The Endophytic Microbiome as a Hotspot of Synergistic Interactions, with Prospects of Plant Growth Promotion
Simple Summary
Abstract
1. Introduction
2. Rhizosphere and Root Endophytes
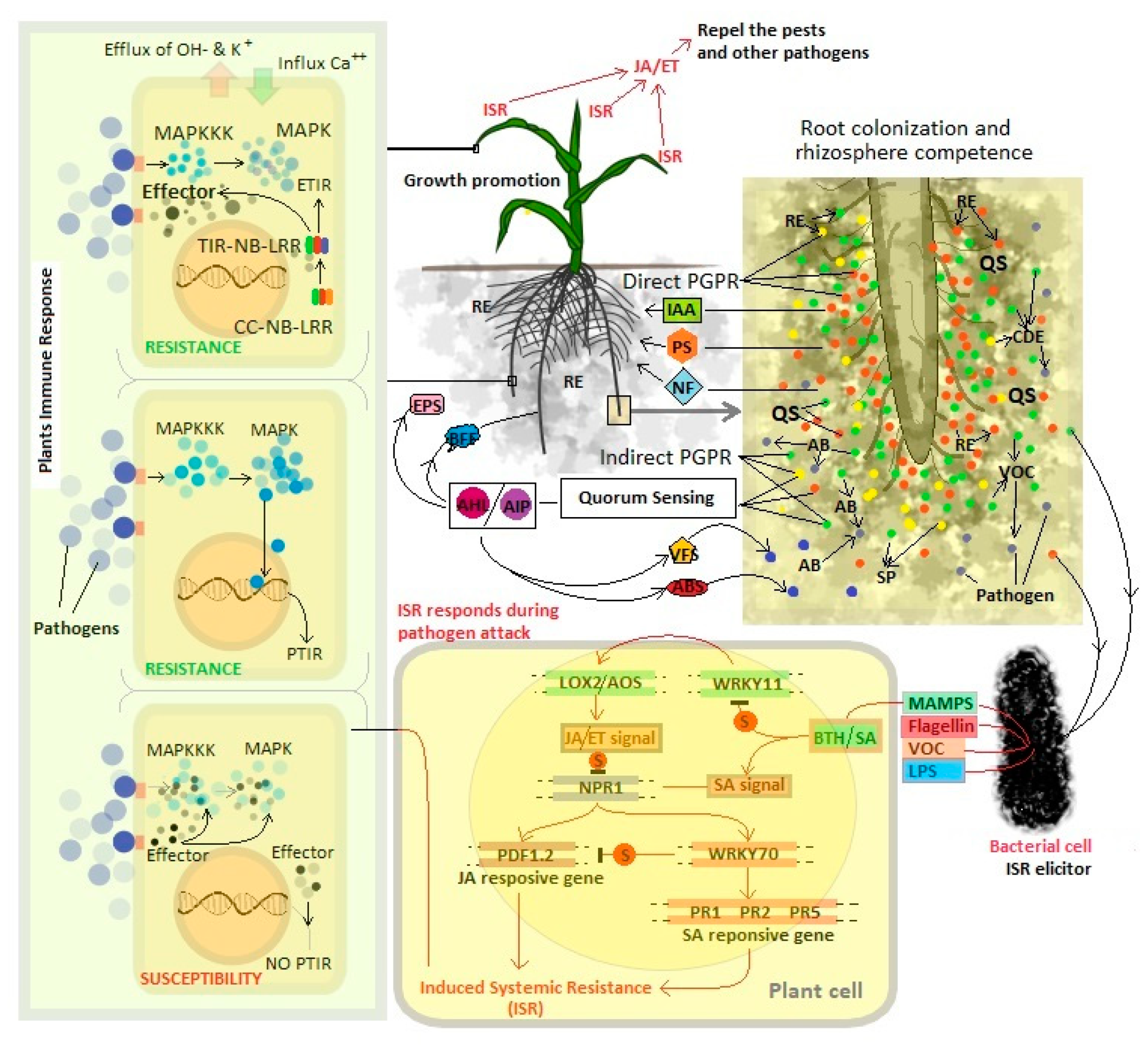
2.1. Root Endophytes as Plant Growth-Promoting Rhizobacteria (PGPR)
2.2. Mode of Action of Endophytic PGPR
2.2.1. Direct Mechanism
Nitrogen-Fixation
Solubilization of Phosphate
IAA Production
1-Aminocyclopropane-1-carboxylate (ACC) Deaminase
Role of Bacteria in Root System Architecture (RSA), and Its Modification
2.2.2. Indirect Mechanisms
Siderophore Production
HCN Production
Cell Wall Degrading Enzymes
Antibiotic Production
Volatile Organic Compounds Production
3. Role of Microbial Signals Modulate PGPR Functions
3.1. Regulation of Quorum Sensing by Plant-Associated Bacteria
3.2. Role of Quorum Sensing in Plant Defense and Biocontrol
4. Root Colonization and Rhizosphere Competence
5. Endophytic Arbascular Mycorhiza (AMF)
6. Endophytic PGPR and “Omics” Technologies
6.1. Effect of Root-Metabolome on Root-Microbiome
6.2. Metagenomes of Root-Associated Endophytes
6.3. Proteome Analysis for the Effect of Endophytes on Host Plants
6.4. PGPR Impact on the Plant Transcriptome
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Bavel, J. The world population explosion: Causes, backgrounds and -projections for the future. Facts Views Vis. ObGyn 2013, 5, 281–291. [Google Scholar] [PubMed]
- Schoneveld, G.C.; Van der Haara, S.; Ekowati, D.; Andrianto, A.; Komarudin, H.; Okarda, B.; Jelsma, I.; Pacheco, P. Certification, good agricultural practice and smallholder heterogeneity: Differentiated pathways for resolving compliance gaps in the Indonesian oil palm sector. Glob. Environ. Chang. 2019, 57, 101933. [Google Scholar] [CrossRef]
- Pervaiz, Z.H.; Iqbal, J.; Zhang, Q.; Chen, D.; Wei, H.; Saleem, M. Continuous Cropping Alters Multiple Biotic and Abiotic Indicators of Soil Health. Soil Syst. 2020, 4, 59. [Google Scholar] [CrossRef]
- Özkara, A.; Akyil, D.; Konuk, M. Pesticides, Environmental Pollution, and Health. In Environmental Health Risk-Hazardous Factors to Living Species; InTech: London, UK, 2016. [Google Scholar]
- Heinen, R.; Biere, A.; Harvey, J.A.; Bezemer, T.M. Effects of SoilOrganisms on Above ground plant-insect interactions in the field: Patterns, mechanisms and the role of methodology. Front. Ecol. Evol. 2018, 6, 106. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 1–15. [Google Scholar] [CrossRef]
- Ortíz-Castro, R.; Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; López-Bucio, J. The role of microbial signals in plant growth and development. Plant Signal. Behav. 2009, 4, 701–712. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar]
- Pervaiza, Z.H.; Contreras, J.; Hupp, B.M.; Lindenberger, J.H.; Chen, D.; Zhang, Q.; Wang, C.; Twigg, P.; Saleem, M. Root microbiome changes with root branching order and root chemistry in peach rhizosphere soil. Rhizosphere 2020, 16, 100249. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emergingtrends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Richardson, A.E.; Baréa, J.M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil. 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Couillerot, O.; Prigent-Combaret, C.; Caballero-Mellado, J.; Moënne-Loccoz, Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett. Appl. Microbiol. 2009, 48, 505–512. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.S.; Patra, J.K. Revitalization of plant growth promoting rhizobacte-ria for sustainable development in agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Misaghi, I.J.; Donndelinger, C.R. Endophytic bacteria in symptom free cotton plants. Phytopathology 1990, 80, 808–811. [Google Scholar] [CrossRef]
- Potshangbam, M.; Devi, S.I.; Sahoo, D.; Strobel, G.A. Functional characterization of endophytic fungal community associated with Oryza sativa L. and Zea mays L. Front. Microbiol. 2017, 8, 325. [Google Scholar] [CrossRef]
- Thomas, P.; Sekhar, A.C. Live cell imaging reveals extensive intracellular cytoplasmic colonization of banana by normally non-cultivable endophytic bacteria. AoB Plants 2014, 6, plu002. [Google Scholar] [CrossRef]
- Thomas, P.; Reddy, M.K. Microscopic elucidation of abundant endophytic bacteria colonizing the cell wall–plasma membrane peri-space in the shoot-tip tissue of banana. AoB Plants 2013, 5, plt011. [Google Scholar] [CrossRef]
- Compant, S.; Clement, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo-and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Strobel, G. Endophytes as sources of bioactive products. Microbes Infect. 2003, 5, 535–544. [Google Scholar] [CrossRef]
- Andreote, F.D.; Lacava, P.T.; Gai, C.S.; Araujo, W.L.; Maccheroni, W.; Overbeek, L.S.V.; Elsas, J.D.V.; Azevedo, J.L. Model plants for studying the interaction between Methylobacterium mesophilicum and Xylella fastidiosa. Can. J. Microbiol. 2006, 52, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Pasternak, J.J.; Glick, B.R. Isolation and characterization of mutants of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2 that overproduce indoleacetic acid. Curr. Microbiol. 1996, 32, 67–71. [Google Scholar]
- James, E.K.; Olivares, F.L. Infection and colonization of sugarcane and other gramineous plants by endophytic diazotrophs. Crit. Rev. Plant Sci. 1998, 17, 77–119. [Google Scholar] [CrossRef]
- Verma, S.K.; Kingsley, K.L.; Bergen, M.S.; Kowalski, K.P.; White, J.F. Fungal disease prevention in seedlings of rice (oryza sativa) and other grasses by growth-promoting seed-associated endophytic bacteria from invasive Phragmites australis. Microorganisms 2018, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Paungfoo-Lonhienne, C.; Rentsch, D.; Robatzek, S.; Webb, R.I.; Sagulenko, E.; Nasholm, T.; Schmidt, S.; Lonhienne, T.G.A. Turning the Table: Plants Consume Microbes as a Source of Nutrients. PLoS ONE 2010, 5, e11915. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.; Agrawal, M.; Bharathkumar, C.B. Diverse cellular colonizing endophytic bacteria in field shoots and in vitro cultured papaya with physiological and functional implications. Physiol. Plant. 2019, 166, 729–747. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.B.; Taghavi, S.; Mergeay, M.; Van der Lelie, D. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- McNear, D.H., Jr. The Rhizosphere-Roots, soil and everything in between. Nat. Educ. 2013, 4, 1. [Google Scholar]
- Rosier, A.; Medeiros, F.H.V.; Bais, H.P. Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 2018, 428, 35–55. [Google Scholar] [CrossRef]
- Oliveira, A.L.M.; Urquiaga, S.; Baldani, J.I. Processos e Mecanismos Envolvidos na Influência de Microrganismos Sobre o Crescimento Vegetal. Seropédica: CNPAB, Ago; Embrapa Agrobiologia-Documentos 161; Embrapa: Brasilia, Brazil, 2003; p. 40. [Google Scholar]
- Xia, Y.; DeBolt, S.; Dreyer, J.; Scott, D.; Williams, M.A. Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 2015, 6, 490. [Google Scholar] [CrossRef]
- Hallmann, J.; Quadt-Hallmann, A.; Mahaffee, W.F.; Kloepper, J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997, 43, 895–914. [Google Scholar] [CrossRef]
- Zakria, M.J.; Njolom, A.; Saeki, Y.; Akao, S. Colonization and nitrogen-fi xing ability of Herbaspirillum sp. strain B501 gfp1 and assessment of its growth-promoting ability in cultivated rice. Microbes Environ. 2007, 22, 197–206. [Google Scholar] [CrossRef]
