetti, L.; Viale, G.; Lanfrancone, L.; Carbone, R.; Alcalay, M. Pirin delocalization in melanoma progression identified by high content immuno-detection based approaches. BMC Cell Biol. 2010, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Licciulli, S.; Luise, C.; Scafetta, G.; Capra, M.; Giardina, G.; Nuciforo, P.; Bosari, S.; Viale, G.; Mazzarol, G.; Tonelli, C.; et al. Pirin Inhibits Cellular Senescence in Melanocytic Cells. Am. J. Pathol. 2011, 178, 2397–2406. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Hirota, A.; Kawachi, Y.; Itoh, K.; Nakamura, Y.; Xu, X.; Banno, T.; Takahashi, T.; Yamamoto, M.; Otsuka, F. Ultraviolet A Irradiation Induces NF-E2-Related Factor 2 Activation in Dermal Fibroblasts: Protective Role in UVA-Induced Apoptosis. J. Investig. Dermatol. 2005, 124, 825–832. [Google Scholar] [CrossRef]
- Sample, A.; Zhao, B.; Wu, C.; Qian, S.; Shi, X.; Aplin, A.; He, Y.-Y. The Autophagy Receptor Adaptor p62 is Up-regulated by UVA Radiation in Melanocytes and in Melanoma Cells. Photochem. Photobiol. 2018, 94, 432–437. [Google Scholar] [CrossRef]
- Thomsen, K.G.; Terp, M.G.; Lund, R.R.; Søkilde, R.; Elias, D.; Bak, M.; Litman, T.; Beck, H.C.; Lyng, M.B.; Ditzel, H.J. miR-155, identified as anti-metastatic by global miRNA profiling of a metastasis model, inhibits cancer cell extravasation and colonization in vivo and causes significant signaling alterations. Oncotarget 2015, 6, 29224–29239. [Google Scholar] [CrossRef]
- Xiang, X.; Zhuang, X.; Ju, S.; Zhang, S.; Jiang, H.; Mu, J.; Zhang, L.; Miller, D.; Grizzle, W.; Zhang, H.-G. miR-155 promotes macroscopic tumor formation yet inhibits tumor dissemination from mammary fat pads to the lung by preventing EMT. Oncogene 2011, 30, 3440–3453. [Google Scholar] [CrossRef]
- Higashi, K.; Tomigahara, Y.; Shiraki, H.; Miyata, K.; Mikami, T.; Kimura, T.; Moro, T.; Inagaki, Y.; Kaneko, H. A Novel Small Compound That Promotes Nuclear Translocation of YB-1 Ameliorates Experimental Hepatic Fibrosis in Mice. J. Biol. Chem. 2011, 286, 4485–4492. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, H.; Sumiyoshi, H.; Higashi, K.; Nakao, S.; Minakawa, K.; Sumida, K.; Saito, K.; Ikoma, N.; Mabuchi, T.; Ozawa, A.; et al. A novel small compound accelerates dermal wound healing by modifying infiltration, proliferation and migration of distinct cellular components in mice. J. Dermatol. Sci. 2014, 74, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, M.D.; Chessum, N.E.A.; Rye, C.S.; Pasqua, A.E.; Tucker, M.J.; Wilding, B.; Evans, L.E.; Lepri, S.; Richards, M.; Sharp, S.Y.; et al. Discovery of a Chemical Probe Bisamide (CCT251236): An Orally Bioavailable Efficacious Pirin Ligand from a Heat Shock Transcription Factor 1 (HSF1) Phenotypic Screen. J. Med. Chem. 2016, 60, 180–201. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Bryan, L.; Jemal, A. Breast cancer statistics, 2013. CA Cancer J. Clin. 2013, 64, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Veer, L.J.V.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.M.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef]
- Karlsson, E.; Delle, U.; Danielsson, A.; Olsson, B.; Abel, F.; Karlsson, P.; Helou, K. Gene expression variation to predict 10-year survival in lymph-node-negative breast cancer. BMC Cancer 2008, 8, 254. [Google Scholar] [CrossRef]
- Shubbar, E.; Helou, K.; Kovács, A.; Nemes, S.; Hajizadeh, S.; Enerbäck, C.; Einbeigi, Z. High levels of γ-glutamyl hydrolase (GGH) are associated with poor prognosis and unfavorable clinical outcomes in invasive breast cancer. BMC Cancer 2013, 13, 47. [Google Scholar] [CrossRef]
- Suleman, M.; Chen, A.; Ma, H.; Wen, S.; Zhao, W.; Lin, D.; Wu, G.; Li, Q. PIR promotes tumorigenesis of breast cancer by upregulating cell cycle activator E2F1. Cell Cycle 2019, 18, 2914–2927. [Google Scholar] [CrossRef]
- DeGregori, J. The genetics of the E2F family of transcription factors: Shared functions and unique roles. Biochim. Biophys. Acta 2002, 1602, 131–150. [Google Scholar] [CrossRef]
- Sun, C.C.; Li, S.-J.; Hu, W.-D.; Zhang, J.; Zhou, Q.; Liu, C.; Li, L.-L.; Songyang, Y.-Y.; Zhang, F.; Chen, Z.-L.; et al. Comprehensive Analysis of the Expression and Prognosis for E2Fs in Human Breast Cancer. Mol. Ther. 2019, 27, 1153–1165. [Google Scholar] [CrossRef] [PubMed]
- Hutter, C.; Zenklusen, J.C. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018, 173, 283–285. [Google Scholar] [CrossRef]
