Simple Summary
The study of cell features has historically been key for the progress of biological sciences and its relevance remains intact. In recent years, mass cytometry has emerged as a promising and powerful technology, capable of studying multiple parameters of cells in the same sample. Mass cytometry has been quickly applied to many research areas, particularly to the study of the immune system for different purposes, in which the simultaneous analysis of a large number of proteins is crucial. However, despite being a technique that is on the rise, its performance in scientific publications has not yet been evaluated. In this work, a bibliometric methodology known as H-Classics was applied to analyse the most relevant articles, known as highly cited papers (HCPs), and determine the main scientific producers (authors, institutions, and countries) and trends around mass cytometry research field. The results confirmed a high interest and application in immunological studies. The identified HCPs came from prestigious institutions and were published in high impact journals. These results may help researchers to expand their knowledge and to establish new valuable collaborative networks around mass cytometry.
Abstract
Mass cytometry (CyTOF) is a relatively novel technique for the multiparametric analysis of single-cell features with an increasing central role in cell biology, immunology, pharmacology, and biomedicine. This technique mixes the fundamentals of flow cytometry with mass spectrometry and is mainly used for in-depth studies of the immune system and diseases with a significant immune load, such as cancer, autoimmune diseases, and viral diseases like HIV or the recently emerged COVID-19, produced by the SARS-CoV-2 coronavirus. The objective of this study was to provide a useful insight into the evolution of the mass cytometry research field, revealing the knowledge structure (conceptual and social) and authors, countries, sources, documents, and organizations that have made the most significant contribution to its development. We retrieved 937 articles from the Web of Science (2010–2019), analysed 71 Highly Cited Papers (HCP) through the H-Classics methodology and computed the data by using Bibliometrix R package. HCP sources corresponded to high-impact journals, such as Nature Biotechnology and Cell, and its production was concentrated in the US, and specifically Stanford University, affiliation of the most relevant authors in the field. HCPs analysis confirmed great interest in the study of the immune system and complex data processing in the mass cytometry research field.
1. Introduction
Studying a broad range of single-cell features has always been of great interest to both researchers and clinicians. For decades, this purpose has been partly fulfilled by flow-cytometry technology [1]. For the study of cellular constituents and biomarkers by flow cytometry, cells are typically stained with monoclonal antibodies conjugated to fluorochromes. The key principle of flow cytometry is that, after the excitation of stained cells in a flow cytometer, a variety of aspects of individual particles can be measured through light scattering and fluorescence emission [2]. Flow cytometry has allowed the study of countless biological features [3] and is also being clinically used for the diagnosis of haematological diseases, such as leukaemia [4].
However, since flow cytometry is not without limitations, more comprehensive approaches to single-cell feature analysis have been developed, especially for multiparametric analyses [5]. Some examples are high dimensional fluorescent flow cytometry, single-cell mRNA sequencing, and mass cytometry [6]. The mass cytometry technology or cytometry by time of flight (CyTOF), is a relatively novel technique that was first described by Bandura et al., as an instrument for real-time analysis of individual biological cells or other microparticles [7]. The CyTOF constitutes a hybrid between flow cytometry and mass spectrometry. Instead of fluorophores, the cells are immunologically stained with antibodies conjugated with unique and stable metallic particles. Then, in the mass cytometer, samples ionised and focused under the same acceleration potential. The ions arrive at the detector in an ion mass-dependent manner, which enables the differentiation of single-cell parameters. For analysing this information, the detector is coupled to a time of flight analyser (TOF), from where the data is acquired and subsequently processed [8,9]. Due to the high availability of different metallic particles, mass cytometry allows the study of more than 40 different parameters simultaneously in the same sample [6].
The applications of mass cytometry are as broad as previous fluorescence-based techniques for studying single-cell features, with the exception that CyTOF does not allow cell recovery by sorting, as cells are destroyed in the process. Through performing deep profiling of samples, mass cytometry experiments have obtained comparable results to those of flow-cytometry, for example, being in the same way capable of quantifying surface and intracellular biomarkers from both non-stimulated [10] and stimulated [11] cells. The most important use of this technique is probably its capacity for immunophenotyping, that its, in-depth study of the characteristics and diversity of the immune system under different conditions [6]. Deep CyTOF profiling has also been applied in several drug pharmacodynamics, pharmacokinetics and discovery studies [12] to help determine better drug combinations and drug targets, especially for autoimmune diseases and cancer [13,14,15], and has also been used to support vaccine evaluation and development processes [16].
The huge potential of this technique suggests a booming trend in the use of CyTOF within the research community. However, limited information on the research evaluation of this new technology in scientific communications is currently available. A widely used approach for analysing scientific publications is the citation classics methodology. Citation classics was first defined by Garfield as a way to retrospectively determine the most highly cited papers (HCP) around a research topic [17]. The HCPs analysis aims to discover trends in the research community and identify relevant authors, institutions, or groups [17,18,19]. This analysis may facilitate comprehension of the research output and provide the basis for developing new theories, techniques, research lines and collaboration networks. However, to perform a good analysis, the criteria used to select the most cited papers should not be arbitrary. For example, some authors have arbitrarily established thresholds in the number of articles selected, limiting the list to the 50 [20] or 100 [21] most cited, or limiting the selection to those articles that have been cited at least 400 times [22].
To improve the selection criteria for HCP, Martinez et al. [23] proposed that the selection of HCPs should be based on two parameters: the H-Index [24] and the H-core concept [25]. The H-Index, defined as the number of papers with citation number > h, as was presented by Hirsch as a way to improve quantitative measures and impact of a researcher’s scientific output in one robust single parameter and avoid the disadvantage of using other indicators, such as the total number of papers, the total number of citations, citations per paper, or the number of significant papers [24]. Therefore, the application of H-index on citation classics reduces unpredictability [23].
Martinez et al. [23] gave the name H-Classics to this new identification method of HCPs based on the H-index. Two great advantages of using H-Classics is that it provides standardization of the criteria and thresholds used for selecting HCPs and includes the collection of papers published in a given field and their impact on a single procedure [26]. The H-Classics methodology has already been applied to medical areas such as paediatrics [27], dentistry [18], rheumatology [28], and microbiology [19].
In this paper, we focus on the study of mass cytometry research by using the H-Classics methodology. This H-Classics study aims to answer the following research questions (RQ):
- RQ1. Which are the most relevant journals, authors, institutions, and countries in mass cytometry research?
- RQ2. Which are the most cited documents in mass cytometry?
- RQ3. Which are the knowledge structures (conceptual and social structure) in mass cytometry?
2. Materials and Methods
2.1. Bibliographic Database and Query Design
The source of information related to scientific production and citations was the Web of Science (WoS) database (Clarivate Analytics). Academic publications indexed in WoS on CyTOF from 2010 to 2019 were obtained. To set and adequately delimit the research area under study, a specific search equation was formulated according to the database search logic of WoS. Table 1 demonstrates the query design, indexes, timespan, and data download date. The retrieved WoS dataset and the citation report are available at Zenodo repository [29].

Table 1.
Details of dataset search strategy for the identification of mass cytometry papers in WoS.
2.2. H-Classics Methodology
After choosing the database and the query design, the following two steps for identifying HCPs were applied for the mass cytometry research field, accordingly to Martinez et al., description of the H- Classics methodology [23]:
- Compute the H-index of the research area. The computation of H-index of the research area is done by establishing a ranking of the papers according to their citations. The WoS database provides filtering tools to easily compute the H-index of the research area.
- Compute the H-core of the research area. This step consists of recovering the highly cited papers that are included in the H-core of the research area [23].
2.3. Bibliometrix (Science Mapping Analysis)
A comprehensive science mapping analysis [30,31] was performed to the data obtained from the HCPs selected. Data were processed and H-Classics scientific production was analysed through a bibliometric workflow supported by the open-source tool Bibliometrix package, which is programmed in R [32,33]. Following Aria et al., methodology, data were extracted from the Clarivate Analytics WoS database, loaded in R, and converted into a bibliographic data frame into which several elements, such as the authors’ names, titles, keywords, and other information, were introduced [33].
The descriptive analysis was performed applying several Bibliometrix functions described by Aria et al., to obtain tables and figures of HCPs production and citations over time, most productive authors, institutions, countries, most globally cited documents and authors production over time. Author productivity and citation impact were also calculated through H-index functions. The next step was the creation of networks in which different connections between attributes of a document were represented through a matrix. These connections were used to represent figures and networks corresponding to the collaboration network between HCPs’ authors (social structure) and co-occurrence network (conceptual structure) [33]. Co-ocurrence networks are generated by connecting pairs of terms (keywords) using a set of criteria defining co-occurrence. Data visualizations of author production and representation of social and conceptual structures were created through the Biblioshiny R package (https://bibliometrix.org/Biblioshiny.html), a Bibliometrix R Package web interface.
3. Results
3.1. Citation Report and Record Count
The total mass cytometry publications retrieved (937) combined a sum of 25,801 times cited, making an average of 27.54 citations per paper. The H-index was 71, which means that 71 studies (Table A1) had received at least 71 citations and were therefore categorised as HCPs (H-Classics publications). Of these, 56 were original articles, 14 were reviews and 1 was a book chapter.
3.2. Distribution of Publications by Year, Average Citations per Year and Record Count
Figure 1 shows the yearly distribution of HCP documents from 2010 to 2019. The main production was concentred in the period 2014–2017. The peak number of HCPs occurred in 2016, with 17 HCP published.
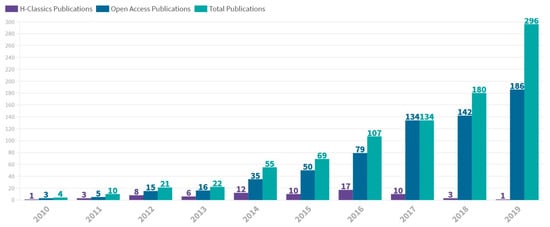
Figure 1.