- Costa, F.E.C.; Melo, I.S. Endophytic and rhizospheric bacteria from Opuntia ficus-indica mill and their ability to promote plant growth in cowpea, Vigna unguiculata (L.) Walp. Afr. J. Microbiol. Res. 2012, 6, 1345–1353. [Google Scholar]
- Olanrewaju, O.S.; Ayangbenro, A.S.; Glick, B.R.; Babalola, O.O. Plant health: Feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019, 103, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root Exudation of Primary Metabolites: Mechanisms and Their Roles in Plant Responses to Environmental Stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [PubMed]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influencesof plant growth-promoting rhizobacteria on nutrient acquisitionprocess. A review. Biol. Fertil. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth-promoting rhizobacteria: Current perspective. J. King Saud. Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Li, Z.; Zu, C.; Wang, C.; Yang, J.; Yu, H.; Wu, H. Different responses of rhizosphere and non-rhizosphere soil microbial communities to consecutive Piper nigrum L. monoculture. Sci. Rep. 2016, 6, 35825. [Google Scholar] [CrossRef] [PubMed]
- Kloepper, J.W.; Schroth, M.N. Plant growth-promoting rhizobacteria on radishes. In Proceedings of the 4th International Conf. on Plant Pathogenic Bacteria, Station de Pathologie Vegetable et Phytobacteriologie, INRA, Angers, France, 27 August–2 September 1978; pp. 879–882. [Google Scholar]
- Verma, J.P.; Yadav, J.; Tiwari, K.N.; Lavakush; Singh, V. Impact of Plant Growth Promoting Rhizobacteria on Crop Production. Int. J. Agric. Res. 2010, 5, 954–983. [Google Scholar] [CrossRef]
- Antoun, H.; Kloepper, J. Plant Growth Promoting Rhizobacteria (PGPR). Encycl. Genet. 2001, 1, 1477–1480. [Google Scholar]
- Barea, J.M. Future challenges and perspectives for applying microbial biotechnology in sustainable agriculture based on a better understanding of plant-microbiome interactions. J. Soil Sci. Plant Nutr. 2015, 15, 261–282. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 1–16. [Google Scholar] [CrossRef]
- Khan, M.S.; Gao, J.; Chen, X.; Zhang, M.; Yang, F.; Du, Y.; Moe, T.S.; Munir, I.; Xue, J.; Zhang, X. Isolation and characterization of plant growth-promoting endophytic bacteria Paenibacillus polymyxa SK1 from Lilium lancifolium. BioMed Res. Int. 2020, 2020, 8650957. [Google Scholar] [CrossRef] [PubMed]
- Ferchichi, N.; Toukabri, W.; Boularess, M.; Smaoui, A.; Mhamdi, R.; Trabelsi, D. Isolation, identification and plant growth promotion ability of endophytic bacteria associated with lupine root nodule grown in Tunisian soil. Arch. Microbiol. 2019, 201, 1333–1349. [Google Scholar] [CrossRef]
- Maggini, V.; Mengoni, A.; Gallo, E.R.; Biffi, S.; Fani, R.; Firenzuoli, F.; Bogani, P. Tissue specificity and differential effects on in vitro plant growth of single bacterial endophytes isolated from the roots, leaves and rhizospheric soil of Echinacea purpurea. BMC Plant Biol. 2019, 19, 1–9. [Google Scholar] [CrossRef]
- Borah, A.; Das, R.; Mazumdar, R.; Thakur, D. Culturable endophytic bacteria of Camellia species endowed with plant growth promoting characteristics. J. Appl. Microbiol. 2019, 127, 1364–5072. [Google Scholar] [CrossRef] [PubMed]
- Abedinzadeh, M.; Etesami, H.; Alikhani, H.A. Characterization of rhizosphere and endophytic bacteria from roots of maize (Zea mays L.) plant irrigated with wastewater with biotechnological potential in agriculture. Biotechnol. Rep. 2018, 20, e00305. [Google Scholar] [CrossRef]
- Chen, X.; Pizzatti, C.; Bonaldi, M.; Saracchi, M.; Erlacher, A.; Kunova, A.; Berg, G.; Cortesi, P. Biological Control of Lettuce Drop and Host Plant Colonization by Rhizospheric and Endophytic Streptomycetes. Front. Microbiol. 2016, 7, 714. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Jabborova, D.; Berg, G. Synergistic interactions between Bradyrhizobium japonicum and the endophyte Stenotrophomonas rhizophila and their effects on growth, and nodulation of soybean under salt stress. Plant Soil 2015, 405, 35–45. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Saxena, A.K.; Singh, D.P. Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biol. 2012, 4, 605–611. [Google Scholar] [CrossRef]
- Shi, Y.; Lou, K.; Li, C. Growth and photosynthetic efficiency promotion of sugar beet (Beta vulgaris L.) by endophytic bacteria. Photosynth. Res. 2010, 105, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Weilharter, A.; Mitter, B.; Shin, M.V.; Chain, P.S.; Nowak, J.; Sessitsch, A. Complete genome sequence of the plant growth-promoting endophyte Burkholderia phytofirmans strain PsJN. J. Bacteriol. 2011, 193, 3383–3384. [Google Scholar] [CrossRef] [PubMed]
- De Torres-Zabala, M.; Bennett, M.H.; Mansfield, J.W.; Egea, R.; Bo, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef]
- Cocking, E.C. Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil 2003, 252, 169–175. [Google Scholar] [CrossRef]
- Yanni, Y.G.; Rizk, R.Y.; Abd El-Fattah, F.K.; Squartini, A.; Corich, V.; Giacomini, A.; De Bruijn, F.; Rademaker, J.; Maya-Flores, J.; Ostrom, P.; et al. The beneficial plant growth promoting association of Rhizobium leguminosarum bv. trifolii with rice roots. Aust. J. Plant Physiol. 2001, 28, 845–870. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda, M.C.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Bekri, M.A.; Desair, J.; Keijers, V.; Proost, P.; Searle-van Leeuwen, M.; Vanderleyden, J.; vande Broek, A. Azospirillum irakense produces a novel type of pectate lyase. J. Bacteriol. 1999, 181, 2440–2447. [Google Scholar] [CrossRef]
- Sekar, C.; Prasad, N.N.; Sundaram, M.D. Enhancement of polygalacturonase activity during auxin induced para nodulation and endorhizosphere colonization of Azospirillum in rice roots. J. Exp. Biol. 2000, 38, 80–83. [Google Scholar]
- Agbodjato, N.A.; Noumavo, P.A.; Baba-Moussa, F.; Salami, H.A.; Sina, H.; Sèzan, A.; Bankolé, H.; Adjanohoun, A.; Baba-Moussa, L. Characterization of Potential Plant Growth Promoting Rhizobacteria Isolated from Maize (Zea mays L.) in Central and Northern Benin (West Africa). Appl. Environ. Soil Sci. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Bruto, M.; Prigent-Combaret, C.; Muller, D.; Moënne-Loccoz, Y. Analysis of genes contributing to plant-beneficial functions in plant growth-promoting rhizobacteria and related Proteobacteria. Sci. Rep. 2014, 4, srep06261. [Google Scholar] [CrossRef]
- Lei, X.; Wang, E.T.; Chen, W.F.; Sui, X.H.; Chen, W.X. Diverse bacteria isolated from root nodules of wild Vicia species grown in temperate region of China. Arch. Microbiol. 2008, 190, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Yang, S.; Tang, F.; Zhu, H. Symbiosis specificity in the legume: Rhizobial mutualism. Cell. Microbiol. 2012, 14, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Muthukumarasamy, R.; Cleenwerck, I.; Revathi, G.; Vadivelu, M.; Janssens, D.; Hoste, B.; Caballero-Mellado, J. Natural association of Gluconacetobacter diazotrophicus and diazotrophic Acetobacter peroxydans with wetland rice. Syst. Appl. Microbiol. 2005, 28, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Hurek, T.; Reinhold-Hurek, B. Azoarcus sp. strain BH72 as a model for nitrogen fixing grass endophytes. J. Biotechnol. 2003, 106, 169–178. [Google Scholar] [CrossRef]
