Comparative Transcriptomic Analysis of Regenerated Skins Provides Insights into Cutaneous Air-Breathing Formation in Fish
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish
2.2. Cutaneous Air-Breathing Confirmation in Loach
2.3. Cutaneous Incision and Morphological Observations of Regenerated Skins
2.4. Cutaneous Incision and Histological Observations of Regenerated Skins
2.5. RNA Isolation and cDNA Library Constructions
2.6. Sequencing, De Novo Assembly and Functional Annotation
2.7. Differentially Expressed Gene (DEG) Analysis and Enrichment Analysis
2.8. Mining of DEGs Related to Cutaneous Air-Breathing
2.9. Validation of Transcriptome Data by qPCR
2.10. Statistical Analysis
3. Results
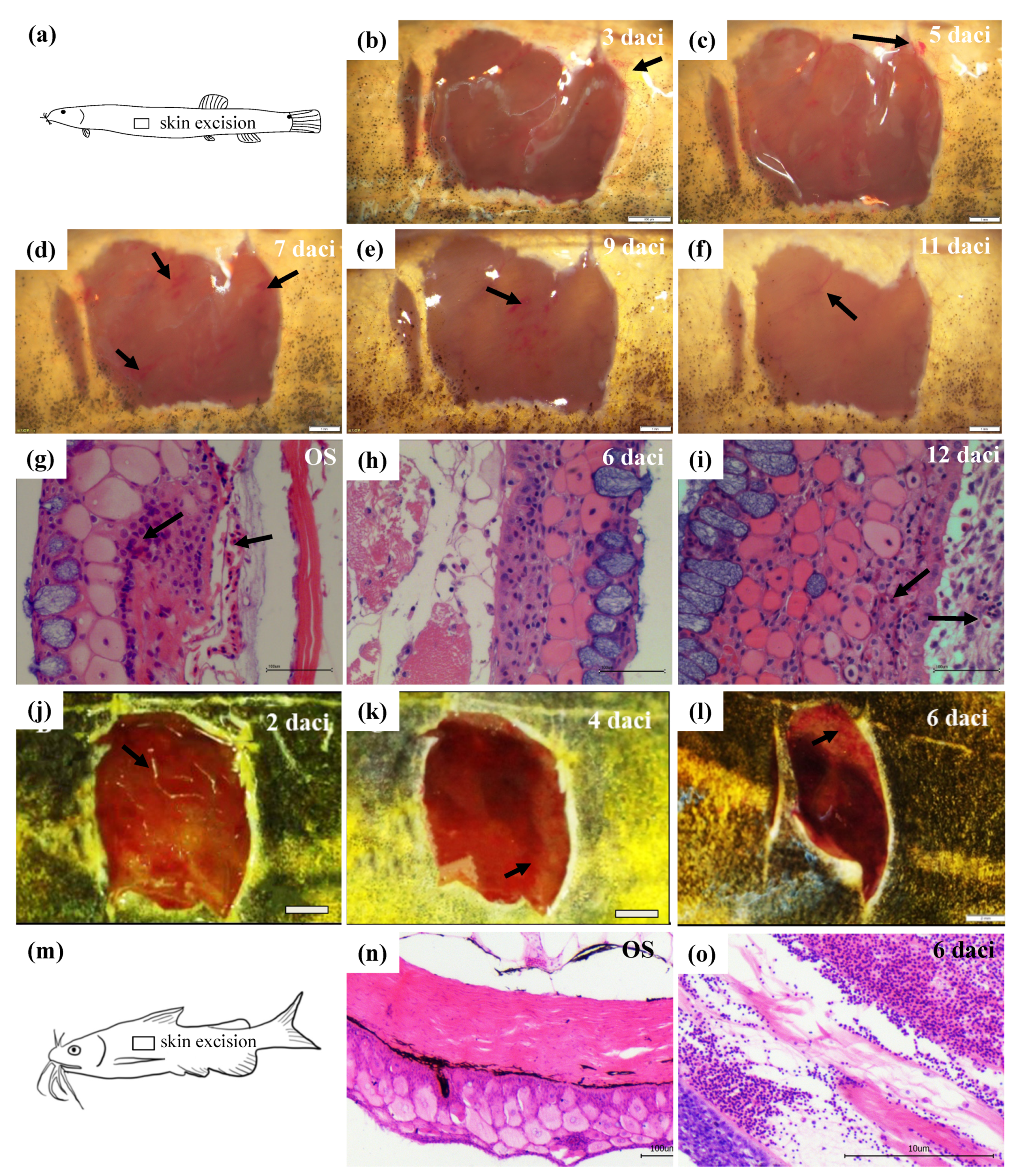