Annual scientific production in mass cytometry between 2010 and 2019. Number of H-Classics publications, open access publications and total publications are represented. The figure was designed with Flourish.
Figure 2 shows the average citations per year of HCPs in the mass cytometry field, remaining fairly constant between 2010 and 2018, before seeing a sharp increase to 208 in 2019.
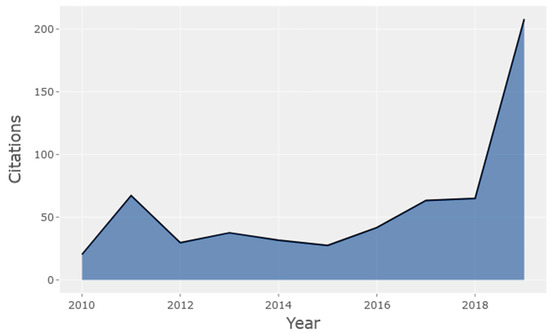
Figure 2.
Average citations per year of HCPs in the mass cytometry research field (2010–2019).
3.3. Journals, Authors, Institutions and Countries
In this section, different social units (journals, authors, institutions, and countries) were analysed. First, the most productive journals were determined in terms of the total number of HCPs in the selected time frame. Table 2 shows journals with two or more HCPs in the mass cytometry research field. Nature Biotechnology with 9 HCPs was the most productive journal, followed by Cell and Cytometry Part A, with 8 and 5 HCPs, respectively.

Table 2.
Distribution of sources with two or more HCPs in the area of mass cytometry.
Table 3 highlights the 20 most relevant authors in mass cytometry according to the number of published HCPs. Nolan GP ranked highest with 23 HCPs followed by Bendall SC (14 HCPs) and Newell EW (12 HCPs). Nolan GP was also one of the authors with the earliest date of first publication and the highest number of total citations (6531). The affiliations of these authors (75%) corresponded mainly to American hospitals, research centers, and universities.

Table 3.
Top 20 most relevant authors and their impact (H-index and TC) in the field of mass cytometry.
Affiliations and countries were also analysed. To determine the most relevant institutions, all authors from each HCP were considered. Table 4 shows the ranking of institutions with 6 or more HCP. The most productive affiliations were compared with two quality and performance indicators of global university ranking: the 2019 Quacquarelli Symonds (QS) World University Rankings and 2019 Academic Ranking of World Universities (ARWU), which allow an approximate measurement of the relative position in which the most influential institutions in CyTOF publications were found. The most productive institution was Stanford University (USA), with 172 registered affiliations in HCPs, and it was also the affiliation of the top 3 authors (Table 3). As shown in Table 4, the 2019 QS/ARWU rankings indicate that top positions were held by Stanford University.

Table 4.
Top number of affiliations registered (institutions) in mass cytometry highly cited papers (HCPs).
Table 5 shows the ranking of countries among producers of HCPs in mass cytometry, based on the affiliation of the corresponding author, and ordered according to the total number of HCPs. For a more global view of HCP contribution, the affiliations of all authors were also determined (Freq SCP) (Table 5). The countries of origin of all authors, and not only of the corresponding authors, were considered when determining the importance of a country in HCPs production.

Table 5.
Corresponding author’s country and country scientific production in the field of mass cytometry’s highly cited papers (HCPs).
Most of the H-Classics were derived from USA (n = 44, 61.97%), followed by Switzerland (n = 8, 11, 27%) and Singapore (n = 6, 8, 45%). Interestingly, only 25% of the USA HCPs were categorised as multiple country publications (MCP), the lowest ratio among the 12 top countries measured, after Canada and Germany. To relate gross domestic product (GDP) with the production of HCPs, Table 5 also displays the ranking of countries ordered by Adjustment Index (AI) based on GDP per capita [28]. The adjustment index (AI) was calculated as AI = ((total number of HCP/GDP per capita of the country) × 100).
3.4. Content Analysis: Highly Cited Papers
The first HCP is from 2010, “Highly multiparametric analysis by mass cytometry” by Olga Ornatsky et al. [8]. They reviewed mass cytometry as a new technology capable of addressing the studies usually run by flow cytometers with better results, emphasizing its multiparametric analysis potential for biological research and drug development. To describe the basic principles of this technique, they used 20 different antibodies for immunophenotyping of human leukaemia cell lines and patient samples; and performed differential cell analysis, proteins identification, and metal-encoded bead arrays. This article pointed out the next steps in CyTOF development, such as improving measurement efficiency, the construction of isotope-binding polymer tags for a wider group of elements, and challenges of multiparametric data processing and representation. Ornatsky’s article, with 203 total citations, is the only HCP from 2010.
The 2011 article “Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum” by Bendall et al. [34] was the most cited mass cytometry HCP, with 1233 total citations (Table 6). In this article, healthy human bone marrow was used to simultaneously measure 34 parameters in single cells. It provided a system-wide view of immune signalling in healthy human haematopoiesis as a powerful way to compare mechanistic and pharmacologic studies. The second highest cited paper in the list (Table 6), with 716 total citations, was the 2012 article entitled “viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukaemia” by Amir et al. In the article, the authors presented and validated a computerised tool that allows correct visualization of multiparametric mass cytometry information [35]. The third most cited paper was published by Ginsen et al., in 2014 and entitled “Highly multiplexed imaging of tumour tissues with subcellular resolution by mass cytometry” (500 total citations). The authors mixed mass cytometry with immunohistochemical techniques to study the heterogeneity of breast cancer tumours in tissues, rather than cell suspensions [36]. In 2011, Qiu et al. published the article “Extracting a cellular hierarchy from high-dimensional cytometry data with sPADE” [37], intending to solve some of the difficulties of analysing huge amounts of multidimensional single-cell data. The authors presented a computational approach that facilitates the analysis of cellular heterogeneity, the identification of cell types, and the comparison of functional markers in response to perturbations. This article was the fourth most cited HCP, with 498 total citations.

Table 6.
Top 10 most highly cited papers in mass cytometry (2010–2019).
Finally, the latest HCP published was “Dimensionality reduction for visualizing single-cell data using UMAP” by Becht et al., (2019) [44], who used a nonlinear dimensionality-reduction technique for analysing mass cytometry biological data. This new algorithm, called UMAP, achieved a more efficient and improved visualization and understanding of single-cell data, which is still a challenge in recent years despite advances in computational methods for visualizing high dimensional data.
3.5. Top Author’s Production Over Time
Figure 3 shows the top authors’ production in mass cytometry HCPs over time. Authors such as Nolan GP or Bendall SC have maintained a consistent scientific production throughout almost the entire decade. Authors such as Simonds EF, Finck R, and Fantil WJ, had a higher contribution during the first half of the study period, while Spitzer MH had a more significant contribution during the second half.
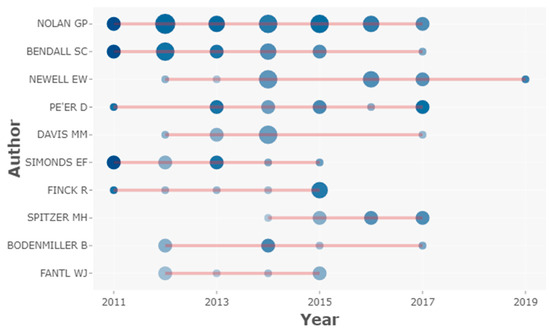
Figure 3.
Top authors’ production over time. The horizontal red lines indicate the scientific production timeline per author from 2010 to 2019. The circle size represents the number of HCPs published per year and the colour intensity (colour coding) is proportional to the sum of total citations per year. The figure was created using Biblioshiny web interface.
3.6. Conceptual and Social Structure
3.6.1. Conceptual Structure: Co-Occurrence Network
To analyse the frequency of keywords associated with the HCPs and study possible relationships between them, a co-occurrence keyword network was generated. Figure 4 shows the most used keywords in the analysed HCPs and predicted interconnections based on paired presence. Despite the presence of differentiated clusters, no noteworthy distinctions of nodes were observed. Frequent terms closely related to each other, such as “flow cytometry” and “mass cytometry”, were found to be interconnected with most other less frequent terms like “network” or “peripheral blood”. Terms such as “expression”, “T-cells, “macrophages”, “homeostasis” or “disease” tended to be interconnected. Similarly, terms like “responses”, “activation”, “populations”, “natural killer cells” were clustered together.
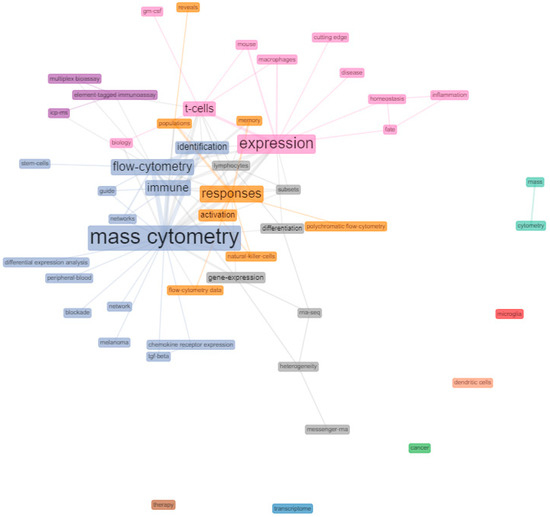
Figure 4.