- Engelhard, M.; Hurek, T.; Reinhold-Hurek, B. Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ. Microbiol. 2000, 2, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Masson-Boivin, C.; Sachs, J.L. Symbiotic nitrogen fixation by rhizobia—The roots of a success story. Curr. Opin. Plant Biol. 2018, 44, 7–15. [Google Scholar] [CrossRef]
- Lindstrom, K.; Mousavi, S.A. Effectiveness of nitrogen fixation in rhizobia. Microb. Biotechnol. 2020, 13, 1314–1335. [Google Scholar] [CrossRef]
- Robledo, M.; Garcia-Tomsig, N.I.; Jimenez-Zurdo, J.I. Riboregulation in Nitrogen-Fixing Endosymbiotic Bacteria. Microorganisms 2020, 8, 384. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.Z.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. Springerplus 2013, 2, 1–14. [Google Scholar] [CrossRef]
- Khan, M.S.; Zaidi, A.; Wani, P.A. Role of phosphate-solubilizing microorganisms in sustainable agriculture-A review. Agron. Sustain. Dev. 2007, 27, 29–43. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant growth-promoting rhizobacteria (PGPR): Emergence in agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Kpomblekou-A, K.; Tabatabai, M.A. Effect of low-molecular weight organic acids on phosphorus release and phytoavailabilty of phosphorus in phosphate rocks added to soils. Agric. Ecosyst. Environ. 2003, 100, 275–284. [Google Scholar] [CrossRef]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic bacteria in sunfl ower (Helianthus annuus L.): Isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Kuklinsky-Sobral, W.L.; Araujo, R.; Mendes, I.O.; Geraldi, A.A.; Pizzirani-Kleiner, J.L. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 2004, 6, 1244–1251. [Google Scholar] [CrossRef]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Spaepen, S.W.; Gocke, D.; Pohl, M.; Steyaert, J.; Vanderleyden, J. Characterization of phenylpyruvate decarboxylase, involved in auxin production of Azospirillum brasilense. J. Bacteriol. 2007, 189, 7626–7633. [Google Scholar] [CrossRef]
- Remans, R.; Beebe, S.; Blair, M.; Manrique, G.; Tovar, E.; Rao, I.; Croonenborghs, A.; Torres-Gutierrez, R.; El-Howeity, M.; Michiels, J.; et al. Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean (Phaseolus vulgaris L.). Plant Soil 2008, 302, 149–161. [Google Scholar] [CrossRef]
- Ji, S.H.; Gururani, M.A.; Chun, S.-C. Isolation and characterization of plant growth promoting endophytic diazotrophicbacteria from Korean rice cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef]
- Jasim, B.; Jimtha, J.C.; Shimil, V.; Jyothis, M.; Radhakrishnan, E.K. Studies on the factors modulating indole-3-acetic acid production in endophytic bacterial isolates from Piper nigrum and molecular analysis of ipdc gene. J. Appl. Microbiol. 2014, 117, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Defez, R.; Esposito, R.; Angelini, A.; Bianco, C. Overproduction of indole-3-acetic acid in free-living rhizobia induces transcriptional changes resembling those occurring inside nodule bacteroids. Mol. Plant Microbe Interact. 2016, 29, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Defez, R.; Andreozzi, A.; Bianco, C. The overproduction of indole-3-acetic acid (IAA) in endophytes upregulates nitrogen fixation in both bacterial cultures and inoculated rice plants. Microb. Ecol. 2017, 74, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Wang, Z.W.; Dong, F.; Lei, G.J.; Shi, Y.Z.; Li, G.X.; Zheng, S.J. Exogenous auxin alleviates cadmium toxicity in Arabidopsis thaliana by stimulating synthesis of hemicellulose 1 and increasing the cadmium fixation capacity of root cell walls. J. Hazard. Mater. 2013, 263, 398–403. [Google Scholar] [CrossRef]
- Bhutani, N.; Maheshwari, R.; Negi, M.; Suneja, P. Optimization of IAA production by endophytic Bacillus spp. from Vigna radiatafor their potential use as plant growth promoters. Isr. J. Plant Sci. 2018, 65, 1–2. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Botina, S.G.; Netrusov, A.I. Bacteria associated with orchid roots and microbial production of auxin. Microbiol. Res. 2007, 162, 69–76. [Google Scholar] [CrossRef]
- Chandra, S.; Askari, K.; Kumari, M. Optimization of indole acetic acid production by isolated bacteria from Stevia rebaudiana rhizosphere and its effects on plant growth. J. Genet. Eng. Biotechnol. 2018, 16, 581–586. [Google Scholar] [CrossRef]
- Ali, B.; Hasnain, S. Potential of bacterial indoleacetic acid to induce adventitious shoots in plant tissue culture. Lett. Appl. Microbiol. 2007, 45, 128–133. [Google Scholar] [CrossRef]
- Luo, S.; Xu, T.; Chen, L.; Chen, J.; Ra, C.; Xiao, X.; Wan, Y.; Zeng, G.; Long, F.; Liu, C.; et al. Endophyte-assisted promotion of biomass production and metal-uptake of energy crop sweet sorghum by plant-growth-promoting endophyte Bacillus sp. SLS18. Appl. Microbiol. Biotechnol. 2012, 93, 1745–1753. [Google Scholar]
- Verma, V.C.; Singh, S.K.; Prakash, S. Bio-control and plant growth promotion potential of siderophore producing endophytic Streptomyces from Azadirachta indica A. Juss. J. Basic Microbiol. 2011, 51, 550–556. [Google Scholar] [CrossRef]
- Walitang, D.I.; Kim, K.; Madhaiyan, M.; Kim, Y.K.; Kang, Y.; Sa, T. Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of Rice. BMC Microbiol. 2017, 17, 209. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Waqas, M.; Kang, S.-M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.-Y.; et al. Bacterial Endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene role in plant growth, development and senescence: Interaction with other phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Cheng, Z.; Czarny, J.; Duan, J. Promotion of plant growth by ACC deaminase-producing soil bacteria. Eur. J. Plant Pathol. 2007, 119, 329–339. [Google Scholar] [CrossRef]
- Gupta, S.; Pandey, S. ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French Bean (Phaseolus vulgaris) plants. Front Microbiol. 2019, 10, 1506. [Google Scholar] [CrossRef]
- Nonaka, S.; Sugawara, M.; Minamisawa, K.; Yuhashi, K.I.; Ezura, H. 1-aminocyclopropane-1-carboxylate deaminase producing Agrobacterium confers higher ability for gene transfer into plant cells. Appl. Environ. Microbiol. 2008, 74, 2526–2528. [Google Scholar] [CrossRef]
- Jasim, B.; Jimtha, C.J.; Jyothis, M.; Radhakrishnan, E.K. Plant growth promoting potential of endophytic bacteria isolated from Piper nigrum. Plant Growth Regul. 2013, 71, 1–11. [Google Scholar] [CrossRef]
- Thanananta, T. Cloning of 1-Aminocyclopropane-1- carboxylate Deaminase Gene from Soil Microorganism. Int. J. Sci. Technol. 1997, 2, 56–60. [Google Scholar]
- Zamioudis, C.; Mastranesti, P.; Dhonukshe, P.; Blilou, I.; Pieterse, C.M. Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 2013, 162, 304–318. [Google Scholar] [CrossRef]
- Rashid, S.; Charles, T.C.; Glick, B.R. Isolation and characterization of new plant growth-promoting bacterial endophytes. Appl. Soil Ecol. 2012, 61, 217–224. [Google Scholar] [CrossRef]
- Scheres, B.; Benfey, P.; Dolan, L. Root Development. Arab. Book. 2002, 1, e0101. [Google Scholar] [CrossRef] [PubMed]
