Immunoautophagy-Related Long Noncoding RNA (IAR-lncRNA) Signature Predicts Survival in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression Data and Clinicopathological Characteristics
2.2. Development of the Prognostic Immuno-Autophagy-Related lncRNAs Signature
2.3. Prognostic Ability of Immuno-Autophagy-Related lncRNAs Signature
2.4. Correlation between Immune Cells and Signature
2.5. GO and KEGG Analysis
2.6. Statistical Analysis
3. Results
3.1. Correlating Autophagy-Related Genes and Immune-Related Genes with lncRNAs
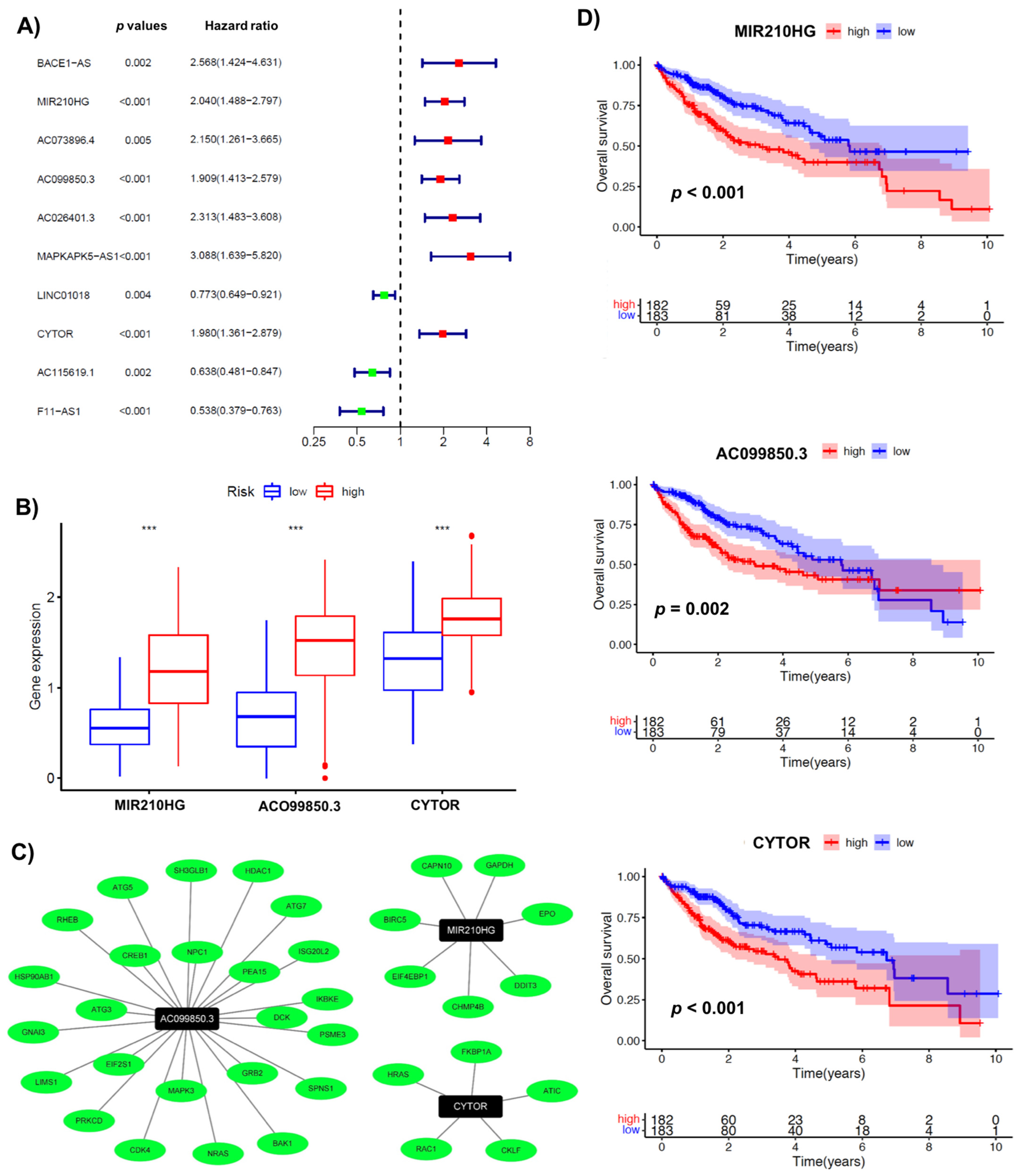
3.2. A Signature Involving 3 Immuno-Autophagy-Related lncRNAs with Prognostic Potential
3.3. Validating the Prognostic Potential of Immuno-Autophagy-Related lncRNA Signature in Low- and High-Risk HCC Groups
3.4. Association of Immuno-Autophagy-Related lncRNA Signature with Clinical Characteristics
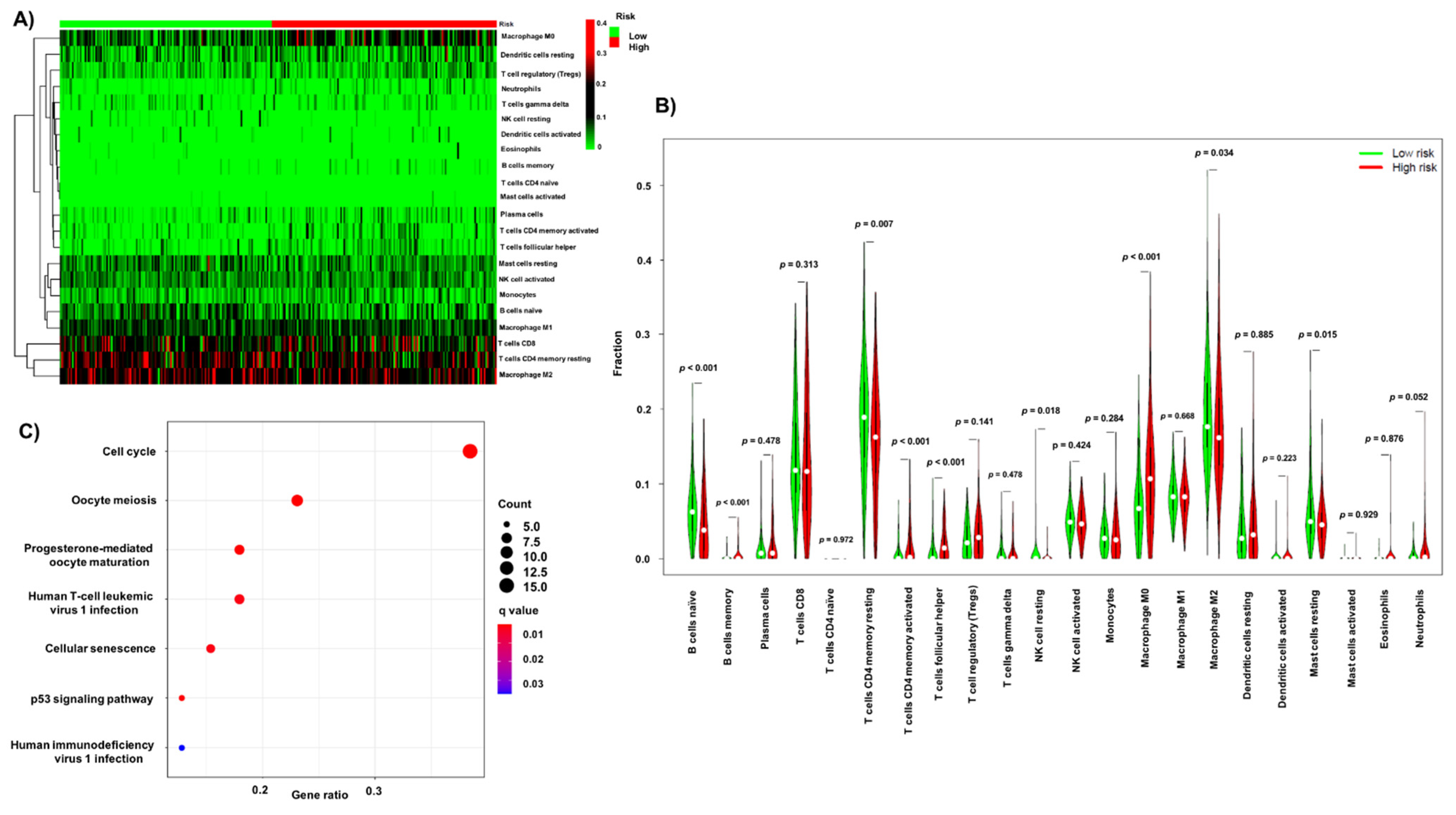
3.5. Association of Infiltrating Immune Cells and Obtained Signature
3.6. GO and KEGG Pathway Enrichment Analysis of the Obtained Signature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chavez-Dominguez, R.; Perez-Medina, M.; Lopez-Gonzalez, J.S.; Galicia-Velasco, M.; Aguilar-Cazares, D. The Double-Edge Sword of Autophagy in Cancer: From Tumor Suppression to Pro-tumor Activity. Front. Oncol. 2020, 10, 578418. [Google Scholar] [CrossRef] [PubMed]
- Nixon, A.R. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef]
- Peng, Y.-F.; Shi, Y.-H.; Shen, Y.-H.; Ding, Z.-B.; Ke, A.-W.; Zhou, J.; Qiu, S.-J.; Fan, J. Promoting Colonization in Metastatic HCC Cells by Modulation of Autophagy. PLoS ONE 2013, 8, e74407. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, S.; Ward, J.W.; Watson, M.; Momin, B.; Richardson, L.C. Hepatocellular carcinoma–United States, 2001–2006. MMWR Morb. Mortal. Wkly. Rep. 2010, 59, 517–520. [Google Scholar]
- Liu, M.; Jiang, L.; Guan, X.-Y. The genetic and epigenetic alterations in human hepatocellular carcinoma: A recent update. Protein Cell 2014, 5, 673–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Li, H.; Luo, K.; Sharma, A.; Sun, X. Prognostic gene expression signature revealed the involvement of mutational pathways in cancer genome. J. Cancer 2020, 11, 4510–4520. [Google Scholar] [CrossRef]