Keyword co-occurrence network of mass cytometry HCPs. Co-occurrence networks are shared interconnection of terms based on their paired presence within a definite unit of text. The size of the terms indicates their greater or lesser frequency with which they coincide with another term in the network throughout the HCPs (co-occurrence). Network parameters: field (keyword plus), network layout (Fruchterman & Reingold), normalization (association), node colour by year (No), clustering algorithm (Louvain), number of nodes (5–50), remove isolated nodes (No), and minimum edges (2). Graphical parameters: opacity (0–0.7), number of labels (0–50), label cex (Yes), label size (0–6), node shape (Box), edge size (0.1–5), and curved edges (No). The figure was created using Biblioshiny web interface.
3.6.2. Social Structure: Collaboration Network
Figure 5 shows the social structure through a collaboration network, where nodes are authors and links are co-authorships. There were two strongly differentiated clusters. On the one hand, the red cluster, led by Nolan GP and Bendall SC had a solid structure of collaborations with other influential authors, for example, Simonds EF, Finck R, and Levine JH.
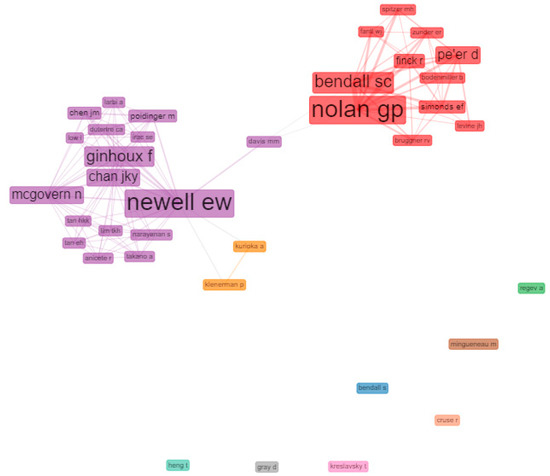
Figure 5.
Clusters of most relevant author collaboration networks in mass cytometry’s HCPs. Network parameters: field (authors), normalization (No), clustering algorithm (Louvain), network Layout (automatic layout), number of nodes (37), remove isolated nodes (No), and minimum edges (2). Graphical parameters: opacity (0–0.7), number of labels (5–50), label cex (Yes), label size (0–6), node shape (Box), edge size (0.1–5), curved edges (No). The figure was created using Biblioshiny web interface.
On the other hand, the violet cluster, led by Newell EW had a structure of collaboration with other impact authors such as Chen JM, Chan JKY, Ginhoux F and Mcgovern N among others. The author who acted as inter-collaborator between groups was Davis MM. The violet cluster appears related to a small orange cluster, with satellite collaborations involving Kurioka A and Kleneman P.
4. Discussion
In the present study, Mass Cytometry highly cited papers have been identified and analysed for the first time using the H-Classics methodology. The analysis of the HCPs allows us to highlight the following findings:
71 Mass Cytometry HCPs were identified in the period 2010–2019, with a predominance of original articles (56) compared to reviews (14). The increasing trend in citations per year presented in this work indicates a robust interest in mass cytometry in recent years, as CyTOF research is currently experiencing a potential growth phase. The HCPs were more frequent in the second half of the study period, with relatively recent publications attracting scientific interest. During the period analysed, 2016 showed the highest number of HCPs in mass cytometry. A peak so close to the end of the analysed time frame indicates that there is a growing interest in mass cytometry in the research community, considering that a minimum citation time window of 5 years is usually needed by a paper to obtain its maximum number of cites [45]. The immediate recognition of mass cytometry articles is probably also a consequence of most of them being published in open access journals, which usually results into a higher number of citations and impact compared to publications in non-open access journals [46]. Having this in mind and knowing that older articles are expected to have higher absolute citation rates, it is expected that the peak of HCPs will be actualised to a more recent date in years to come.
Compared with other H-Classics studies on other disciplines, a smaller number of HCPs was identified in the present work. This is an expected result, considering that H-Classics studies usually focus on more general thematic areas and tend to analyse longer periods. For example, in the study by Moral-Muñoz et al., 645 HCPs in the microbiology area were analysed, covering a period of more than 100 years [19]. In our study, the timespan analysed was chosen considering that it covers most of the scientific production in mass cytometry. The rationale for choosing WoS as a source of the HCPs instead of other databases such as Scopus or Google Scholar, was that it provides numerous analysis tools for processing the data and offers highly reliable research information, which is particularly good for the study of biological sciences [47].
The HCP with the highest citations count, about differential immune and drug responses, was authored by Bendall et al., (2011). The reason that this article is the most cited in the field of mass cytometry may be due to two facts: first, it is one of the first works to successfully apply this novel technology (2011). Secondly, the paper highlights the utility of CyTOF in drug action and mechanistic studies and therefore could attract the attention of numerous pharmacology studies. The capacity of CyTOF for the evaluation of the effects of drugs both in the clinical and pre-clinical setting has been indicated in several studies [12,48,49]. The first HCP was published by Ornatsky et al., (2010) about immunophenotyping potential of the technique and it occupies position 24 out of 71. This article helps to understand how, from the beginning, this technique has been focused on the study of the immune system [8]. Interestingly, among the most cited papers, the second and the fourth-ranked were based on computer strategies to achieve adequate visualization and analysis of the results obtained by mass cytometry. This topic is repeated among HCPs throughout the entire decade studied, and was observed even in the most recently published HCP corresponding to 2019 [44]. The consistent interest in these articles suggests that the treatment of multiparametric data continues to be a challenge for this technique.
Nature Biotechnology was the most productive journal with 9 HCPs, followed by Cell with 8 HCPs. It is expected that the most cited articles of modern technology such as CyTOF, used in medicine and biology, will appear in more influential journals in this field. Nature Biotechnology is a journal belonging to Journal Citation Report from Web of Science, found in Q1 Quartile and ranked 02/156 (2019) in the area of Biotechnology & Applied Microbiology, with a Journal Impact Factor (JIF) of 36.56 (2019). High JIF journals are usually considered as a standard of quality and reliability in the scientific community. Equally applicable to our findings, high journal impact factors helped explain the high number of citations of HCPs studied in the field of paediatrics [27].
Nolan GP and Bendall SC were the authors present in the highest number of HCPs, 23 and 14, respectively. The high participation of these authors in HCPs consolidates them as world leaders in this technique and demonstrates the predominance of USA in global mass cytometry production, from which Standford University (172 affiliations registered) and Harvard University (28 affiliations registered) stand out as the most relevant institutions. According to ARWU/QS 2019, these institutions stand out for their high funding, internationalization of research, and quality of teaching staff/researchers. USA appears to have a low MCP ratio but, considering that the largest collaboration networks and the highest production of HCP occur in this territory, it is to be expected that the possibilities of international collaboration will be limited. This high scientific production is also possible thanks to a high number of collaborative works between the two main authors as they appear to cluster together in the collaboration network and have similar HCP production over time. Nolan GP leads the research group known as Nolan Lab (Stanford University), focused on the study of haematopoiesis, cancer and leukaemia, autoimmunity and inflammation, and computational approaches for network and systems immunology, with special knowledge in the mass cytometry technology [35].
Another collaboration cluster is that led by Newell EW, who is also the third author with regards to HCPs in mass cytometry, with 2044 total citations. Newell EW, affiliated to Stanford University & Singapore Immunology Network (SIGN), leads the Newell Lab research laboratory. This group innovatively applies mass cytometry to T-cell responses. They focus on unravelling the roles of extremely diverse immune cells with clinical implications in cancer and infectious diseases [50].
Although CyTOF has quickly emerged as a very useful and powerful technology for multidimensional analysis, the costs of the equipment and reagents (mass cytometer and metal-conjugated antibodies) are still much higher than those of conventional flow cytometry. This would explain why only groups and institutions with high funding and accessibility to a mass cytometer, such as Standford University, produce most of the HCPs in mass cytometry. The USA is the country with the highest funding in mass cytometry according to the Web of Science (Funding Agencies). American dominance in the production of H-Classics is also observed in other medical-related areas [27,28]. If we consider the value of AI as a strategy to adjust the production of HCPs to GDP, the ranking of the most productive countries in HCP would not change significantly. USA remains the country with the highest HCP and AI value (0.067). The only country for which the ranking would change is China. Despite having the lowest GDP in the list ($10,261.68), it has a considerable HCP production reflected in its AI (0.010).
Co-occurrence is a concept which refers to the common presence, frequency of occurrence, and close proximity of similar keywords associated with several articles [51]. Looking at the co-occurrence network (keywords) as an indicator of the conceptual structure of mass cytometry research field, keywords like “flow cytometry” and “mass cytometry” appear quite frequently linked and closely related to the rest of the network terms. The co-occurrence of these two terms was highly expected, as they are often complementary techniques or are under constant comparisons throughout the HCPs. These comparisons often highlight the advantages of one technology over the other. Probably the most mentioned issue is that flow cytometry has its multiplexing capacity limited by the number of markers (fluorochromes) that can be simultaneously analysed due to spectral overlap and autofluorescence [52], while that mass cytometry can accurately quantify more than 40 parameters on single cells due to the broadest spectrum of heavy-metals isotopes available [53].
Other topics that appear clustered together are those related to immune cells (“T-cells”, “macrophages”) or the ones related to the “activation” “responses” or “natural killer cells”. These frequently repeated topics were also expected among CyTOF HCPs as one of the most notable strengths of mass cytometry is its great immunophenotyping power for a vast diversity of study purposes. In the field of oncology research, the characterization of immune cell subsets through mass cytometry have pushed new knowledge in different types of cancer, such as melanoma, glioblastoma, pancreatic, breast or colon [54] and have also contributed to the study of less frequent diseases, such as the lungs Löfgren’s syndrome [55] or common ones like type I diabetes [56].
Other nodes that appear linked together are those related to data analysis (“differential expression analysis” or “network”). These results on the conceptual structure are further supported by what was observed in the content analysis mentioned above. The complexity of the multiparametric analysis involved in CyTOF has been reviewed in various publications [57,58].