- Lauter, F.R.; Ninnemann, O.; Bucher, M.; Riesmeier, J.W.; Frommer, W.B. Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 8139–8144. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.J.; Shin, R.; Schachtma, D.P. Expression ofKT/KUPgenes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 2004, 134, 1135–1145. [Google Scholar] [CrossRef] [PubMed]
- Pothier, J.F.; Wisniewski-Dyé, F.; Weiss-Gayet, M.; Mënne-Loccoz, Y.; Prigent-Combaret, C. Promoter-trap identification of wheat seed extract-induced genes in the plant-growth-promoting rhizobacterium Azospirillum brasilense Sp245. Microbiology 2007, 153, 3608–3622. [Google Scholar] [CrossRef][Green Version]
- Niranjan-Raj, S.; Lavanya, S.; Amruthesh, K.; Niranjana, S.; Shetty, H.S. Comparative evaluation of Pseudomonas fluorescens and their lipopolysaccharides as implicated in induction of resistance against pearl millet downy mildew. Arch. Phytopathol. Plant Prot. 2011, 44, 1285–1299. [Google Scholar] [CrossRef]
- Ryu, C.M.; Farag, M.A.; Hu, C.-H.; Reddy, M.S.; Wei, H.-X.; Pare, P.W.; Kloepper, J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 4927–4932. [Google Scholar] [CrossRef]
- Dodd, I.C.; Zinovkina, N.Y.; Safronova, V.I.; Belimov, A.A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef]
- Bakker, P.A.; Pieterse, C.M.; Van Loon, L.C. Induced systemic resistance by fluorescent Pseudomonas spp. Phytopathology 2007, 97, 239–243. [Google Scholar] [CrossRef]
- Phillips, D.A.; Fox, T.C.; King, M.D.; Bhuvaneswari, T.V.; Teuber, L.R. Microbial products trigger amino acid exudation from plant roots. Plant Physiol. 2004, 136, 2887–2894. [Google Scholar] [CrossRef]
- Contesto, C.; Milesi, S.; Mantelin, S.; Zancarini, A.; Desbrosses, G.; Varoquaux, F.; Bellini, C.; Kowalczyk, M.; Touraine, B. The auxin-signaling pathway is required for the lateral root response of Arabidopsis to the rhizobacterium Phyllobacterium brassicacearum. Planta 2010, 232, 1455–1470. [Google Scholar]
- Cacciari, I.; Lippi, D.; Pietrosanti, T.; Pietrosanti, W. Phytohormone-like substances produced by single and mixed diazotrophic cultures of Azospirillum and Arthrobacter. Plant Soil 1989, 115, 151–153. [Google Scholar] [CrossRef]
- Hussain, A.; Hasnain, S. Cytokinin production by some bacteria: Its impact on cell division in cucumber cotyledons. Afr. J. Microbiol. Res. 2009, 3, 704–712. [Google Scholar]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- Wilson, M.K.; Abergel, R.J.; Arceneaux, J.E.; Raymond, K.N.; Byers, B.R. Temporal production of the two Bacillus anthracis siderophores, petrobactin and bacillibactin. Biometals 2010, 23, 129–134. [Google Scholar] [CrossRef]
- Ferreira, M.J.; Silva, H.; Cunha, A. Siderophore-producing rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: A review. Pedosphere 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Alías-Villegas, C.; Bellogín, R.A.; del Cerro, P.; Espuny, M.R.; Jimenez-Guerrero, I.; Lopez-Baena, F.J.; Ollero, F.J.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef]
- Burd, G.I.; Dixon, D.G.; Glick, B.R. Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can. J. Microbiol. 2000, 46, 237–245. [Google Scholar] [CrossRef]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef]
- Idris, R.; Trifonova, R.; Puschenreiter, M.; Wenzel, W.W.; Sessitsch, A. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. Environ. Microb. 2004, 70, 2667–2677. [Google Scholar] [CrossRef]
- Sheng, X.F.; Xia, J.J.; Jiang, C.Y.; He, L.Y.; Qian, M. Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ. Pollut. 2008, 156, 1164–1170. [Google Scholar] [CrossRef]
- Schalk, I.J. Metal trafficking via siderophores in Gram negative bacteria: Specificities and characteristics of the pyoverdine pathway. J. Inorg. Biochem. 2008, 102, 1159–1169. [Google Scholar] [CrossRef]
- El-Tarabily, K.A.; Nassar, A.H.; Hardy, G.E.S.J.; Sivasithamparam, K. Plant growth promotion and biological control of Pythium aphanidermatum, a pathogen of cucumber, by endophytic actinomycetes. J. Appl. Microbiol. 2009, 106, 13–26. [Google Scholar] [CrossRef]
- Franco, C.M.M.; Michelsen, P.; Conn, V.M.; Loria, R.; Moll, S. Endophytic actinomycetes: Effective biocontrolagents for cereal root diseases. Phytopathology 2006, 96, S37. [Google Scholar]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Pseudomonas siderophores: A mechanism explaining disease-suppressive soils. Curr. Microbiol. 1980, 4, 317–320. [Google Scholar] [CrossRef]
- Li, J.H.; Wang, E.T.; Chen, W.F.; Chen, W.X. Genetic diversity and potential for promotion of plant growth detected in nodule endophytic bacteria of soybean grown in Heilongjiang province of China. Soil Biol. Biochem. 2008, 40, 238–246. [Google Scholar] [CrossRef]
- Budzikiewicz, H. Secondary metabolites from fluorescent Pseudomonas. FEMS Microbiol. Rev. 1993, 104, 209–228. [Google Scholar] [CrossRef]
- Ryan, R.P.; Monchy, S.; Cardinale, M.; Taghavi, S.; Crossman, L.; Avison, M.B.; Berg, G.; Van der Lelie, D.; Dow, J.M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 2009, 7, 514–525. [Google Scholar] [CrossRef]
- Rijavec, T.; Lapanje, A. Hydrogen cyanide in the rhizosphere: Not suppressing plant pathogens, but rather regulating availability of phosphate. Front Microbiol. 2016, 7, 1785. [Google Scholar] [CrossRef]
- Prasad, M.P.; Dagar, S. Identification and characterization of endophytic bacteria from fruits like avocado and black grapes. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 937–947. [Google Scholar]
- Rudrappa, T.; Splaine, R.E.; Biedrzycki, M.L.; Bais, H.P. Cyanogenic pseudomonads influence multitrophic interactions in the rhizosphere. PLoS ONE 2008, 3, e2073. [Google Scholar] [CrossRef]
- Pal, K.K.; Gardener, B.M. Biological control of plant pathogens. Plant Health Instr. 2006, 1–25. [Google Scholar] [CrossRef]
- Sakiyama, C.C.H.; Paula, E.M.; Pereira, P.C.; Borges, A.C.; Silva, D.O. Characterization of pectin lyase producedby an endophytic strain isolated from coffee cherries. Lett. Appl. Microbiol. 2001, 33, 117–121. [Google Scholar] [CrossRef]
- Tripathi, S.; Kamal, S.; Sheramati, I.; Oelmuller, R.; Varma, A. Mycorrhizal Fungi and other Root Endophytes as Biocontrol Agents Against Root Pathogens, in Mycorrhiza; Varma, A., Ed.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 281–306. [Google Scholar]
- Jadhav, H.P.; Shaikh, S.S.; Sayyed, R.Z. Role of hydrolytic enzymes of rhizoflora in biocontrol of fungal phytopathogens: An overview. In Rhizotrophs: Plant Growth Promotion to Bioremediation; Springer: Singapore, 2017; pp. 183–203. [Google Scholar]
- Badreddine, I.; Lafitte, C.; Heux, L.; Skandalis, N.; Spanou, Z.; Martinez, Y.; Esquerre-Tugaye, M.-T.; Bulone, V.; Dumas, B.; Bottin, A. Cell wall chitosaccharides are essential components and exposed patterns of the phytopathogenic oomycete Aphanomyces euteiches. Eukaryot. Cell 2008, 7, 1980–1993. [Google Scholar] [CrossRef]
- Khare, E.; Yadav, A. The Role of Microbial Enzyme Systems in Plant Growth Promotion. Clim. Chang. Environ. Sustain. 2017, 5, 122. [Google Scholar] [CrossRef]
- Jadhav, H.P.; Sayyed, R.Z. Hydrolytic Enzymes of Rhizospheric Microbes in Crop Protection. MOJ Cell Sci. Rep. 2016, 3, 135–136. [Google Scholar]
- Mercado-Blanco, J.; Bakker, P.A.H.M. Interactions between plantsand beneficial Pseudomonas spp.: Exploiting bacterial traits for crop protection. Antonie Leeuwenhoek 2007, 92, 367–389. [Google Scholar] [CrossRef]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef]