- Sharma, A.; Biswas, A.; Liu, H.; Sen, S.; Paruchuri, A.; Katsonis, P.; Lichtarge, O.; Dakal, T.C.; Maulik, U.; Gromiha, M.M.; et al. Mutational Landscape of the BAP1 Locus Reveals an Intrinsic Control to Regulate the miRNA Network and the Binding of Protein Complexes in Uveal Melanoma. Cancers 2019, 11, 1600. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-Y.; Lan, S.-H.; Wu, S.-R.; Chiu, Y.-C.; Lin, X.-Z.; Su, I.-J.; Tsai, T.-F.; Yen, C.-J.; Lu, T.-H.; Liang, F.-W.; et al. Hepatocellular carcinoma-related cyclin D1 is selectively regulated by autophagy degradation system. Hepatology 2018, 68, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Lan, S.; Wu, S.; Zuchini, R.; Lin, X.; Su, I.; Tsai, T.; Lin, Y.; Wu, C.; Liu, H. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology 2014, 59, 505–517. [Google Scholar] [CrossRef] [Green Version]
- Chu, Y.-L.; Ho, C.-T.; Chung, J.-G.; Rajasekaran, R.; Sheen, L.-Y. Allicin Induces p53-Mediated Autophagy in Hep G2 Human Liver Cancer Cells. J. Agric. Food Chem. 2012, 60, 8363–8371. [Google Scholar] [CrossRef]
- Liu, X.-W.; Cai, T.-Y.; Zhu, H.; Cao, J.; Su, Y.; Hu, Y.-Z.; He, Q.-J.; Yang, B. Q6, a novel hypoxia-targeted drug, regulates hypoxia-inducible factor signaling via an autophagy-dependent mechanism in hepatocellular carcinoma. Autophagy 2013, 10, 111–122. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, S.-L.; Chen, C.-T.; Wang, J.-J.; Kuo, Y.-H.; Li, C.-C.; Wu, C.-C. Sedanolide induces autophagy through the PI3K, p53 and NF-?B signaling pathways in human liver cancer cells. Int. J. Oncol. 2015, 47, 2240–2246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.-H.; Ding, Z.-B.; Zhou, J.; Qiu, S.-J.; Fan, J. Prognostic significance of Beclin 1-dependent apoptotic activity in hepatocellular carcinoma. Autophagy 2009, 5, 380–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.-B.; Shi, Y.-H.; Zhou, J.; Qiu, S.-J.; Xu, Y.; Dai, Z.; Shi, G.-M.; Wang, X.-Y.; Ke, A.-W.; Wu, B.; et al. Association of Autophagy Defect with a Malignant Phenotype and Poor Prognosis of Hepatocellular Carcinoma. Cancer Res. 2008, 68, 9167–9175. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Liu, F.; Han, S.; Qi, Y.; Hu, X.; Zhou, C.; Liang, H.; Zhang, Z. Autophagy-Related Gene Pairs Signature for the Prognosis of Hepatocellular Carcinoma. Front. Mol. Biosci. 