It is important to highlight the impact that this technique is likely to have in areas such as clinical medicine, where it already has been proposed as a powerful tool for the diagnosis of autoimmune and complex hematologic diseases [59,60]. Similarly, a recent expansion of CyTOF, known as Imaging Mass Cytometry (IMC), has been proposed in the field of personalised medicine [61,62]. Finally, it is highly expected that CyTOF will contribute to the research on the recently emerged COVID-19 disease, caused by the SARS-COV2 coronavirus, which can cause serious immunological responses, in a similar way as it has contributed to the study of HIV [63]. In fact, this technology has already been used to elucidate the characteristics of the immune system in patients with COVID-19 during infection and recovery [64,65]. It would not be surprising if this technology gained more prominence in drug and vaccine development processes, as it has already supported the development and evaluation of hepatitis C and influenza vaccines [66,67].
5. Conclusions
In conclusion, this study shows for the first time the main protagonists and predominant trends in the area of mass cytometry, reflected in the most cited articles in the discipline. Likewise with studies in other areas, we think that this bibliometric information can help researchers, in particular those with an interest in immunology, to expand knowledge in this field and set valuable collaborations in the mass cytometry research field.
Author Contributions
Conceptualization, D.E.D.Z.-S.; methodology, P.S.-N.; software, P.S.-N.; validation, C.S. and M.I.L.; formal analysis, D.E.D.Z.-S.; investigation, D.E.D.Z.-S.; resources, D.E.D.Z.-S. and P.S.-N.; data curation, P.S.-N.; writing—original draft preparation, D.E.D.Z.-S.; writing—review and editing, D.E.D.Z.-S.; visualization, P.S.-N.; supervision, C.S. and M.I.L.; project administration, M.I.L.; funding acquisition, M.I.L. All authors have read and agreed to the published version of the manuscript.
Funding
The present study has been supported by grants of Instituto de Salud Carlos III cofounded by Fondo Europeo de Desarrollo Regional—FEDER (PI19-00883, PT 20/00127)) as well as P18-RT-3364 and “Plan Propio de I+D+I 2020” (IBIMA). CIBERehd and Plataforma ISCiii de Investigación Clínica are funded by Instituto de Salud Carlos III.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in Zenodo repository at http://doi.org/10.5281/zenodo.4462149.
Acknowledgments
The authors thank IBIMA for facilitating the publication of this article.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Appendix A

Table A1.
H-Classics collection in mass cytometry (71 HCP).
Table A1.
H-Classics collection in mass cytometry (71 HCP).
| Title | Authors | DOI | Total Citations |
|---|---|---|---|
| Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum | Bendall, Sean C.; Simonds, Erin F.; Qiu, Peng; Amir, El-ad D.; Krutzik, Peter O.; Finck, Rachel; Bruggner, Robert V.; Melamed, Rachel; Trejo, Angelica; Ornatsky, Olga I.; Balderas, Robert S.; Plevritis, Sylvia K.; Sachs, Karen; Pe’er, Dana; Tanner, Scott D.; Nolan, Garry P. | 10.1126/science.1198704 | 1233 |
| viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukaemia | Amir, El-ad David; Davis, Kara L.; Tadmor, Michelle D.; Simonds, Erin F.; Levine, Jacob H.; Bendall, Sean C.; Shenfeld, Daniel K.; Krishnaswamy, Smita; Nolan, Garry P.; Pe’er, Dana | 10.1038/nbt.2594 | 716 |
| Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry | Giesen, Charlotte; Wang, Hao A. O.; Schapiro, Denis; Zivanovic, Nevena; Jacobs, Andrea; Hattendorf, Bodo; Schueffler, Peter J.; Grolimund, Daniel; Buhmann, Joachim M.; Brandt, Simone; Varga, Zsuzsanna; Wild, Peter J.; Guenther, Detlef; Bodenmiller, Bernd | 10.1038/NMETH.2869 | 500 |
| Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE | Qiu, Peng; Simonds, Erin F.; Bendall, Sean C.; Gibbs, Kenneth D., Jr.; Bruggner, Robert V.; Linderman, Michael D.; Sachs, Karen; Nolan, Garry P.; Plevritis, Sylvia K. | 10.1038/nbt.1991 | 498 |
| Standardizing immunophenotyping for the Human Immunology Project | Maecker, Holden T.; McCoy, J. Philip; Nussenblatt, Robert | 10.1038/nri3158 | 450 |
| A polarizing question: do M1 and M2 microglia exist? | Ransohoff, Richard M. | 10.1038/nn.4338 | 435 |
| Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis | Levine, Jacob H.; Simonds, Erin F.; Bendall, Sean C.; Davis, Kara L.; Amir, El-ad D.; Tadmor, Michelle D.; Litvin, Oren; Fienberg, Harris G.; Jager, Astraea; Zunder, Eli R.; Finck, Rachel; Gedman, Amanda L.; Radtke, Ina; Downing, James R.; Pe’er, Dana; Nolan, Garry P. | 10.1016/j.cell.2015.05.047 | 413 |
| Single-Cell Trajectory Detection Uncovers Progression and Regulatory Coordination in Human B Cell Development | Bendall, Sean C.; Davis, Kara L.; Amir, El-ad David; Tadmor, Michelle D.; Simonds, Erin F.; Chen, Tiffany J.; Shenfeld, Daniel K.; Nolan, Garry P.; Pe’er, Dana | 10.1016/j.cell.2014.04.005 | 377 |
| Cytometry by Time-of-Flight Shows Combinatorial Cytokine Expression and Virus-Specific Cell Niches within a Continuum of CD8( + ) T Cell Phenotypes | Newell, Evan W.; Sigal, Natalia; Bendall, Sean C.; Nolan, Garry P.; Davis, Mark M. | 10.1016/j.immuni.2012.01.002 | 360 |
| A deep profiler’s guide to cytometry | Bendall, Sean C.; Nolan, Garry P.; Roederer, Mario; Chattopadhyay, Pratip K. | 10.1016/j.it.2012.02.010 | 351 |
| Mass Cytometry: Single Cells, Many Features | Spitzer, Matthew H.; Nolan, Garry P. | 10.1016/j.cell.2016.04.019 | 341 |
| Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators | Bodenmiller, Bernd; Zunder, Eli R.; Finck, Rachel; Chen, Tiffany J.; Savig, Erica S.; Bruggner, Robert V.; Simonds, Erin F.; Bendall, Sean C.; Sachs, Karen; Krutzik, Peter O.; Nolan, Garry P. | 10.1038/nbt.2317 | 292 |
| Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade | Wei, Spencer C.; Levine, Jacob H.; Cogdill, Alexandria P.; Zhao, Yang; Anang, Nana-Ama A. S.; Andrews, Miles C.; Sharma, Padmanee; Wang, Jing; Wargo, Jennifer A.; Pe’er, Dana; Allison, James P. | 10.1016/j.cell.2017.07.024 | 285 |
| Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species | Guilliams, Martin; Dutertre, Charles-Antoine; Scott, Charlotte L.; McGovern, Naomi; Sichien, Dorine; Chakarov, Svetoslav; Van Gassen, Sofie; Chen, Jinmiao; Poidinger, Michael; De Prijck, Sofie; Tavernier, Simon J.; Low, Ivy; Irac, Sergio Erdal; Mattar, Citra Nurfarah; Sumatoh, Hermi Rizal; Low, Gillian Hui Ling; Chung, Tam John Kit; Chan, Dedrick Kok Hong; Tan, Ker Kan; Hon, Tony Lim Kiat; Fossum, Even; Bogen, Bjame; Choolani, Mahesh; Chan, Jerry Kok Yen; Larbi, Anis; Luche, Herve; Henri, Sandrine; Saeys, Yvan; Newell, Evan William; Lambrecht, Bart N.; Malissen, Bernard; Ginhoux, Florent | 10.1016/j.immuni.2016.08.015 | 274 |
| Normalization of mass cytometry data with bead standards | Finck, Rachel; Simonds, Erin F.; Jager, Astraea; Krishnaswamy, Smita; Sachs, Karen; Fantl, Wendy; Pe’er, Dana; Nolan, Garry P.; Bendall, Sean C. | 10.1002/cyto.a.22271 | 254 |
| Genetic and Environmental Determinants of Human NK Cell Diversity Revealed by Mass Cytometry | Horowitz, Amir; Strauss-Albee, Dara M.; Leipold, Michael; Kubo, Jessica; Nemat-Gorgani, Neda; Dogan, Ozge C.; Dekker, Cornelia L.; Mackey, Sally; Maecker, Holden; Swan, Gary E.; Davis, Mark M.; Norman, Paul J.; Guethlein, Lisbeth A.; Desai, Manisha; Parham, Peter; Blish, Catherine A. | 10.1126/scitranslmed.3006702 | 229 |