- Someya, N.; Kataoka, N.; Komagata, T.; Hirayae, K.; Hibi, T.; Akutsu, K. Biological control of cyclamen soil borne diseases by Serratia marcescens strain B2. Plant Dis. 2000, 84, 334–340. [Google Scholar] [CrossRef]
- Fridlender, M.; Inbar, J.; Chet, I. Biological control of soil borne plant pathogens by a ß–1,3–glucanase producing Pseudomonas cepacia. Soil Biol. Biochem. 1993, 25, 1211–1221. [Google Scholar] [CrossRef]
- Budi, S.W.; Van Tuinen, D.; Arnould, C.; Dumas-Gaudot, E.; Gianinazzi-Pearson, V.; Gianinazzi, S. Hydrolytic enzyme activity of Paenibacillus sp. strain B2 and effects of the antagonistic bacterium on cell integrity of two soil borne pathogenic fungi. Appl. Soil Ecol. 2000, 15, 191–199. [Google Scholar] [CrossRef]
- Garbeva, P.; Van Veen, J.A.; Van Elas, J.D. Assessment of the diversity, and antagonism towards Rhizoctonia solani AG3, of Pseudomonas species in soil from different agricultural regimes. FEMS Microbiol. Ecol. 2004, 47, 51–64. [Google Scholar]
- Loqman, S.; Ait Barka, E.; Clément, C.; Ouhdouch, Y. Antagonistic actinomycetes from Moroccan soil to control the grapevine gray mold. World J. Microbiol. Biotechnol. 2009, 25, 81–91. [Google Scholar] [CrossRef]
- Palumbo, J.D.; Yuen, G.Y.; Jochum, C.C.; Tatum, K.; Kobayashi, D.Y. Mutagenesis of beta-1,3-Glucanase Genes in Lysobacter enzymogenes Strain C3 Results in Reduced Biological Control Activity Toward Bipolaris Leaf Spot of Tall Fescue and Pythium Damping-Off of Sugar Beet. Phytopathology 2005, 95, 701–707. [Google Scholar] [CrossRef]
- Macagnan, D.; Romeiro, R.S.; Pomella, A.W.V.; De Souza, J.T. Production of lytic enzymes and siderophores, and inhibition of germination of basidiospores of Moniliophthora (ex Crinipellis) perniciosaby phylloplane actinomycetes. Biol. Control 2008, 47, 309–314. [Google Scholar] [CrossRef]
- Verma, S.C.; Ladha, J.K.; Tripathi, A.K. Evaluation ofplant growth promoting and colonization abilityof endophyti cdiazotrophs from deep water rice. J. Biotechnol. 2001, 91, 127–141. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant Growth Promoting and Biocontrol Activity of Streptomyces spp. as Endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef]
- El-Banna, N.; Winkelmann, G. Pyrrolnitrin from Burkholderia cepacia: Antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 1998, 85, 69–78. [Google Scholar] [CrossRef]
- Velusamy, P.; Immanuel, J.E.; Gnanamanickam, S.S.; Thomashow, L. Biological control of rice bacterial blight by plant-associated bacteria producing 2,4-diacetylphloroglucinol. Can. J. Microbiol. 2006, 52, 56–65. [Google Scholar] [CrossRef]
- Andreote, F.D.; Rossetto, P.B.; Mendes, R.; Avila, L.A.; Labate, C.A.; Pizzirani Kleiner, A.A.; Azevedo, J.L.; Araujo, W.L. Bacterial community in the rhizosphere and rhizoplane of wild type and transgenic eucalyptus. World J. Microbiol. Biotechnol. 2009, 25, 1065–1073. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, X.; Kuang, S.; Liu, G.; Zhang, C.; Sun, C. Antagonistic activity and mode of action of phenazine-1-carboxylic acid, produced by marine bacterium Pseudomonas aeruginosa PA31x, against Vibrio anguillarum in vitro and in a zebrafish in vivo model. Front Microbiol. 2017, 8, 289. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Wang, Y.; Sun, X.; Tang, K. Bioactive natural products from endophytes: A review. Appl. Biochem. Microbiol. 2008, 44, 136–142. [Google Scholar] [CrossRef]
- Cho, K.M.; Hong, S.Y.; Lee, S.M.; Kim, Y.H.; Kahng, G.G.; Lim, Y.P.; Kim, H.; Yun, H.D. Endophytic bacterial communities in ginseng and their antifungal activity against pathogens. Microb. Ecol. 2007, 54, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Ezra, D.; Castillo, U.F.; Strobel, G.A.; Hess, W.M.; Porter, H.; Jensen, J.B.; Condron, M.A.M.; Teplow, D.B.; Sears, J.; Maranta, M.; et al. Coronamycins, peptide antibioticsproduced by a verticillate Streptomyces sp. (MSU-2110) endophytic on Monsterasp. Microbiology 2004, 150, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Asraful Islam, S.; Math, R.K.; Kim, J.; Yun, M.G.; Cho, J.J.; Kim, E.J.; Lee, Y.H.; Yun, H.D. Effect of Plant Age on Endophytic Bacterial Diversity of Balloon Flower (Platycodon grandiflorum) Root and Their Antimicrobial Activities. Curr. Microbiol. 2010, 61, 346–356. [Google Scholar] [CrossRef]
- Farag, M.A.; Zhang, H.; Ryu, C.-M. Dynamic chemical communication between plants and bacteria through airborne signals: Induced resistance by bacterial volatiles. J. Chem. Ecol. 2013, 39, 1007–1018. [Google Scholar] [CrossRef]
- De Vrieze, M.; Pandey, P.; Bucheli, T.D.; Varadarajan, A.R.; Ahrens, C.H.; Weisskopf, L.; Bailly, A. Volatile Organic Compounds from Native Potato-associated Pseudomonas as Potential Anti-oomycete Agents. Front. Microbiol. 2015, 6, 1295. [Google Scholar] [CrossRef]
- Del Rosario Cappellari, L.; Chiappero, J.; Banchio, E. Invisible signals from the underground: A practical method to investigate the effect of microbial volatile organic compounds emitted by rhizobacteria on plant growth. Biochem. Mol. Biol. Educ. 2019, 47, 388–393. [Google Scholar] [CrossRef]
- Sumayo, M.; Hahm, M.S.; Ghim, S.Y. Determinants of plant growth-promoting Ochrobactrum lupini KUDC1013 involved in induction of systemic resistance against Pectobacterium carotovorum subsp. Carotovorum in tobacco leaves. Plant Pathol. J. 2013, 29, 174–181. [Google Scholar] [CrossRef]
- Tahir, H.A.S.; Qin, G.; Wu, H.; Niu, Y.; Rong, H.; Gao, X. Bacillus volatiles adversely affect the physiology and ultrastructure of Ralstonia solanacearum and induce systemic resistance in tobacco against Bacillus wilt. Sci. Rep. 2017, 7, 40481. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Luna, F.M.; López-Bucio, J.; Altamirano-Hernández, J.; Valencia-Cantero, E.; De la Cruz, H.R.; Macías-Rodríguez, L. Plant growth-promoting rhizobacteria modulate root-system architecture in Arabidopsis thaliana through volatile organic compound emission. Symbiosis 2010, 51, 75–83. [Google Scholar] [CrossRef]
- D’alessandro, M.; Erb, M.; Ton, J.; Brandenburg, A.; Karlen, D.; Zopfi, J.; Turlings, T.C. Volatiles produced by soil-borne endophytic bacteria increase plant pathogen resistance and affect tritrophic interactions. Plant Cell Environ. 2014, 37, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Rudrappa, T.; Biedrzycki, M.L.; Kunjeti, S.G.; Donofrio, N.M.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. The rhizobacterial elicitor acetoin induces systemic resistance in Arabidopsis thaliana. Commun. Integr. Biol. 2010, 3, 130–138. [Google Scholar] [CrossRef]
- Xie, S.; Liu, J.; Gu, S.; Chen, X.; Jiang, H.; Ding, T. Antifungal activity of volatile compounds produced by endophytic Bacillus subtilis DZSY21 against Curvularia lunata. Ann. Microbiol. 2020, 70, 1–10. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, B.; Liu, H.; Han, J.; Zhang, Y. Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol. Control 2017, 105, 27–39. [Google Scholar] [CrossRef]
- Atmosukarto, I.; Castillo, U.; Hess, W.M.; Sears, J.; Strobel, G. Isolation and characterization of Muscodor albus I-41.3 s, a volatile antibiotic producing fungus. Plant Sci. 2005, 169, 854–861. [Google Scholar] [CrossRef]