3.1. Morphological and Histological Observations of Skin Regeneration
3.2. De Novo Assembly and Functional Annotation
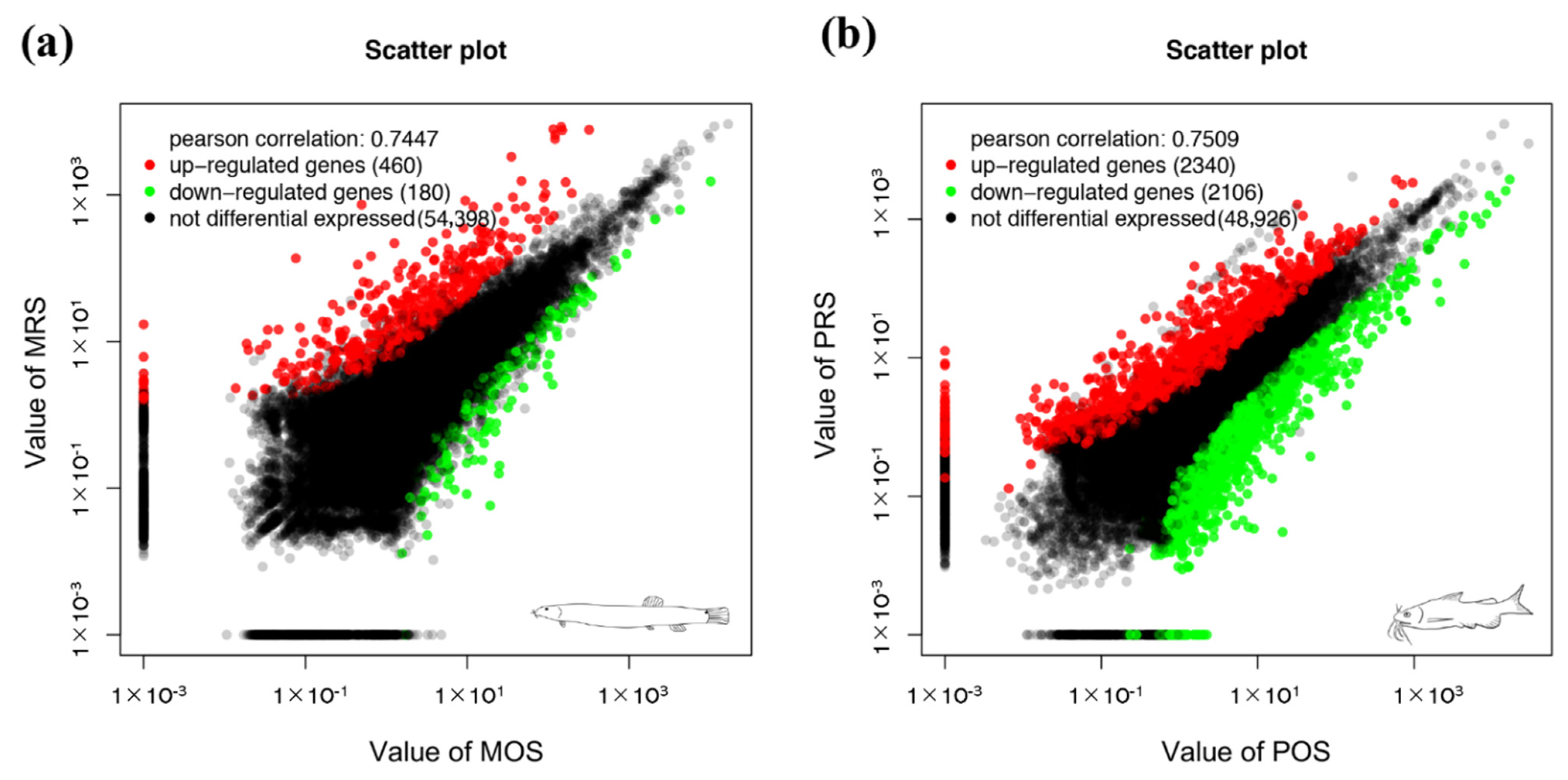
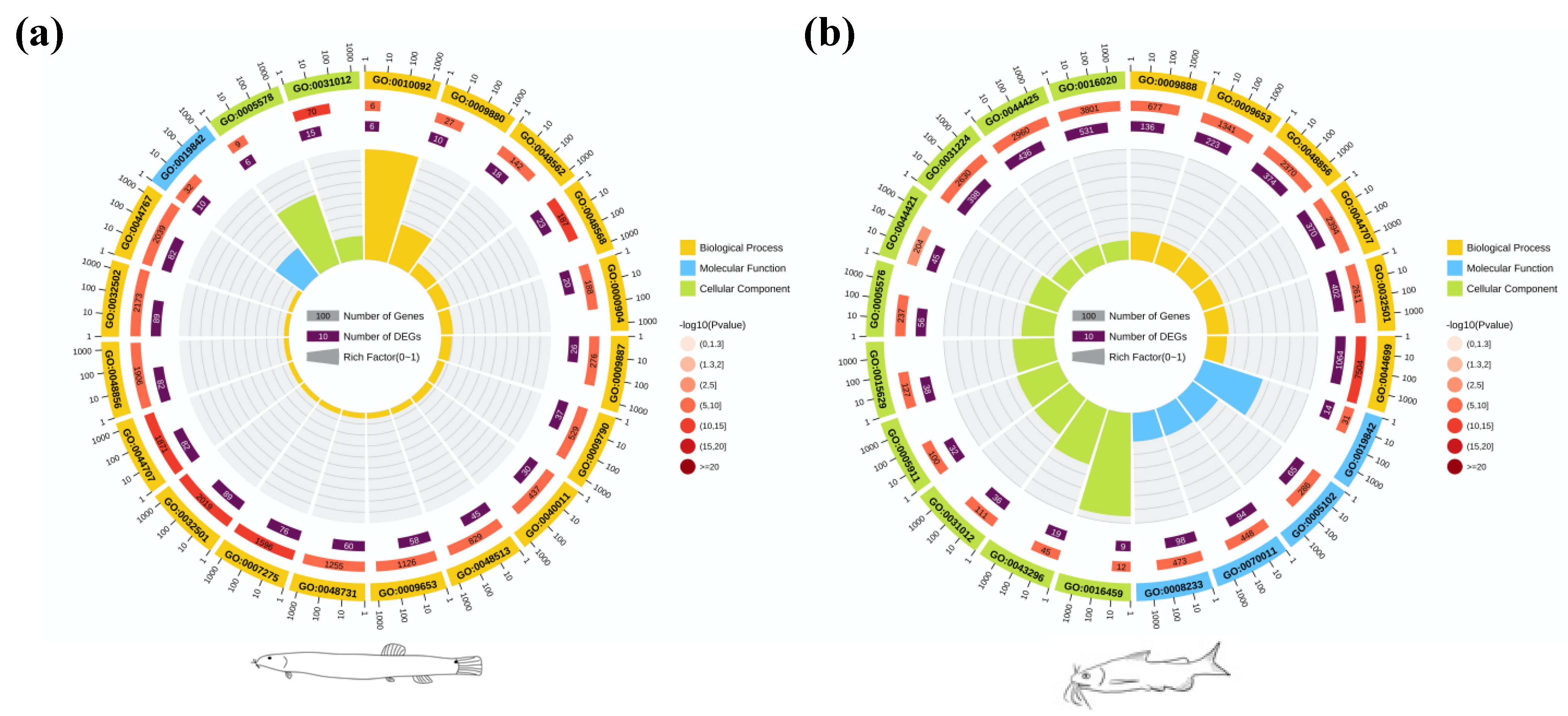
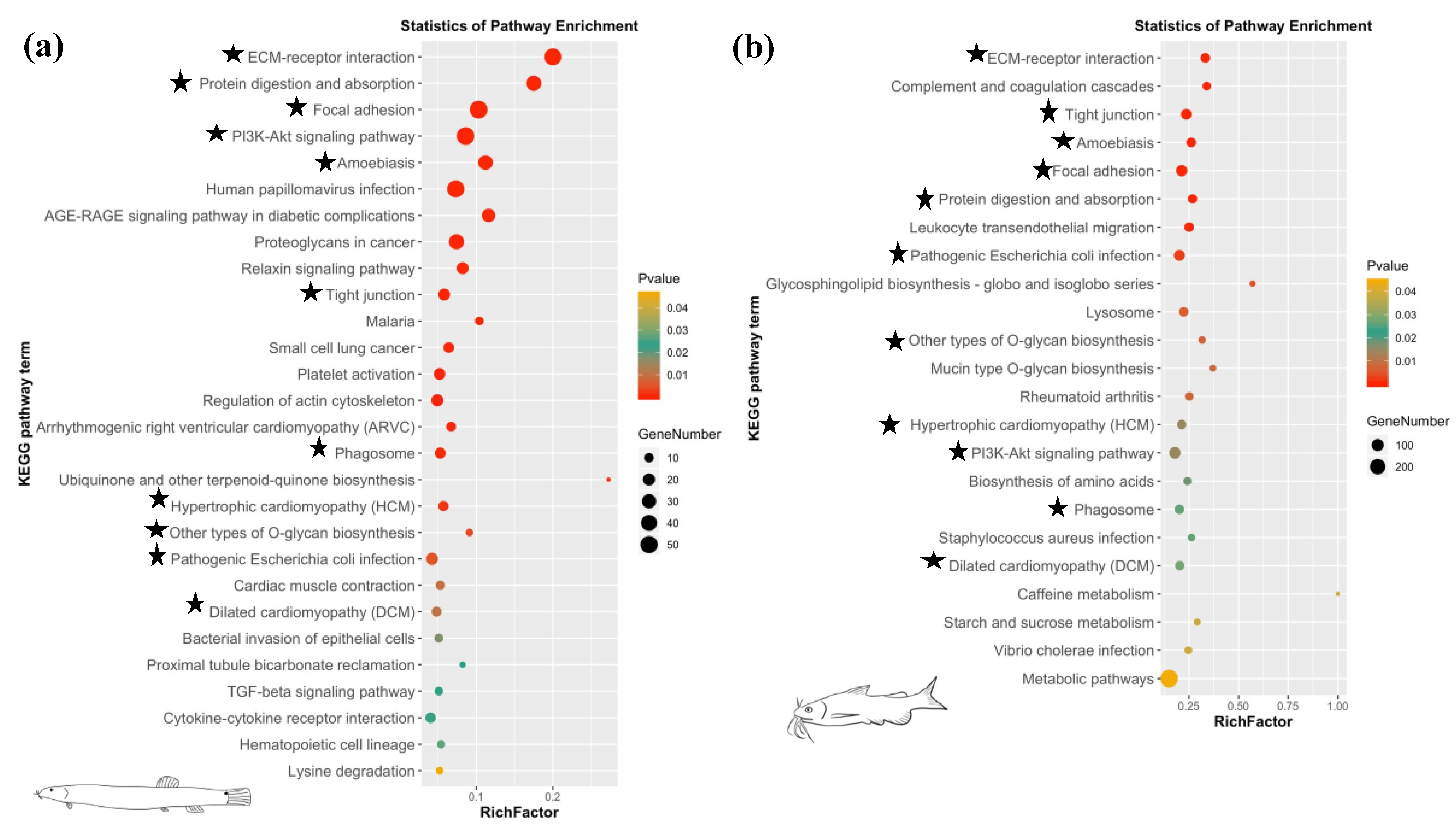