2021, 8, 670241. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, G.; Yu, Y.; Li, S. A Prognostic Autophagy-Related Gene Pair Signature and Small-Molecule Drugs for Hepatocellular Carcinoma. Front. Genet. 2021, 12, 689801. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Lu, Z.; Li, R.; Shao, W.; Zheng, Y.; Shi, X.; Li, Y.; Song, J. Construction of a Prognostic Model for Hepatocellular Carcinoma Based on Immunoautophagy-Related Genes and Tumor Microenvironment. Int. J. Gen. Med. 2021, 14, 5461–5473. [Google Scholar] [CrossRef]
- Xin, X.; Wu, M.; Meng, Q.; Wang, C.; Lu, Y.; Yang, Y.; Li, X.; Zheng, Q.; Pu, H.; Gui, X.; et al. Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer 2018, 17, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ying, L.; Huang, Y.; Chen, H.; Wang, Y.; Xia, L.; Chen, Y.; Liu, Y.; Qiu, F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. BioSyst. 2012, 9, 407–411. [Google Scholar] [CrossRef]
- Xiong, H.; Ni, Z.; He, J.; Jiang, S.; Li, X.; Gong, W.; Zheng, L.; Chen, S.; Li, B.; Zhang, N.; et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017, 36, 3528–3540. [Google Scholar] [CrossRef]
- Li, W.; Dong, X.; He, C.; Tan, G.; Li, Z.; Zhai, B.; Feng, J.; Jiang, X.; Liu, C.; Jiang, H.; et al. LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 2019, 38, 1–13. [Google Scholar] [CrossRef]
- Yang, S.; Zhou, Y.; Zhang, X.; Wang, L.; Fu, J.; Zhao, X.; Yang, L. The prognostic value of an autophagy-related lncRNA signature in hepatocellular carcinoma. BMC Bioinform. 2021, 22, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Liu, H.; Herwig-Carl, M.C.; Dakal, T.C.; Schmidt-Wolf, I.G.H. Epigenetic Regulatory Enzymes: Mutation Prevalence and Coexistence in Cancers. Cancer Investig. 2021, 39, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.; Zhang, Z.; Zhao, X.; Dong, X. Autophagy-related genes prognosis signature as potential predictive markers for immunotherapy in hepatocellular carcinoma. PeerJ 2020, 8, e8383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Xia, S.; Shi, X.; Chen, X.; Shang, D. The novel immune-related genes predict the prognosis of patients with hepatocellular carcinoma. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Dai, Y.; Qiang, W.; Lin, K.; Gui, Y.; Lan, X.; Wang, D. An immune-related gene signature for predicting survival and immunotherapy efficacy in hepatocellular carcinoma. Cancer Immunol. Immunother. 2020, 70, 967–979. [Google Scholar] [CrossRef]