- Chan, K.-G.; Atkinson, S.; Mathee, K.; Sam, C.-K.; Chhabra, S.R.; Camara, M.; Koh, C.-L.; Williams, P. Characterization of N-acylhomoserine lactone-degrading bacteria associated with the Zingiber officinale (ginger) rhizosphere: Co-existence of quorum quenching and quorum sensing in Acinetobacter and Burkholderia. BMC Microbiol. 2011, 11, 51. [Google Scholar] [CrossRef]
- Zuniga, A.; Donoso, R.A.; Ruiz, D.; Ruz, G.A.; González, B. Quorum-sensing systems in the plant growth-promoting bacterium Paraburkholderia phytofirmans PsJN exhibit cross-regulation and are involved in biofilm formation. Mol. Plant-Microbe Interact. 2017, 30, 557–565. [Google Scholar] [CrossRef]
- Steindler, L.; Venturi, V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 2007, 266, 1–9. [Google Scholar] [CrossRef]
- Fuqua, W.C.; Winans, S.C. A LuxR-LuxI type regulatory system activates Agrobacterium Tiplasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 1994, 176, 2796–2806. [Google Scholar] [CrossRef] [PubMed]
- Elasri, M.; Delorme, S.; Lemanceau, P.; Stewart, G.; Laue, B.; Glickmann, E.; Oger, P.M.; Dessaux, Y. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 2001, 67, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.; Lamosa, P.; Russell, C. Quorum-sensing signal autoinducer-2 (AI-2) characterization of phospho-(S)-4, 5-dihydroxy-2,3-pentanedione isomerization by LsrG protein. J. Biol. Chem. 2011, 286, 18331–18343. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jia, J.; Popat, R.; Ortori, C.A.; Li, J.; Diggle, S.P.; Gao, K.; Camara, M. Characterisation of two quorum sensing systems in the endophytic Serratia plymuthica strain G3: Differential control of motility and biofilm formation according to life-style. BMC Microbiol. 2011, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Vial, L.; Cuny, C.; Gluchoff-Fiasson, K.; Comte, G.; Oger, P.M.; Faure, D.; Dessaux, Y.; Bally, R.; Wisniewski-Dyé, F. N-acyl-homoserine lactone -mediated quorum-sensing inAzospirillum: An exception rather than a rule. FEMS Microbiol. Ecol. 2006, 58, 155–168. [Google Scholar] [CrossRef]
- Boyer, M.; Bally, R.; Perrotto, S.; Chaintreuil, C.; Wisniewski-Dyé, F. A quorum-quenching approach to identify quorum-sensing-regulated functions in Azospirillum lipoferum. Res. Microbiol. 2008, 159, 699–708. [Google Scholar] [CrossRef]
- Steidle, A.; Sigl, K.; Schuhegger, R.; Ihring, A.; Schmid, M.; Gantner, S.; Stoffels, M.; Riedel, K.; Givskov, M.; Hartmann, A.; et al. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing thetomato rhizosphere. Appl. Environ. Microbiol. 2001, 67, 5761–5770. [Google Scholar] [CrossRef]
- Vandeputte, O.M.; Kiendrebeogo, M.; Rajaonson, S.; Diallo, B.; Mol, A.; El Jaziri, M.; Baucher, M. Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 2010, 76, 243–253. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Diggle, S.P.; Cornelis, P.; Williams, P.; Camara, M. 4-Quinolone signalling in Pseudomonas aeruginosa: Old molecules, new perspectives. Int. J. Med. Microbiol. 2006, 296, 83–91. [Google Scholar] [CrossRef]
- Suarez-Moreno, Z.R.; Devescovi, G.; Myers, M.; Hallack, L.; Mendonca-Previato, L.; Caballero-Mellado, J.; Venturi, V. Commonalities and differences in regulation of n-acyl homoserine lactone quorum sensing in the beneficial plant-associated burkholderia species cluster. Appl. Environ. Microbiol. 2010, 6, 4302–4317. [Google Scholar] [CrossRef]
- Ryu, C.M.; Choi, H.K.; Lee, C.H.; Murphy, J.F.; Lee, J.K.; Kloepper, J.W. Modulation of Quorum Sensing in Acylhomoserine Lactone-Producing or -Degrading Tobacco Plants Leads to Alteration of Induced Systemic Resistance Elicited by the Rhizobacterium Serratia marcescens 90–166. Plant Pathol. J. 2013, 29, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Puopolo, G.; Cimmino, A.; Palmieri, M.C.; Giovannini, O.; Evidente, A.; Pertot, I. Lysobacter capsici AZ78 produces cyclo(L-Pro-L-Tyr), a 2,5-diketopiperazine with toxic activity against sporangia of Phytophthora infestans and Plasmopara viticola. J. Appl. Microbiol. 2014, 117, 1168–1180. [Google Scholar] [CrossRef] [PubMed]
- Schuhegger, R.; Ihring, A.; Gantner, S.; Bahnweg, G.; Knappe, C.; Vogg, G.; Hutzler, P.; Schmid, M.; Breusegem, F.V.; Eberl, L.; et al. Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 2006, 29, 909–918. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jia, J.; Atkinson, S.; Camara, M.; Gao, K.; Li, H.; Cao, J. Biocontrol potential of an endophytic Serratia sp. G3 and its modeof action. World J. Microbiol. Biotechnol. 2010, 26, 1465–1471. [Google Scholar] [CrossRef]
- Benhamou, N.; Kloepper, J.W.; Quadt-Hallmann, A.; Tuzun, S. Induction of defense-related ultrastructural modifications in pea root tissues inoculated with endophytic bacteria. Plant Physiol. 1996, 112, 919–929. [Google Scholar] [CrossRef]
- Schenk, S.T.; Hernandez-Reyes, C.; Samans, B.; Stein, E.; Neumann, C.; Schikora, M.; Reichelt, M.; Mithofer, A.; Becker, A.; Kogel, K.-H.; et al. N-Acyl-Homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell. 2014, 26, 2708–2723. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef]
- Schroth, M.N.; Hancock, J.G. Disease-suppressive soil and root-colonizing bacteria. Science 1982, 216, 1376–1381. [Google Scholar] [CrossRef]
- Iniguez, A.L.; Dong, Y.; Carter, H.D.; Ahmer, B.M.; Stone, J.M.; Triplett, E.W. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant Microbe Interact. 2005, 18, 169–178. [Google Scholar] [CrossRef]
- Cavalcante, J.J.; Vargas, C.; Nogueira, E.M.; Vinagre, F.; Schwarcz, K.; Baldani, J.I.; Ferreira, P.C.; Hemerly, A.S. Members of the ethylene signalling pathway are regulated in sugarcane during the association with nitrogen-fixing endophytic bacteria. J. Exp. Bot. 2007, 58, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Frommel, M.I.; Nowak, J.; Lazarovits, G. Growth enhance-ment and developmental modifications of in vitro grown potato (Solanum tuberosum ssp. tuberosum) as affected by a nonfluorescent Pseudomonas sp. Plant Physiol. 1991, 96, 928–936. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Coenye, T.; Sturz, A.V.; Vandamme, P.; Barka, E.A.; Salles, J.F.; Van Elsas, J.D.; Faure, D.; Reiter, B.; Glick, R.B.; et al. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 2005, 55, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Kaplan, H.; Sessitsch, A.; Nowak, J.; Barka, E.A.; Clement, C. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: From the rhizosphere to inflorescence tissues. FEMS Microbiol. Ecol. 2008, 63, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed]
- McCully, M.E. Niches for bacterial endophytes in crop plants: A plant biologist’s view. Aust. J. Plant Physiol. 2001, 28, 983–990. [Google Scholar] [CrossRef]
- Pankievicz, V.C.; Camilios-Neto, D.; Bonato, P.; Balsanelli, E.; Tadra-Sfeir, M.Z.; Faoro, H.; Chubatsu, L.S.; Donatti, L.; Wajnberg, G.; Passetti, F.; et al. RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol. Biol. 2016, 90, 589–603. [Google Scholar] [CrossRef]
- Alqueres, S.; Meneses, C.; Rouws, L.; Rothballer, M.; Baldani, I.; Schmid, M.; Hartmann, A. The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol. Plant Microbe Interact. 2013, 26, 937–945. [Google Scholar] [CrossRef]
- Gehring, C.A.; Johnson, N.C. Beyond ICOM8: Perspectives on advances in mycorrhizal research from 2015 to 2017. Mycorrhiza 2018, 28, 197–201. [Google Scholar] [CrossRef]