| An Immune Atlas of Clear Cell Renal Cell Carcinoma | Chevrier, Stephane; Levine, Jacob Harrison; Zanotelli, Vito Riccardo Tomaso; Silina, Karina; Schulz, Daniel; Bacac, Marina; Ries, Carola Hermine; Ailles, Laurie; Jewett, Michael Alexander Spencer; Moch, Holger; van den Broek, Maries; Beisel, Christian; Stadler, Michael Beda; Gedye, Craig; Reis, Bernhard; Pe’er, Dana; Bodenmiller, Bernd | 10.1016/j.cell.2017.04.016 | 227 |
| Systemic Immunity Is Required for Effective Cancer Immunotherapy | Spitzer, Matthew H.; Carmi, Yaron; Reticker-Flynn, Nathan E.; Kwek, Serena S.; Madhireddy, Deepthi; Martins, Maria M.; Gherardini, Pier Federico; Prestwood, Tyler R.; Chabon, Jonathan; Bendall, Sean C.; Fong, Lawrence; Nolan, Garry P.; Engleman, Edgar G. | 10.1016/j.cell.2016.12.022 | 224 |
| Wishbone identifies bifurcating developmental trajectories from single-cell data | Setty, Manu; Tadmor, Michelle D.; Reich-Zeliger, Shlomit; Ange, Omer; Salame, Tomer Meir; Kathail, Pooja; Choi, Kristy; Bendall, Sean; Friedman, Nir; Pe’er, Dana | 10.1038/nbt.3569 | 218 |
| Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis | Rao, Deepak A.; Gurish, Michael F.; Marshall, Jennifer L.; Slowikowski, Kamil; Fonseka, Chamith Y.; Liu, Yanyan; Donlin, Laura T.; Henderson, Lauren A.; Wei, Kevin; Mizoguchi, Fumitaka; Teslovich, Nikola C.; Weinblatt, Michael E.; Massarotti, Elena M.; Coblyn, Jonathan S.; Helfgott, Simon M.; Lee, Yvonne C.; Todd, Derrick J.; Ykerk, Vivian P. B.; Goodman, Susan M.; Pernis, Alessandra B.; Ivashkiv, Lionel B.; Karlson, Elizabeth W.; Nigrovic, Peter A.; Filer, Andrew; Buckley, Christopher D.; Lederer, James A.; Raychaudhuri, Soumya; Renner, Michael B. B. | 10.1038/nature20810 | 214 |
| Dimensionality reduction for visualizing single-cell data using UMAP | Becht, Etienne; McInnes, Leland; Healy, John; Dutertre, Charles-Antoine; Kwok, Immanuel W. H.; Ng, Lai Guan; Ginhoux, Florent; Newell, Evan W. | 10.1038/nbt.4314 | 208 |
| Single-cell technologies for monitoring immune systems | Chattopadhyay, Pratip K.; Gierahn, Todd M.; Roederer, Mario; Love, J. Christopher | 10.1038/ni.2796 | 206 |
| Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukaemia | Romee, Rizwan; Rosario, Maximillian; Berrien-Elliott, Melissa M.; Wagner, Julia A.; Jewell, Brea A.; Schappe, Timothy; Leong, Jeffrey W.; Abdel-Latif, Sara; Schneider, Stephanie E.; Willey, Sarah; Neal, Carly C.; Yu, Liyang; Oh, Stephen T.; Lee, Yi-Shan; Mulder, Arend; Claas, Frans; Cooper, Megan A.; Fehniger, Todd A. | 10.1126/scitranslmed.aaf2341 | 204 |
| Highly multiparametric analysis by mass cytometry | Ornatsky, Olga; Bandura, Dmitry; Baranov, Vladimir; Nitz, Mark; Winnik, Mitchell A.; Tanner, Scott | 10.1016/j.jim.2010.07.002 | 203 |
| High-dimensional analysis of the murine myeloid cell system | Becher, Burkhard; Schlitzer, Andreas; Chen, Jinmiao; Mair, Florian; Sumatoh, Hermi R.; Teng, Karen Wei Weng; Low, Donovan; Ruedl, Christiane; Riccardi-Castagnoli, Paola; Poidinger, Michael; Greter, Melanie; Ginhoux, Florent; Newell, Evan W. | 10.1038/ni.3006 | 198 |
| The Spectrum and Regulatory Landscape of Intestinal Innate Lymphoid Cells Are Shaped by the Microbiome | Gury-BenAri, Meital; Thaiss, Christoph A.; Serafini, Nicolas; Winter, Deborah R.; Giladi, Amir; Lara-Astiaso, David; Levy, Maayan; Salame, Tomer Meir; Weiner, Assaf; David, Eyal; Shapiro, Hagit; Dori-Bachash, Mally; Pevsner-Fischer, Meirav; Lorenzo-Vivas, Erika; Keren-Shaul, Hadas; Paul, Franziska; Harmelin, Alon; Eberl, Gerard; Itzkovitz, Shalev; Tanay, Amos; Di Santo, James P.; Elinav, Eran; Amit, Ido | 10.1016/j.cell.2016.07.043 | 194 |
| PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression | Jiao, Shiping; Xia, Weiya; Yamaguchi, Hirohito; Wei, Yongkun; Chen, Mei-Kuang; Hsu, Jung-Mao; Hsu, Jennifer L.; Yu, Wen-Hsuan; Du, Yi; Lee, Heng-Huan; Li, Chia-Wei; Chou, Chao-Kai; Lim, Seung-Oe; Chang, Shih-Shin; Litton, Jennifer; Arun, Banu; Hortobagyi, Gabriel N.; Hung, Mien-Chie | 10.1158/1078-0432.CCR-16-3215 | 183 |
| Revealing the vectors of cellular identity with single-cell genomics | Wagner, Allon; Regev, Aviv; Yosef, Nir | 10.1038/nbt.3711 | 181 |
| FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data | Van Gassen, Sofie; Callebaut, Britt; Van Helden, Mary J.; Lambrecht, Bart N.; Demeester, Piet; Dhaene, Tom; Saeys, Yvan | 10.1002/cyto.a.22625 | 180 |
| Mapping the human DC lineage through the integration of high-dimensional techniques | See, Peter; Dutertre, Charles-Antoine; Chen, Jinmiao; Gunther, Patrick; McGovern, Naomi; Irac, Sergio Erdal; Gunawan, Merry; Beyer, Marc; Handler, Kristian; Duan, Kaibo; Bin Sumatoh, Hermi Rizal; Ruffin, Nicolas; Jouve, Mabel; Gea-Mallorqui, Ester; Hennekam, Raoul C. M.; Lim, Tony; Yip, Chan Chung; Wen, Ming; Malleret, Benoit; Low, Ivy; Shadan, Nurhidaya Binte; Fen, Charlene Foong Shu; Tay, Alicia; Lum, Josephine; Zolezzi, Francesca; Larbi, Anis; Poidinger, Michael; Chan, Jerry K. Y.; Chen, Qingfeng; Renia, Laurent; Haniffa, Muzlifah; Benaroch, Philippe; Schlitzer, Andreas; Schultze, Joachim L.; Newell, Evan W.; Ginhoux, Florent | 10.1126/science.aag3009 | 179 |
| High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy | Krieg, Carsten; Nowicka, Malgorzata; Guglietta, Silvia; Schindler, Sabrina; Hartmann, Felix J.; Weber, Lukas M.; Dummer, Reinhard; Robinson, Mark D.; Levesque, Mitchell P.; Becher, Burkhard | 10.1038/nm.4466 | 178 |
| Automated identification of stratifying signatures in cellular subpopulations | Bruggner, Robert V.; Bodenmiller, Bernd; Dill, David L.; Tibshirani, Robert J.; Nolan, Garry P. | 10.1073/pnas.1408792111 | 176 |
| Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency | Simoni, Yannick; Fehlings, Michael; Kloverpris, Henrik N.; McGovern, Naomi; Koo, Si-Lin; Loh, Chiew Yee; Lim, Shawn; Kurioka, Ayako; Fergusson, Joannah R.; Tang, Choong-Leong; Kam, Ming Hian; Dennis, Koh; Lim, Tony Kiat Hon; Fui, Alexander Chung Yaw; Hoong, Chan Weng; Chan, Jerry Kok Yen; de Lafaille, Maria Curotto; Narayanan, Sriram; Baig, Sonia; Shabeer, Muhammad; Toh, Sue-Anne Ee Shiow; Tan, Henry Kun Kiaang; Anicete, Rosslyn; Tan, Eng-Huat; Takano, Angela; Klenerman, Paul; Leslie, Alasdair; Tan, Daniel S. W.; Tan, Iain Beehuat; Ginhoux, Florent; Newell, Evan W. | 10.1016/j.immuni.2016.11.005 | 164 |
| Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm | Zunder, Eli R.; Finck, Rachel; Behbehani, Gregory K.; Amir, El-ad D.; Krishnaswamy, Smita; Gonzalez, Veronica D.; Lorang, Cynthia G.; Bjornson, Zach; Spitzer, Matthew H.; Bodenmiller, Bernd; Fantl, Wendy J.; Pe’er, Dana; Nolan, Garry P. | 10.1038/nprot.2015.020 | 162 |
| A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory | Swadling, Leo; Capone, Stefania; Antrobus, Richard D.; Brown, Anthony; Richardson, Rachel; Newell, Evan W.; Halliday, John; Kelly, Christabel; Bowen, Dan; Fergusson, Joannah; Kurioka, Ayako; Ammendola, Virginia; Del Sorbo, Mariarosaria; Grazioli, Fabiana; Esposito, Maria Luisa; Siani, Loredana; Traboni, Cinzia; Hill, Adrian; Colloca, Stefano; Davis, Mark; Nicosia, Alfredo; Cortese, Riccardo; Folgori, Antonella; Klenerman, Paul; Barnes, Eleanor | 10.1126/scitranslmed.3009185 | 158 |
| Clinical recovery from surgery correlates with single-cell immune signatures | Gaudilliere, Brice; Fragiadakis, Gabriela K.; Bruggner, Robert V.; Nicolau, Monica; Finck, Rachel; Tingle, Martha; Silva, Julian; Ganio, Edward A.; Yeh, Christine G.; Maloney, William J.; Huddleston, James I.; Goodman, Stuart B.; Davis, Mark M.; Bendall, Sean C.; Fantl, Wendy J.; Angst, Martin S.; Nolan, Garry P. | 10.1126/scitranslmed.3009701 | 157 |
| Single-Cell Multiomics: Multiple Measurements from Single Cells | Macaulay, Iain C.; Ponting, Chris P.; Voet, Thierry | 10.1016/j.tig.2016.12.003 | 150 |
| Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization | Newell, Evan W.; Sigal, Natalia; Nair, Nitya; Kidd, Brian A.; Greenberg, Harry B.; Davis, Mark M. | 10.1038/nbt.2593 | 141 |
| Human immune system variation | Brodin, Petter; Davis, Mark M. | 10.1038/nri.2016.125 | 139 |
| A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites | Dodd, Dylan; Spitzer, Matthew H.; Van Treuren, William; Merrill, Bryan D.; Hryckowian, Andrew J.; Higginbottom, Steven K.; Le, Anthony; Cowan, Tina M.; Nolan, Garry P.; Fischbach, Michael A.; Sonnenburg, Justin L. | 10.1038/nature24661 | 135 |
| The transcriptional landscape of alpha beta T cell differentiation | Mingueneau, Michael; Kreslavsky, Taras; Gray, Daniel; Heng, Tracy; Cruse, Richard; Ericson, Jeffrey; Bendall, Sean; Spitzer, Matt; Nolan, Garry; Kobayashi, Koichi; von Boehmer, Harald; Mathis, Diane; Benoist, Christophe; Best, Adam J.; Knell, Jamie; Goldrath, Ananda; Jojic, Vladimir; Koller, Daphne; Shay, Tal; Regev, Aviv; Cohen, Nadia; Brennan, Patrick; Brenner, Michael; Kim, Francis; Rao, Tata Nageswara; Wagers, Amy; Heng, Tracy; Ericson, Jeffrey; Rothamel, Katherine; Ortiz-Lopez, Adriana; Mathis, Diane; Bezman, Natalie A.; Sun, Joseph C.; Min-Oo, Gundula; Kim, Charlie C.; Lanier, Lewis L.; Miller, Jennifer; Brown, Brian; Merad, Miriam; Gautier, Emmanuel L.; Jakubzick, Claudia; Randolph, Gwendalyn J.; Monach, Paul; Blair, David A.; Dustin, Michael L.; Shinton, Susan A.; Hardy, Richard R.; Laidlaw, David; Collins, Jim; Gazit, Roi; Rossi, Derrick J.; Malhotra, Nidhi; Sylvia, Katelyn; Kang, Joonsoo; Kreslavsky, Taras; Fletcher, Anne; Elpek, Kutlu; Bellemare-Pelletier, Angelique; Malhotra, Deepali; Turley, Shannon | 10.1038/ni.2590 | 134 |
| Automatic Classification of Cellular Expression by Nonlinear Stochastic Embedding (ACCENSE) | Shekhar, Karthik; Brodin, Petter; Davis, Mark M.; Chakraborty, Arup K. | 10.1073/pnas.1321405111 | 130 |
| Highly multiplexed simultaneous detection of RNAs and proteins in single cells | Frei, Andreas P.; Bava, Felice-Alessio; Zunder, Eli R.; Hsieh, Elena W. Y.; Chen, Shih-Yu; Nolan, Garry P.; Gherardini, Pier Federico | 10.1038/NMETH.3742 | 129 |
| High-Dimensional Single-Cell Mapping of Central Nervous System Immune Cells Reveals Distinct Myeloid Subsets in Health, Aging, and Disease | Mrdjen, Dunja; Pavlovic, Anto; Hartmann, Felix J.; Schreiner, Bettina; Utz, Sebastian G.; Leung, Brian P.; Lelios, Iva; Heppner, Frank L.; Kipnis, Jonathan; Merkler, Doron; Greter, Melanie; Becher, Burkhard | 10.1016/j.immuni.2018.01.011 | 128 |
| Heterogeneity of Human CD4 + T Cells Against Microbes | Sallusto, Federica | 10.1146/annurev-immunol-032414-112056 | 117 |
| Mass spectrometry imaging and profiling of single cells | Lanni, Eric J.; Rubakhin, Stanislav S.; Sweedler, Jonathan V. | 10.1016/j.jprot.2012.03.017 | 117 |
| An interactive reference framework for modelling a dynamic immune system | Spitzer, Matthew H.; Gherardini, Pier Federico; Fragiadakis, Gabriela K.; Bhattacharya, Nupur; Yuan, Robert T.; Hotson, Andrew N.; Finck, Rachel; Carmi, Yaron; Zunder, Eli R.; Fantl, Wendy J.; Bendall, Sean C.; Engleman, Edgar G.; Nolan, Garry P. | 10.1126/science.1259425 | 115 |
| destiny: diffusion maps for large-scale single cell data in R | Angerer, Philipp; Haghverdi, Laleh; Buettner, Maren; Theis, Fabian J.; Marr, Carsten; Buettner, Florian | 10.1093/bioinformatics/btv715 | 114 |
| Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method | Lin, Jia-Ren; Fallahi-Sichani, Mohammad; Sorger, Peter K. | 10.1038/ncomms9390 | 114 |
| Single-cell mass cytometry adapted to measurements of the cell cycle | Behbehani, Gregory K.; Bendall, Sean C.; Clutter, Matthew R.; Fantl, Wendy J.; Nolan, Garry P. | 10.1002/cyto.a.22075 | 114 |
| CD161 Defines a Transcriptional and Functional Phenotype across Distinct Human T Cell Lineages | Fergusson, Joannah R.; Smith, Kira E.; Fleming, Vicki M.; Rajoriya, Neil; Newell, Evan W.; Simmons, Ruth; Marchi, Emanuele; Bjorkander, Sophia; Kang, Yu-Hoi; Swadling, Leo; Kurioka, Ayako; Sahgal, Natasha; Lockstone, Helen; Baban, Dilair; Freeman, Gordon J.; Sverremark-Ekstrom, Eva; Davis, Mark M.; Davenport, Miles P.; Venturi, Vanessa; Ussher, James E.; Willberg, Christian B.; Klenerman, Paul | 10.1016/j.celrep.2014.09.045 | 113 |
| A platinum-based covalent viability reagent for single-cell mass cytometry | Fienberg, Harris G.; Simonds, Erin F.; Fantl, Wendy J.; Nolan, Garry P.; Bodenmiller, Bernd | 10.1002/cyto.a.22067 | 108 |
| Single-cell mass cytometry for analysis of immune system functional states | Bjornson, Zach B.; Nolan, Garry P.; Fantl, Wendy J. | 10.1016/j.coi.2013.07.004 | 103 |
| Single-cell protein analysis | Wu, Meiye; Singh, Anup K. | 10.1016/j.copbio.2011.11.023 | 102 |
| Comparison of Clustering Methods for High-Dimensional Single-Cell Flow and Mass Cytometry Data | Weber, Lukas M.; Robinson, Mark D. | 10.1002/cyto.a.23030 | 100 |
| A Continuous Molecular Roadmap to iPSC Reprogramming through Progression Analysis of Single-Cell Mass Cytometry | Zunder, Eli R.; Lujan, Ernesto; Goltsev, Yury; Wernig, Marius; Nolan, Garry P. | 10.1016/j.stem.2015.01.015 | 95 |
| Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes | Bentzen, Amalie Kai; Marquard, Andrea Marion; Lyngaa, Rikke; Saini, Sunil Kumar; Ramskov, Sofie; Donia, Marco; Such, Lina; Furness, Andrew J. S.; McGranahan, Nicholas; Rosenthal, Rachel; Straten, Per Thor; Szallasi, Zoltan; Svane, Inge Marie; Swanton, Charles; Quezada, Sergio A.; Jakobsen, Soren Nyboe; Eklund, Aron Charles; Hadrup, Sine Reker | 10.1038/nbt.3662 | 93 |
| A High-Dimensional Atlas of Human T Cell Diversity Reveals Tissue-Specific Trafficking and Cytokine Signatures | Wong, Michael Thomas; Ong, David Eng Hui; Lim, Frances Sheau Huei; Teng, Karen Wei Weng; McGovern, Naomi; Narayanan, Sriram; Ho, Wen Qi; Cerny, Daniela; Tan, Henry Kun Kiaang; Anicete, Rosslyn; Tan, Bien Keem; Lim, Tony Kiat Hon; Chan, Chung Yip; Cheow, Peng Chung; Lee, Ser Yee; Takano, Angela; Tan, Eng-Huat; Tam, John Kit Chung; Tan, Ern Yu; Chan, Jerry Kok Yen; Fink, Katja; Bertoletti, Antonio; Ginhoux, Florent; De Lafaille, Maria Alicia Curotto; Newell, Evan William | 10.1016/j.immuni.2016.07.007 | 93 |
| Conditional density-based analysis of T cell signaling in single-cell data | Krishnaswamy, Smita; Spitzer, Matthew H.; Mingueneau, Michael; Bendall, Sean C.; Litvin, Oren; Stone, Erica; Pe’er, Dana; Nolan, Garry P. | 10.1126/science.1250689 | 93 |
| The end of gating? An introduction to automated analysis of high dimensional cytometry data | Mair, Florian; Hartmann, Felix J.; Mrdjen, Dunja; Tosevski, Vinko; Krieg, Carsten; Becher, Burkhard | 10.1002/eji.201545774 | 91 |
| Unifying immunology with informatics and multiscale biology | Kidd, Brian A.; Peters, Lauren A.; Schadt, Eric E.; Dudley, Joel T. | 10.1038/ni.2787 | 91 |
| Cell-of-Origin-Specific 3D Genome Structure Acquired during Somatic Cell Reprogramming | Krijger, Peter Hugo Lodewijk; Di Stefano, Bruno; de Wit, Elzo; Limone, Francesco; van Oevelen, Chris; de Laat, Wouter; Graf, Thomas | 10.1016/j.stem.2016.01.007 | 87 |
| Automated mapping of phenotype space with single-cell data | Samusik, Nikolay; Good, Zinaida; Spitzer, Matthew H.; Davis, Kara L.; Nolan, Garry P. | 10.1038/NMETH.3863 | 85 |
| Crowdsourcing Network Inference: The DREAM Predictive Signaling Network Challenge | Prill, Robert J.; Saez-Rodriguez, Julio; Alexopoulos, Leonidas G.; Sorger, Peter K.; Stolovitzky, Gustavo | 10.1126/scisignal.2002212 | 85 |