- Deng, X.; Bi, Q.; Chen, S.; Chen, X.; Li, S.; Zhong, Z.; Guo, W.; Li, X.; Deng, Y.; Yang, Y. Identification of a Five-Autophagy-Related-lncRNA Signature as a Novel Prognostic Biomarker for Hepatocellular Carcinoma. Front. Mol. Biosci. 2021, 7. [Google Scholar] [CrossRef]
- Xu, Q.; Wang, Y.; Huang, W. Identification of immune-related lncRNA signature for predicting immune checkpoint blockade and prognosis in hepatocellular carcinoma. Int. Immunopharmacol. 2021, 92, 107333. [Google Scholar] [CrossRef]
- Wang, Y.; Li, W.; Chen, X.; Li, Y.; Wen, P.; Xu, F. MIR210HG predicts poor prognosis and functions as an oncogenic lncRNA in hepatocellular carcinoma. Biomed. Pharmacother. 2019, 111, 1297–1301. [Google Scholar] [CrossRef]
- Hu, B.; Yang, X.-B.; Yang, X.; Sang, X.-T. LncRNA CYTOR affects the proliferation, cell cycle and apoptosis of hepatocellular carcinoma cells by regulating the miR-125b-5p/KIAA1522 axis. Aging 2020, 13, 2626–2639. [Google Scholar] [CrossRef]
- Tian, Q.; Yan, X.; Yang, L.; Liu, Z.; Yuan, Z.; Zhang, Y. lncRNA CYTOR promotes cell proliferation and tumor growth via miR-125b/SEMA4C axis in hepatocellular carcinoma. Oncol. Lett. 2021, 22, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Wei, H.; Liu, G.; Zhang, Y. Bioinformatics Profiling of Five Immune-Related lncRNAs for a Prognostic Model of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 667904. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liang, M.; Lan, T.; Wu, X.; Xie, W.; Wang, T.; Chen, Z.; Shen, S.; Peng, B. Four Immune-Related Long Non-coding RNAs for Prognosis Prediction in Patients with Hepatocellular Carcinoma. Front. Mol. Biosci. 2020, 7, 566491. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Reutter, H.; Ellinger, J. DNA Methylation and Bladder Cancer: Where Genotype does not Predict Phenotype. Curr. Genom. 2020, 21, 34–36. [Google Scholar] [CrossRef]

| Risk | Total | High Risk | Low Risk | t | p Value | |
|---|---|---|---|---|---|---|
| Age | <65 | 95 (58.28%) | 46 (63.89%) | 49 (53.85%) | 1.280 | 0.258 |