- Martin, F.M.; Uroz, S.; Barker, D.G. Ancestral alliances: Plant mutualistic symbioses with fungi and bacteria. Science 2017, 356, eaad4501. [Google Scholar] [CrossRef]
- Wezowicz, K.; Rozpadek, P.; Turna, K. Interactions of arbuscular mycorrhizal and endophytic fungiimprove seedling survival and growth in post-mining waste. Mycorrhiza 2017, 27, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.; Abd-Allah, E.F.; Alqarawi, A.A.; Al-Huqail, A.A.; Wirth Sand Egamberdieva, D. The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances Plant growth of Acacia gerrardii under salt stress. Front. Microbiol. 2016, 7, 1089. [Google Scholar] [CrossRef] [PubMed]
- Deveau, A.; Antony-Babu, S.; Le Tacon, F.; Robin, C.; Frey-Klett, P.; Uroz, S. Temporal changes of bacterial communities in the Tuber melanosporum ectomycorhizosphere during ascocarp development. Mycorrhiza 2016, 26, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Marupakula, S.; Mahmood, S.; Finlay, R.D. Analysis of single root tip microbiomes suggests that distinctive bacterial communities are selected by Pinus sylvestris roots colonized by different ectomycorrhizal fungi. Environ. Microbiol. 2016, 18, 1470–1483. [Google Scholar]
- Nguyen, N.H.; Bruns, T.D. The microbiome of Pinus muricata ectomycorrhizae: Community assemblages, fungal species effects, and Burkholderia as important bacteria in multi-partnered symbioses. Microb. Ecol. 2015, 69, 914–921. [Google Scholar] [CrossRef]
- Akyol, T.Y.; Niwa, R.; Hirakawa, H.; Maruyama, H.; Sato, T.; Suzuki, T.; Fukunaga, A.; Sato, T.; Yoshida, S.; Tawaraya, K.; et al. Impact of introduction of arbuscular mycorrhizal fungi on the root microbial community in Agricultural Fields. Microbes Environ. 2019, 34, 23–32. [Google Scholar] [CrossRef]
- Vannini, C.; Carpentieri, A.; Salvioli, A.; Novero, M.; Marsoni, M.; Testa, L.; De Pinto, M.C.; Amoresano, A.; Ortolani, F.; Bracale, M.; et al. An interdomain network: The endobacterium of a mycorrhizal fungus promotes antioxidative responses in both fungal and plant hosts. N. Phytol. 2016, 211, 265–275. [Google Scholar] [CrossRef]
- Lavania, M.; Chauhan, P.S.; Chauhan, S.V.; Singh, H.B.; Nautiyal, C.S. Induction of plant defense enzymes and phenolics by treatment with plant growth-promoting rhizobacteria Serratia marcescens NBRI1213. Curr. Microbiol. 2006, 52, 363–368. [Google Scholar] [CrossRef]
- Dardanelli, M.S.; Manyani, H.; Gonzalez-Barroso, S.; Rodriguez-Carvajal, M.A.; Gil-Serrano, A.M.; Espuny, M.R.; López-Baena, F.J.; Bellogín, R.A.; Megías, M.; Ollero, F.J. Effect of the presence of the plant growth promoting rhizobacterium (PGPR) Chryseobacterium balustinum Aur9 and salt stress in the pattern of flavonoids exuded by soybean roots. Plant Soil 2010, 328, 483–493. [Google Scholar] [CrossRef]
- Burdman, S.; Volpin, H.; Kigel, J.; Kapulnik, Y.; Okon, Y. Promotion of nod gene inducers and nodulation in common bean (Phaseolus vulgaris) roots inoculated with Azospirillum brasilense Cd. Appl. Environ. Microbiol. 1996, 62, 3030–3033. [Google Scholar] [CrossRef]
- Curzi, M.J.; Ribaudo, C.M.; Trinchero, G.D.; Cura, J.A.; Pagano, E.A. Changes in the content of organic and amino acids and ethylene production of rice plants in response to the inoculation with Herbaspirillum seropedicae. J. Plant Interact. 2008, 3, 163–173. [Google Scholar] [CrossRef]
- Bharti, N.; Yadav, D.; Barnawal, D.; Maji, D.; Kalra, A. Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J. Microbiol. Biotechnol. 2013, 29, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Chamam, A.; Sanguin, H.; Bellvert, F.; Meiffren, G.; Comte, G.; Wisniewski-Dyé, F.; Bertrand, C.; Pringent-Combaret, C. Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum-Oryza sativa association. Phytochemistry 2013, 87, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Rozpadek, P.K.; Wężowicz, M.; Nosek, R.; Ważny, K.; Tokarz, M.; Lembicz, M.; Miszalski, Z.; Turnau, K. The fungal endophyte Epichloë typhina improves photosynthesis efficiency of its host orchard grass (Dactylis glomerata). Planta 2015, 242, 1025–1035. [Google Scholar] [CrossRef]
- Schliemann, W.; Ammer, C.; Strack, D. Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 2008, 69, 112–146. [Google Scholar] [CrossRef]
- Rasmussen, S.; Parsons, A.J.; Fraser, K.; Xue, H.; Newman, J.A. Metabolic profiles of Lolium perenne are differentially affected by nitrogen supply, carbohydrate content, and fungal endophyte infection. Plant Physiol. 2008, 146, 1440–1453. [Google Scholar] [CrossRef]
- Bacilio-Jiménez, M.; Aguilar-Flores, S.; Ventura-Zapata, E.; Pérez-Campos, E.; Bouquelet, S.; Zenteno, E. Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 2003, 249, 271–277. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.; Patel, S. Combination of endophytic and rhizospheric plant growth promoting rhizobacteria in Oryza sativa shows higher accumulation of osmoprotectant against saline stress. Acta Physiol. Plant. 2011, 33, 797–802. [Google Scholar] [CrossRef]
- Hiruma, K.; Gerlach, N.; Sacristán, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramirez, D.; Bucher, M.; O’Connell, R.J.; et al. Root Endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef]
- Amrani, A.E.; Dumas, A.; Wick, L.Y.; Yergeau, E.; Berthomee, R. Omics insights into PAH degradation toward improved Green remediation biotechnologies. Environ. Sci. Technol. 2015, 49, 11281–11291. [Google Scholar] [CrossRef]
- Tian, B.-Y.; Cao, Y.; Zhang, K.-Q. Metagenomic insights into communities, functions of endophytes, and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci. Rep. 2015, 5, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Maropola, M.K.A.; Ramond, J.-B.; Trindade, M. Impact of metagenomic DNA extraction procedures on the identifiable endophytic bacterial diversity in Sorghum bicolor (L. Moench). J. Microbiol. Methods 2015, 112, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Ardanov, P.; Sessitsch, A.; Haggman, H.; Kozyrovska, N.; Pirttila, A.M. Methylobacterium -induced endophyte community changes correspond with protection of plants against pathogen attack. PLoS ONE 2012, 7, e46802. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Hardoim, P.; Doring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional Characteristics of an Endophyte Community Colonizing Rice Roots as Revealed by Metagenomic Analysis. MPMI 2012, 25, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-J.; Wang, L.-L.; Li, Q.; Shang, Q.-M. Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS ONE 2019, 14, e022384. [Google Scholar] [CrossRef]
- Long, H.H.; Sonntag, D.G.; Schmidt, D.D.; Baldwin, I.T. The structure of the culturable root bacterial endophyte community of Nicotiana attenuata is organized by soil composition and host plant ethylene production and perception. N. Phytol. 2010, 185, 554–567. [Google Scholar] [CrossRef]
- Puri, R.R.; Adachi, F.; Omichi, M.; Saeki, Y.; Yamamoto, A.; Hayashi, S.; Ali, M.A.; Itoh, K. Metagenomic study of endophytic bacterial community of sweet potato (Ipomoea batatas) cultivated in different soil and climatic conditions. World J. Microbiol. Biotechnol. 2019, 35, 176. [Google Scholar] [CrossRef]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; De Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef]