- Bergman, A.-C.; Alaiya, A.A.; Wendler, W.; Binétruy, B.; Shoshan, M.; Sakaguchi, K.; Bergman, T.; Kronenwett, U.; Auer, G.; Appella, E.; et al. Protein kinase-dependent overexpression of the nuclear protein pirin in c-JUN and RAS transformed fibroblasts. Cell. Mol. Life Sci. 1999, 55, 467–471. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.W.; Oh, S. Comparative analysis of NRF2-responsive gene expression in AcPC-1 pancreatic cancer cell line. Genes Genom. 2015, 37, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, W.; Li, H.; Deng, Q.; Yang, M.; Li, X.; Liang, Z. Identification of key genes and pathways associated with cholangiocarcinoma development based on weighted gene correlation network analysis. PeerJ 2019, 7, e7968. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.; Craft, B.; Brooks, A.; Zhu, J.; Haussler, D. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. BioRxiv 2019, 1–16. [Google Scholar] [CrossRef]
- Panchal, H.D.; Vranizan, K.; Lee, C.Y.; Ho, J.; Ngai, J.; Timiras, P.S. Early Anti-Oxidative and Anti-Proliferative Curcumin Effects on Neuroglioma Cells Suggest Therapeutic Targets. Neurochem. Res. 2008, 33, 1701–1710. [Google Scholar] [CrossRef]
- Edwards, H.; Rubenstein, M.; Dombkowski, A.A.; Caldwell, J.T.; Chu, R.; Xavier, A.C.; Thummel, R.; Neely, M.; Matherly, L.H.; Ge, Y.; et al. Gene Signature of High White Blood Cell Count in B-Precursor Acute Lymphoblastic Leukemia. PLoS ONE 2016, 11, e0161539. [Google Scholar] [CrossRef]
- Teachey, D.T.; Hunger, S.P. Predicting relapse risk in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 2013, 162, 606–620. [Google Scholar] [CrossRef]
| Model | Function | Description | Reference |
|---|---|---|---|
| Human | Enzymatic | Quercetinase activity | [13] |
| Prokaryote | Co-Enzymatic | Inhibition of acetyl-CoA catabolism | [15] |
| Human | Transcriptional regulator | Interaction with NF-I/BCL-3/NF-κB p50 | [11] |
| Human | Transcriptional regulator/Redox sensor | Binding to NF-κB p65 in oxidative conditions | [22] |
| Human | Transcriptional regulator | Fe active form favors its binding and regulation to NF-κB/DNA | [23,24] |
| Plants | Transcriptional regulator/Redox sensor | Regulation of oxidative pathways and cell death and redox sensor | [26] |
| Animal | Redox sensor | Activation in superoxide dismutase (Sod1)-deficient mice | [27] |
| Cancer | Factors | Regulation | Comments | Ref. |
|---|---|---|---|---|
| Lung | TS | Activation | Pirin levels are increased in airway epithelium of chronic smokers | [46] |
| TS | Activation | Pirin overexpression occurs in a dose-dependent manner | [47] | |
| TS | Activation | Interaction with NF-κB resulting in a pro-apoptotic response | [47] | |
| TS | Activation | Pirin overexpression is accompanied by ferroptosis markers upregulation | [49] | |
| TS | Activation | Interaction with NRF2 in smoke-exposed airway epithelial cells | [50] | |
| Cervical | E7 (HPV16) | Activation | Pirin regulates EMT and migration by interacting with NF-κB | [63] |
| Curcumin | Suppression | Curcumin decreases Pirin expression, and consequently EMT and cell migration | [70] | |
| Skin | TPh A | Suppression | Interferes the Pirin interaction with BCL-3, and consequently inhibits cell migration | [38] |
| miR-155 | Suppression | Pirin may mediate metastasis development | [79] | |
| CCG | Suppression | Inhibition of carcinogenic signaling pathways | [43] | |
| Oral | E7 (HPV16) | Activation | EGFR/MEK/ERK and PI3K/AKT pathways are involved in Pirin activation by HPV16 E7 | [63] |
| E7 (HPV16) | Activation | Upregulation of c-Rel and p65 through an interplay with Pirin, promotes cell migration and EMT | [37] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perez-Dominguez, F.; Carrillo-Beltrán, D.; Blanco, R.; Muñoz, J.P.; León-Cruz, G.; Corvalan, A.H.; Urzúa, U.; Calaf, G.M.; Aguayo, F. Role of Pirin, an Oxidative Stress Sensor Protein, in Epithelial Carcinogenesis. Biology 2021, 10, 116. https://doi.org/10.3390/biology10020116
Perez-Dominguez F, Carrillo-Beltrán D, Blanco R, Muñoz JP, León-Cruz G, Corvalan AH, Urzúa U, Calaf GM, Aguayo F. Role of Pirin, an Oxidative Stress Sensor Protein, in Epithelial Carcinogenesis. Biology. 2021; 10(2):116. https://doi.org/10.3390/biology10020116
Chicago/Turabian StylePerez-Dominguez, Francisco, Diego Carrillo-Beltrán, Rancés Blanco, Juan P. Muñoz, Grettell León-Cruz, Alejandro H. Corvalan, Ulises Urzúa, Gloria M. Calaf, and Francisco Aguayo. 2021. "Role of Pirin, an Oxidative Stress Sensor Protein, in Epithelial Carcinogenesis" Biology 10, no. 2: 116. https://doi.org/10.3390/biology10020116
APA StylePerez-Dominguez, F., Carrillo-Beltrán, D., Blanco, R., Muñoz, J. P., León-Cruz, G., Corvalan, A. H., Urzúa, U., Calaf, G. M., & Aguayo, F. (2021). Role of Pirin, an Oxidative Stress Sensor Protein, in Epithelial Carcinogenesis. Biology, 10(2), 116. https://doi.org/10.3390/biology10020116