| ≥65 | 68 (41.72%) | 26 (36.11%) | 42 (46.15%) | |||
| Gender | Female | 50 (30.67%) | 24 (33.33%) | 26 (28.57%) | 0.234 | 0.629 |
| Male | 113 (69.33%) | 48 (66.67%) | 65 (71.43%) | |||
| Child–Pugh | A | 147 (90.18%) | 64 (88.89%) | 83 (91.21%) | 0.053 | 0.819 |
| B + C | 16 (9.82%) | 8 (11.11%) | 8 (8.79%) | |||
| AFP | ≥400 | 30 (18.4%) | 17 (23.61%) | 13 (14.29%) | 1.748 | 0.186 |
| <400 | 133 (81.6%) | 55 (76.39%) | 78 (85.71%) | |||
| Fibrosis | Fibrosis | 113 (69.33%) | 50 (69.44%) | 63 (69.23%) | 0 | 1 |
| No Fibrosis | 50 (30.67%) | 22 (30.56%) | 28 (30.77%) | |||
| Grade | G1–G2 | 99 (60.74%) | 33 (45.83%) | 66 (72.53%) | 10.918 | 0.001 ** |
| G3–G4 | 64 (39.26%) | 39 (54.17%) | 25 (27.47%) | |||
| Stage | Stage I–II | 131 (80.37%) | 57 (79.17%) | 74 (81.32%) | 0.021 | 0.885 |
| Stage III–IV | 32 (19.63%) | 15 (20.83%) | 17 (18.68%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Ge, F.; Sharma, A.; Rudan, O.; Setiawan, M.F.; Gonzalez-Carmona, M.A.; Kornek, M.T.; Strassburg, C.P.; Schmid, M.; Schmidt-Wolf, I.G.H. Immunoautophagy-Related Long Noncoding RNA (IAR-lncRNA) Signature Predicts Survival in Hepatocellular Carcinoma. Biology 2021, 10, 1301. https://doi.org/10.3390/biology10121301
Wang Y, Ge F, Sharma A, Rudan O, Setiawan MF, Gonzalez-Carmona MA, Kornek MT, Strassburg CP, Schmid M, Schmidt-Wolf IGH. Immunoautophagy-Related Long Noncoding RNA (IAR-lncRNA) Signature Predicts Survival in Hepatocellular Carcinoma. Biology. 2021; 10(12):1301. https://doi.org/10.3390/biology10121301
Chicago/Turabian StyleWang, Yulu, Fangfang Ge, Amit Sharma, Oliver Rudan, Maria F. Setiawan, Maria A. Gonzalez-Carmona, Miroslaw T. Kornek, Christian P. Strassburg, Matthias Schmid, and Ingo G. H. Schmidt-Wolf. 2021. "Immunoautophagy-Related Long Noncoding RNA (IAR-lncRNA) Signature Predicts Survival in Hepatocellular Carcinoma" Biology 10, no. 12: 1301. https://doi.org/10.3390/biology10121301
APA StyleWang, Y., Ge, F., Sharma, A., Rudan, O., Setiawan, M. F., Gonzalez-Carmona, M. A., Kornek, M. T., Strassburg, C. P., Schmid, M., & Schmidt-Wolf, I. G. H. (2021). Immunoautophagy-Related Long Noncoding RNA (IAR-lncRNA) Signature Predicts Survival in Hepatocellular Carcinoma. Biology, 10(12), 1301. https://doi.org/10.3390/biology10121301