- Pinto-Carbó, M.; Sieber, S.; Dessein, S.; Wicker, T.; Verstraete, B.; Gademann, K.; Eberl, L.; Carlier, A. Evidence of horizontal gene transfer between obligate leaf nodule symbionts. ISME J. 2016, 10, 2092–2105. [Google Scholar] [CrossRef]
- Cheng, Z.; Mcconkey, B.J.; Glick, B.R. Proteomic studies of plant–bacterial interactions. Soil Biol. Biochem. 2010, 42, 1673–1684. [Google Scholar] [CrossRef]
- Maron, P.; Ranjard, L.; Mougel, C.; Lemanceau, P. Metaproteomics: A new approach for studying functional microbial ecology. Microb. Ecol. 2007, 53, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Witzel, K.; Üstün, S.; Schreiner, M.; Grosch, R.; Börnke, F.; Ruppel, S. A proteomic approach suggests unbalanced proteasome functioning induced by the growth-promoting bacterium Kosakonia radicincitans in Arabidopsis. Front. Plant Sci. 2017, 8, 661. [Google Scholar] [CrossRef] [PubMed]
- Chi, F.; Yang, P.; Han, F.; Jing, Y.; Shen, S. Proteomic analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics 2010, 10, 1861–1874. [Google Scholar] [PubMed]
- Lery, L.M.S.; Hemerly, A.S.; Nogueira, E.M.; Von, K.; Ruger, W.M.A.; Bisch, P.M. Quantitative proteomic analysis of the interaction between the endophytic plant-growth-promoting bacterium Gluconacetobacter diazotrophicus and sugarcane. Mol. Plant-Microbe Interact. 2011, 24, 562–576. [Google Scholar] [CrossRef]
- Miche, L.; Battistoni, F.; Gemmer, S.; Belghazi, M.; Reinhold-Hurek, B. Upregulation of jasmonate-inducible defense proteins and differential colonization of roots of Oryza sativa cultivars with the endophyte Azoarcus sp. Mol. Plant-Microbe Interact. 2006, 19, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Chaudhry, V.; Mishra, A.; Chauhan, P.S.; Rehman, A.; Yadav, A.; Tuteja, N.; Nautiyal, C.S. Gene expression profiling through microarray analysis in Arabidopsis thaliana colonized by Pseudomonas putida MTCC5279, a plant growth promoting rhizobacterium. Plant Signal. Behav. 2012, 7, 235–245. [Google Scholar] [CrossRef]
- Vargas, L.; Gurjao de Carvalho, T.L.; Gomes Ferreira, P.C.; Baldani, V.L.; Baldani, J.I.; Hemerly, A.S. Early responses of rice (Oryza sativa L.) seedlings to inoculation with beneficial diazotrophic bacteria are dependent on plant and bacterial genotypes. Plant Soil 2012, 356, 127–137. [Google Scholar] [CrossRef]
- Brusamarello-Santos, L.C.C.; Pacheco, F.; Aljanabi, S.M.M.; Monteiro, R.A.; Cruz, L.M.; Baura, V.A.; Pedrosa, F.O.; Souza, E.M.; Wassem, R. Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae. Plant Soil 2012, 356, 113–125. [Google Scholar] [CrossRef]
- Verhagen, B.W.; Glazebrook, J.; Zhu, T.; Chang, H.S.; Van Loon, L.C.; Pieterse, C.M. The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol. Plant-Microbe Interact. 2004, 17, 895–908. [Google Scholar] [CrossRef]
- Van de Mortel, J.E.; De Vos, R.C.; Dekkers, E.; Pineda, A.; Guillod, L.; Bouwmeester, K.; van Loon, J.J.A.; Dicke, M.; Raaijmakers, J.M. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012, 160, 2173–2188. [Google Scholar] [CrossRef]
- Maketon, C.; Fortuna, A.M.; Okubara, P.A. Cultivar-dependent transcript accumulation in wheat roots colonized by Pseudomonas fluorescens Q8r1-96 wild type and mutant strains. Biol. Control 2012, 60, 216–224. [Google Scholar] [CrossRef]
- Yi, Y.; De Jong, A.; Frenzel, E.; Kuipers, O.P. Comparative transcriptomics of Bacillus mycoides strains in response to potato-root exudates reveals different genetic adaptation of endophytic and soil isolates. Front. Microbiol. 2017, 8, 1487. [Google Scholar] [CrossRef] [PubMed]
| Sl. No. | Endophyte | Host-Plant | PGP-Attributes | Reference |
|---|---|---|---|---|
| 1. | Paenibacillus polymyxa SK1 | Lilium lancifolium | 1-Aminocycloprapane-1-carboxylic acid (ACC), deaminase, indole-3-acetic acid (IAA), siderophores, nitrogen fixation, and phosphate solubilisation, showed antifungal activities against plant pathogens | [46] |
| 2. | Paenibacillus glycanilyticus LJ121 and Pseudomonas brenneri LJ215 | Lupine root | Increase shoot Dry weight, number of nodules per plant, photosynthetic assimilation rate and chlorophyll a and b content and shoot nitrogen and phosphorus content | [47] |
| 3. | Arthrobacter sp. EpS/L16 | Echinacea purpurea | IAA production, increase in the number of leaves | [48] |
| 4. | Lysinibacillus sp. S24 Brevibacterium sp. S91 | Tea (Camellia sinensis L.) | Highest phosphate solubilisation, IAA and ammonia production | [49] |
| 5. | Enterobacter cloacae R7 | Maize roots | IAA (35.4 mg mL_1), ACC deaminase (+), siderophore (+), and phosphate solubilization (+), alleviating heavy metal stress | [50] |
| 6. | Bacillus cereus N5 | Maize roots | IAA (47.3 mg mL_1), ACC deaminase (+), siderophore (+), and phosphate solubilization (+), tolerance of this plant to environmental stresses | [50] |
| 7. | Streptomyces exfoliatus FT05W | Lettuce roots | solubilize phosphates and to synthesize IAA, active against other soil borne fungal pathogens | [51] |
| 8. | Stenotrophomonas rhizophila ep-17 | Soybean | Beneficial association with Bradyrhizobium in the rhizosphere and promote plant growth, nutrient uptake and grown soybean under salt stress condition. | [52] |
| 9. | Bacillus subtilis SU47 and Arthrobacter sp. SU18 | Wheat | Showed an increase in dry biomass, total soluble sugars and proline content | [53] |
| 10. | Bacillus pumilus 2-1, Chryseobacterium indologene 2-2, and Acinetobacter johnsonii 3-1 | Sugar beet | Increased photosynthetic capacity, increased concentration of carbohydrates | [54] |
| 11. | Burkholderia phytofirmans strain PsJN | Potato, tomato, Onion, maize, barley | ACC deaminase activity, IAA synthesis | [55] |
| 12. | Pseudomonas syringae | Arabidopsis thaliana | IAA and abscisic acid biosynthesis | [56] |
| 13. | Gluconacetobacter diazotrophicus | Sugarcane | Nitrogen fixation | [57] |
| 14. | Rhizobium leguminosarum bv. trifolii | Rice roots | Biological N2 fixation | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vandana, U.K.; Rajkumari, J.; Singha, L.P.; Satish, L.; Alavilli, H.; Sudheer, P.D.V.N.; Chauhan, S.; Ratnala, R.; Satturu, V.; Mazumder, P.B.; et al. The Endophytic Microbiome as a Hotspot of Synergistic Interactions, with Prospects of Plant Growth Promotion. Biology 2021, 10, 101. https://doi.org/10.3390/biology10020101
Vandana UK, Rajkumari J, Singha LP, Satish L, Alavilli H, Sudheer PDVN, Chauhan S, Ratnala R, Satturu V, Mazumder PB, et al. The Endophytic Microbiome as a Hotspot of Synergistic Interactions, with Prospects of Plant Growth Promotion. Biology. 2021; 10(2):101. https://doi.org/10.3390/biology10020101
Chicago/Turabian StyleVandana, Udaya Kumar, Jina Rajkumari, L. Paikhomba Singha, Lakkakula Satish, Hemasundar Alavilli, Pamidimarri D.V.N. Sudheer, Sushma Chauhan, Rambabu Ratnala, Vanisri Satturu, Pranab Behari Mazumder, and et al. 2021. "The Endophytic Microbiome as a Hotspot of Synergistic Interactions, with Prospects of Plant Growth Promotion" Biology 10, no. 2: 101. https://doi.org/10.3390/biology10020101
APA StyleVandana, U. K., Rajkumari, J., Singha, L. P., Satish, L., Alavilli, H., Sudheer, P. D. V. N., Chauhan, S., Ratnala, R., Satturu, V., Mazumder, P. B., & Pandey, P. (2021). The Endophytic Microbiome as a Hotspot of Synergistic Interactions, with Prospects of Plant Growth Promotion. Biology, 10(2), 101. https://doi.org/10.3390/biology10020101