| Stereotypic Immune System Development in Newborn Children | Olin, Axel; Henckel, Ewa; Chen, Yang; Lakshmikanth, Tadepally; Pou, Christian; Mikes, Jaromir; Gustafsson, Anna; Bernhardsson, Anna Karin; Zhang, Cheng; Bohlin, Kajsa; Brodin, Petter | 10.1016/j.cell.2018.06.045 | 84 |
| Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline | Chen, Hao; Lau, Mai Chan; Wong, Michael Thomas; Newell, Evan W.; Poidinger, Michael; Chen, Jinmiao | 10.1371/journal.pcbi.1005112 | 78 |
| Beyond model antigens: high-dimensional methods for the analysis of antigen-specific T cells | Newell, Evan W.; Davis, Mark M. | 10.1038/nbt.2783 | 78 |
| Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility | Strauss-Albee, Dara M.; Fukuyama, Julia; Liang, Emily C.; Yao, Yi; Jarrell, Justin A.; Drake, Alison L.; Kinuthia, John; Montgomery, Ruth R.; John-Stewart, Grace; Holmes, Susan; Blish, Catherine A. | 10.1126/scitranslmed.aac5722 | 77 |
| Single-Cell Analysis in Cancer Genomics | Saadatpour, Assieh; Lai, Shujing; Guo, Guoji; Yuan, Guo-Cheng | 10.1016/j.tig.2015.07.003 | 74 |
| Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data | Diggins, Kirsten E.; Ferrell, P. Brent, Jr.; Irish, Jonathan M. | 10.1016/j.ymeth.2015.05.008 | 72 |
| Chikungunya Viral Arthritis in the United States A Mimic of Seronegative Rheumatoid Arthritis | Miner, Jonathan J.; Yeang, Han Xian Aw; Fox, Julie M.; Taffner, Samantha; Malkova, Olga N.; Oh, Stephen T.; Kim, Alfred H. J.; Diamond, Michael S.; Lenschow, Deborah J.; Yokoyama, Wayne M. | 10.1002/art.39027 | 72 |
References
- Robinson, J.P.; Roederer, M. Flow cytometry strikes gold. Science 2015, 350, 739–740. [Google Scholar] [CrossRef] [PubMed]
- Picot, J.; Guerin, C.L.; Le Van Kim, C.; Boulanger, C.M. Flow cytometry: Retrospective, fundamentals and recent instrumentation. Cytotechnology 2012, 64, 109–130. [Google Scholar] [CrossRef] [PubMed]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow cytometry: Basic principles and applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Pedreira, C.E.; Costa, E.S.; Lecrevisse, Q.; van Dongen, J.J.M.; Orfao, A. Overview of clinical flow cytometry data analysis: Recent advances and future challenges. Trends Biotechnol. 2013, 31, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, D.; Wang, J.; Chen, J. Advances of Single-Cell Protein Analysis. Cells 2020, 9, 1271. [Google Scholar] [CrossRef]
- Simoni, Y.; Chng, M.H.Y.; Li, S.; Fehlings, M.; Newell, E.W. Mass cytometry: A powerful tool for dissecting the immune landscape. Curr. Opin. Immunol. 2018, 51, 187–196. [Google Scholar] [CrossRef]
- Bandura, D.R.; Baranov, V.I.; Ornatsky, O.I.; Antonov, A.; Kinach, R.; Lou, X.; Pavlov, S.; Vorobiev, S.; Dick, J.E.; Tanner, S.D. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009, 81, 6813–6822. [Google Scholar] [CrossRef]
- Ornatsky, O.; Bandura, D.; Baranov, V.; Nitz, M.; Winnik, M.A.; Tanner, S. Highly multiparametric analysis by mass cytometry. J. Immunol. Methods 2010, 361, 1–20. [Google Scholar] [CrossRef]
- Tanner, S.D.; Baranov, V.I.; Ornatsky, O.I.; Bandura, D.R.; George, T.C. An introduction to mass cytometry: Fundamentals and applications. Cancer Immunol. Immunother. 2013, 62, 955–965. [Google Scholar] [CrossRef]
- Dzangué-Tchoupou, G.; Corneau, A.; Blanc, C.; Benveniste, O.; Allenbach, Y. Analysis of cell surface and intranuclear markers on non-stimulated human PBMC using mass cytometry. PLoS ONE 2018, 13, e0194593. [Google Scholar] [CrossRef]
- Nicholas, K.J.; Greenplate, A.R.; Flaherty, D.K.; Matlock, B.K.; Juan, J.S.; Smith, R.M.; Irish, J.M.; Kalams, S.A. Multiparameter analysis of stimulated human peripheral blood mononuclear cells: A comparison of mass and fluorescence cytometry. Cytom. Part A 2016, 89, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Atkuri, K.R.; Stevens, J.C.; Neubert, H. Mass cytometry: A highly multiplexed single-cell technology for advancing drug development. Drug Metab. Dispos. 2015, 43, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Teh, C.E.; Gong, J.N.; Segal, D.; Tan, T.; Vandenberg, C.J.; Fedele, P.L.; Low, M.S.Y.; Grigoriadis, G.; Harrison, S.J.; Strasser, A.; et al. Deep profiling of apoptotic pathways with mass cytometry identifies a synergistic drug combination for killing myeloma cells. Cell Death Differ. 2020, 27. [Google Scholar] [CrossRef] [PubMed]
- Lotsberg, M.L.; Wnuk-Lipinska, K.; Terry, S.; Tan, T.Z.; Lu, N.; Trachsel-Moncho, L.; Røsland, G.V.; Siraji, M.I.; Hellesøy, M.; Rayford, A.; et al. AXL Targeting Abrogates Autophagic Flux and Induces Immunogenic Cell Death in Drug-Resistant Cancer Cells. J. Thorac. Oncol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, T. Proteomic approaches for novel systemic lupus erythematosus (SLE) drug discovery. Expert Opin. Drug Discov. 2018, 13, 765–777. [Google Scholar] [CrossRef]
- Reeves, P.M.; Sluder, A.E.; Paul, S.R.; Scholzen, A.; Kashiwagi, S.; Poznansky, M.C. Application and utility of mass cytometry in vaccine development. FASEB J. 2018, 32, 5–15. [Google Scholar] [CrossRef]
- Garfield, E. Introducing Citation Classics: The Human Side of Scientific Reports. Essays Inf. Sci. 1977, 3, 1–2. [Google Scholar]
- De la Flor-Martínez, M.; Galindo-Moreno, P.; Sánchez-Fernández, E.; Piattelli, A.; Cobo, M.J.; Herrera-Viedma, E. H-classic: A new method to identify classic articles in Implant Dentistry, Periodontics, and Oral Surgery. Clin. Oral Implants Res. 2016, 27, 1317–1330. [Google Scholar] [CrossRef]
- Moral-Munoz, J.A.; Lucena-Antón, D.; Perez-Cabezas, V.; Carmona-Barrientos, I.; González-Medina, G.; Ruiz-Molinero, C. Highly cited papers in Microbiology: Identification and conceptual analysis. FEMS Microbiol. Lett. 2018, 365, e00146. [Google Scholar] [CrossRef]
- Baldwin, K.D.; Kovatch, K.; Namdari, S.; Sankar, W.; Flynn, J.M.; Dormans, J.P. The 50 most cited articles in pediatric orthopedic surgery. J. Pediatr. Orthop. Part B 2012, 21, 463–468. [Google Scholar] [CrossRef]
- Lefaivre, K.A.; Shadgan, B.; O’Brien, P.J. 100 Most cited articles in orthopaedic surgery. Clin. Orthop. Relat. Res. 2011, 469, 1487–1497. [Google Scholar] [CrossRef] [PubMed]
- Ponce, F.A.; Lozano, A.M. The most cited works in Parkinson’s disease. Mov. Disord. 2011, 26, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.A.; Herrera, M.; López-Gijón, J.; Herrera-Viedma, E. H-Classics: Characterizing the concept of citation classics through H-index. Scientometrics 2014, 98, 1971–1983. [Google Scholar] [CrossRef]
- Hirsch, J.E. An index to quantify an individual’s scientific research output. Proc. Natl. Acad. Sci. USA 2005, 102, 16569–16572. [Google Scholar] [CrossRef]
- Rousseau, R. Robert Fairthorne and the empirical power laws. J. Doc. 2005, 61, 194–202. [Google Scholar] [CrossRef]
- Martínez, M.A.; Herrera, M.; Contreras, E.; Ruíz, A.; Herrera-Viedma, E. Characterizing highly cited papers in Social Work through H-Classics. Scientometrics 2015, 102, 1713–1729. [Google Scholar] [CrossRef]
- Chhapola, V.; Tiwari, S.; Deepthi, B.; Kanwal, S.K. Citation classics in pediatrics: A bibliometric analysis. World J. Pediatr. 2018, 14, 607–614. [Google Scholar] [CrossRef]
- Perez-Cabezas, V.; Ruiz-Molinero, C.; Carmona-Barrientos, I.; Herrera-Viedma, E.; Cobo, M.J.; Moral-Munoz, J.A. Highly cited papers in rheumatology: Identification and conceptual analysis. Scientometrics 2018, 116, 555–568. [Google Scholar] [CrossRef]
- Di Zeo-Sánchez, D.E.; Sánchez-Núñez, P.; Stephens, C.; Lucena, M.I. Characterizing Highly Cited Papers in Mass Cytometry through H-Classics: WoS Dataset and Citation Report. 2021. Available online: https://zenodo.org/record/4462149#.YBi7-JMRXIW (accessed on 29 January 2021).
- Chinchilla-Rodríguez, Z.; Vargas-Quesada, B.; Hassan-Montero, Y.; González-Molina, A.; Moya-Anegón, F. New approach to the visualization of international scientific collaboration. Inf. Vis. 2010, 9, 277–287. [Google Scholar] [CrossRef]
- Vargas-Quesada, B.; de Moya-Anegón, F. Visualizing the Structure of Science; Springer: Berlin/Heidelberg, Germany, 2003. [Google Scholar]
- R core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.r-project.org/ (accessed on 12 November 2020).
- Aria, M.; Cuccurullo, C. bibliometrix: An R-tool for comprehensive science mapping analysis. J. Informetr. 2017, 11, 959–975. [Google Scholar] [CrossRef]
- Bendall, S.C.; Simonds, E.F.; Qiu, P.; Amir, E.A.D.; Krutzik, P.O.; Finck, R.; Bruggner, R.V.; Melamed, R.; Trejo, A.; Ornatsky, O.I.; et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 2011, 332, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Amir, E.A.D.; Davis, K.L.; Tadmor, M.D.; Simonds, E.F.; Levine, J.H.; Bendall, S.C.; Shenfeld, D.K.; Krishnaswamy, S.; Nolan, G.P.; Pe’Er, D. ViSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 2013, 31, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Giesen, C.; Wang, H.A.O.; Schapiro, D.; Zivanovic, N.; Jacobs, A.; Hattendorf, B.; Schüffler, P.J.; Grolimund, D.; Buhmann, J.M.; Brandt, S.; et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Simonds, E.F.; Bendall, S.C.; Gibbs, K.D.; Bruggner, R.V.; Linderman, M.D.; Sachs, K.; Nolan, G.P.; Plevritis, S.K. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011, 29, 886–893. [Google Scholar] [CrossRef]
- Maecker, H.T.; McCoy, J.P.; Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 2012, 12, 191–200. [Google Scholar] [CrossRef]
- Ransohoff, R.M. A polarizing question: Do M1 and M2 microglia exist. Nat. Neurosci. 2016, 19, 987–991. [Google Scholar] [CrossRef]
- Levine, J.H.; Simonds, E.F.; Bendall, S.C.; Davis, K.L.; Amir, E.A.D.; Tadmor, M.D.; Litvin, O.; Fienberg, H.G.; Jager, A.; Zunder, E.R.; et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 2015, 162, 184–197. [Google Scholar] [CrossRef]
- Bendall, S.C.; Davis, K.L.; Amir, E.A.D.; Tadmor, M.D.; Simonds, E.F.; Chen, T.J.; Shenfeld, D.K.; Nolan, G.P.; Pe’Er, D. Single-cell trajectory detection uncovers progression and regulatory coordination in human b cell development. Cell 2014, 157, 714–725. [Google Scholar] [CrossRef]
- Newell, E.W.; Sigal, N.; Bendall, S.C.; Nolan, G.P.; Davis, M.M. Cytometry by Time-of-Flight Shows Combinatorial Cytokine Expression and Virus-Specific Cell Niches within a Continuum of CD8+T Cell Phenotypes. Immunity 2012, 36, 142–152. [Google Scholar] [CrossRef]
- Bendall, S.C.; Nolan, G.P.; Roederer, M.; Chattopadhyay, P.K. A deep profiler’s guide to cytometry. Trends Immunol. 2012, 33, 323–332. [Google Scholar] [CrossRef]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019, 37, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Citation time window choice for research impact evaluation. Scientometrics 2013, 94, 851–872. [Google Scholar] [CrossRef]
- Eysenbach, G. Citation advantage of open access articles. PLoS Biol. 2006, 4, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Moed, H.F. New developments in the use of citation analysis in research evaluation. Arch. Immunol. Ther. Exp. 2009, 57, 13–18. [Google Scholar] [CrossRef]
- Nassar, A.F.; Wisnewski, A.V.; Raddassi, K. Progress in automation of mass cytometry barcoding for drug development. Bioanalysis 2016, 8, 1429–1435. [Google Scholar] [CrossRef]
- Nassar, A.F.; Ogura, H.; Wisnewski, A.V. Impact of recent innovations in the use of mass cytometry in support of drug development. Drug Discov. Today 2015, 20, 1169–1175. [Google Scholar] [CrossRef][Green Version]
- Simoni, Y.; Becht, E.; Fehlings, M.; Loh, C.Y.; Koo, S.; Teng, K.W.W.; Yeong, J.P.S.; Nahar, R.; Zhang, T.; Kared, H.; et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018, 557, 575–579. [Google Scholar] [CrossRef]
- Montero Díaz, J.; Cobo, M.; Gutiérrez, M.; Segado Boj, F.; Herrera Viedma, E. Mapeo científico de la Categoría «Comunicación» en WoS (1980–2013). Comun. Rev. Científica Iberoam. Comun. y Educ. 2018, 26, 81–91. [Google Scholar] [CrossRef]
- Perfetto, S.P.; Ambrozak, D.; Nguyen, R.; Chattopadhyay, P.K.; Roederer, M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat. Protoc. 2012, 7, 2067–2079. [Google Scholar] [CrossRef]
- Spitzer, M.H.; Nolan, G.P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780–791. [Google Scholar] [CrossRef]
- Allen, B.M.; Hiam, K.J.; Burnett, C.E.; Venida, A.; DeBarge, R.; Tenvooren, I.; Marquez, D.M.; Cho, N.W.; Carmi, Y.; Spitzer, M.H. Systemic dysfunction and plasticity of the immune macroenvironment in cancer models. Nat. Med. 2020, 26. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, Y.; Lakshmikanth, T.; Chen, Y.; Mikes, J.; Eklund, A.; Brodin, P.; Achour, A.; Grunewald, J. Mass cytometry identifies distinct lung CD4+ T cell patterns in Löfgren’s syndrome and non-Löfgren’s syndrome sarcoidosis. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Barcenilla, H.; Åkerman, L.; Pihl, M.; Ludvigsson, J.; Casas, R. Mass cytometry identifies distinct subsets of regulatory T cells and natural killer cells associated with high risk for type 1 diabetes. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Kimball, A.K.; Oko, L.M.; Bullock, B.L.; Nemenoff, R.A.; van Dyk, L.F.; Clambey, E.T. A Beginner’s Guide to Analyzing and Visualizing Mass Cytometry Data. J. Immunol. 2018, 200, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.M.; Robinson, M.D. Comparison of clustering methods for high-dimensional single-cell flow and mass cytometry data. Cytom. Part A 2016, 89, 1084–1096. [Google Scholar] [CrossRef]
- Tsai, A.G.; Glass, D.R.; Juntilla, M.; Hartmann, F.J.; Oak, J.S.; Fernandez-Pol, S.; Ohgami, R.S.; Bendall, S.C. Multiplexed single-cell morphometry for hematopathology diagnostics. Nat. Med. 2020, 26, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Behbehani, G.K. Applications of Mass Cytometry in Clinical Medicine: The Promise and Perils of Clinical CyTOF. Clin. Lab. Med. 2017, 37, 945–964. [Google Scholar] [CrossRef]
- Chang, Q.; Ornatsky, O.I.; Siddiqui, I.; Loboda, A.; Baranov, V.I.; Hedley, D.W. Imaging Mass Cytometry. Cytom. Part A 2017, 91, 160–169. [Google Scholar] [CrossRef]
- Schulz, D.; Zanotelli, V.R.T.; Fischer, J.R.; Schapiro, D.; Engler, S.; Lun, X.K.; Jackson, H.W.; Bodenmiller, B. Simultaneous Multiplexed Imaging of mRNA and Proteins with Subcellular Resolution in Breast Cancer Tissue Samples by Mass Cytometry. Cell Syst. 2018, 6, 25–36. [Google Scholar] [CrossRef]
- Coindre, S.; Tchitchek, N.; Alaoui, L.; Vaslin, B.; Bourgeois, C.; Goujard, C.; Lecuroux, C.; Bruhns, P.; Le Grand, R.; Beignon, A.S.; et al. Mass Cytometry Analysis Reveals Complex Cell-State Modifications of Blood Myeloid Cells During HIV Infection. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Shi, W.; Liu, X.; Cao, Q.; Ma, P.; Le, W.; Xie, L.; Ye, J.; Wen, W.; Tang, H.; Su, W.; et al. High-dimensional single-cell analysis reveals the immune characteristics of COVID-19. Am. J. Physiol. Cell. Mol. Physiol. 2020, 320, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, L.; Pekkarinen, P.T.; Lakshmikanth, T.; Tan, Z.; Consiglio, C.R.; Pou, C.; Chen, Y.; Mugabo, C.H.; Nguyen, N.A.; Nowlan, K.; et al. Systems-Level Immunomonitoring from Acute to Recovery Phase of Severe COVID-19. Cell Rep. Med. 2020, 1, 100078. [Google Scholar] [CrossRef] [PubMed]
- Alimam, S.; Timms, J.A.; Dillon, R.; Mare, T.; Radia, D.; De Lavallade, H.; Harrison, C.N.; Kordasti, S.; McLornan, D.P. Attenuated Immune Responses to the Annual Influenza A Vaccine in Patients with Myeloproliferative Neoplasms. Blood 2019, 134, 1673. [Google Scholar] [CrossRef]
- Swadling, L.; Capone, S.; Antrobus, R.D.; Brown, A.; Richardson, R.; Newell, E.W.; Halliday, J.; Kelly, C.; Bowen, D.; Fergusson, J.; et al. A human vaccine strategy based on chimpanzee adenoviral and MVA vectors that primes, boosts, and sustains functional HCV-specific T cell memory. Sci. Transl. Med. 2014, 6, 261ra153. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).