3.3. DEG Analysis and Enrichment Analysis
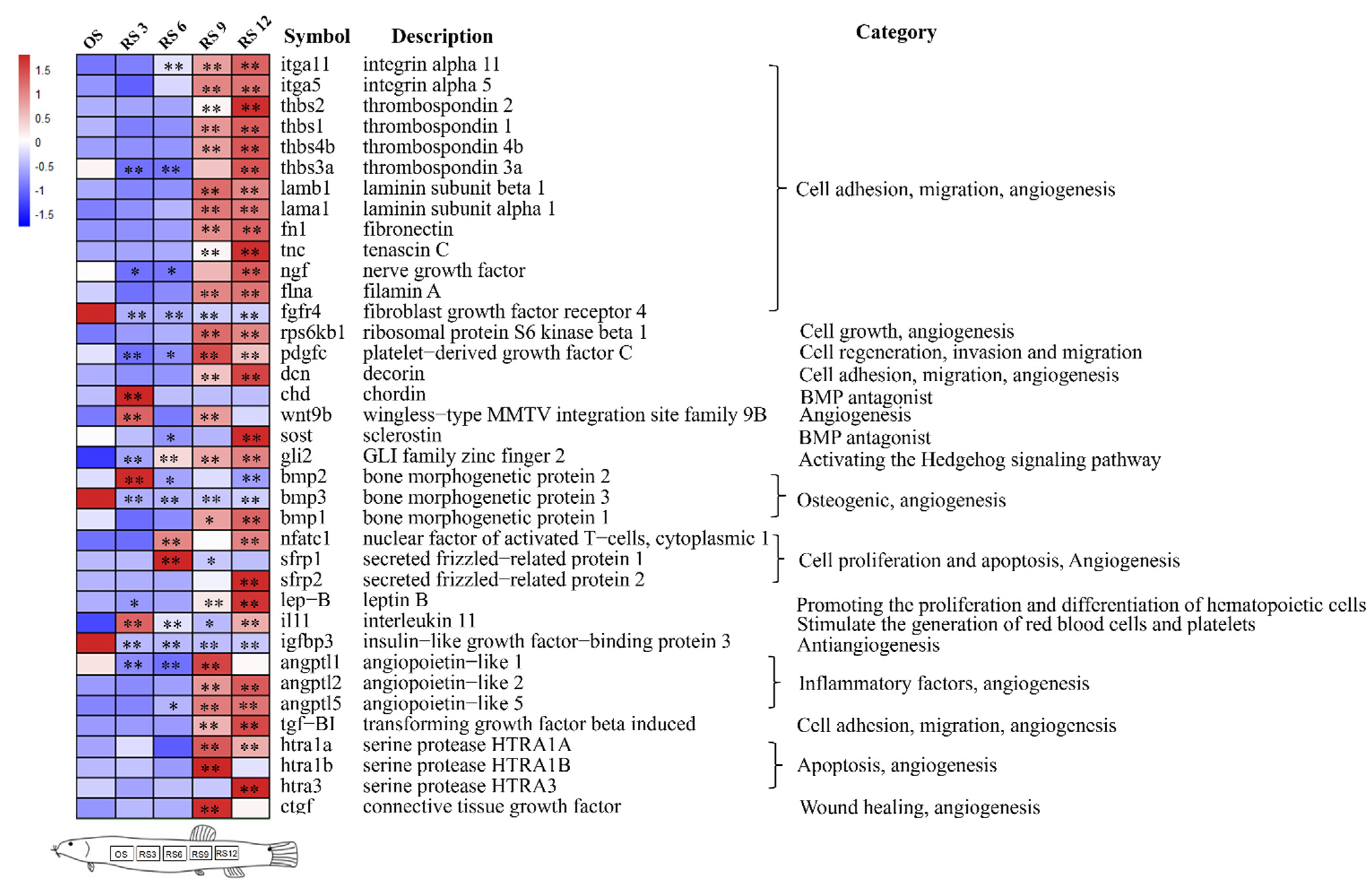
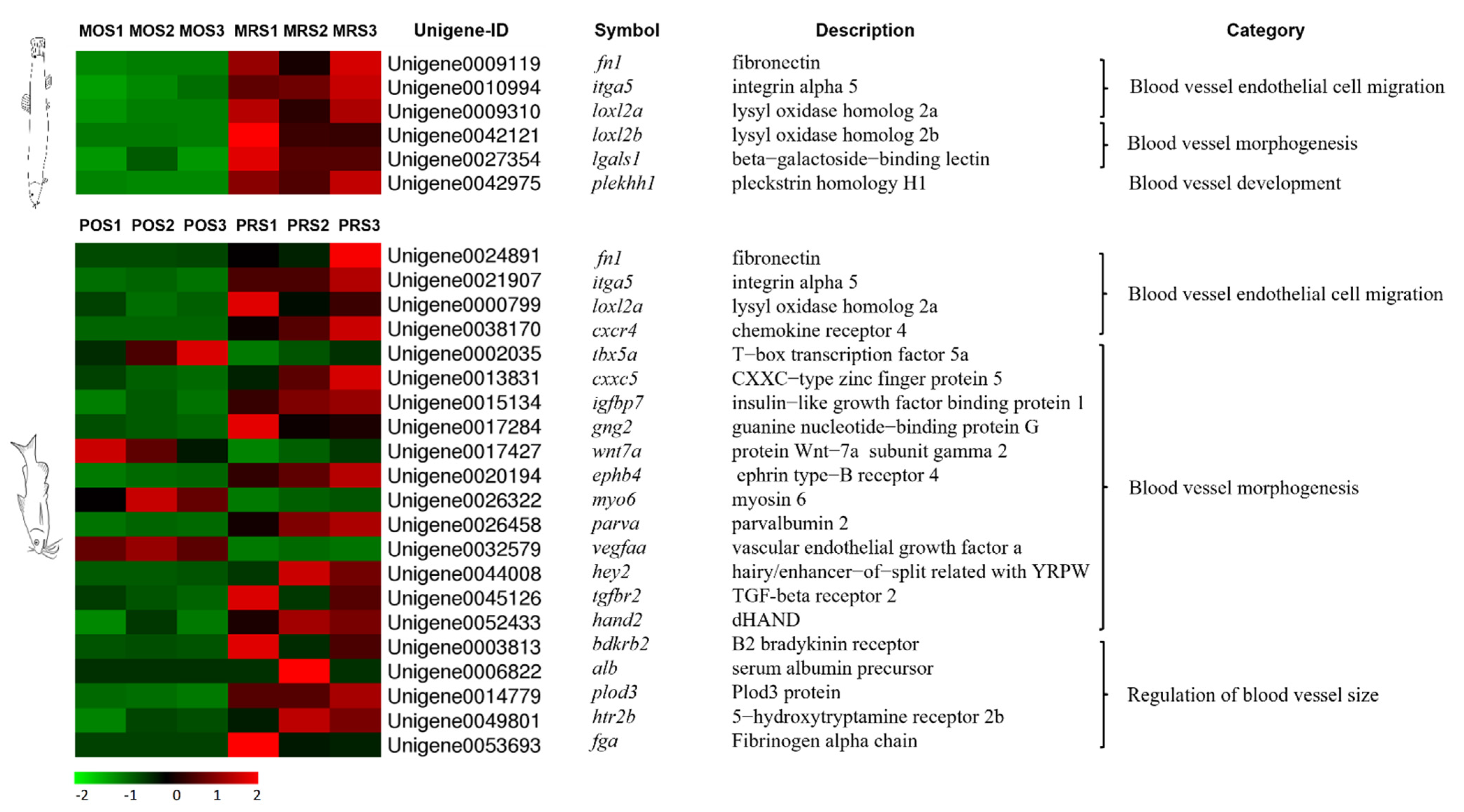
3.4. Mining of Cutaneous Air-Breathing Related Genes in Loach
3.5. Validation of DEGs by qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lefevre, S.; Bayley, M.; McKenzie, D.J.; Craig, J.F. Air-breathing fishes. J. Fish Biol. 2014, 84, 547–553. [Google Scholar] [CrossRef]
- Damsgaard, C.; Baliga, V.B.; Bates, E.; Burggren, W.; McKenzie, D.J.; Taylor, E.; Wright, P.A. Evolutionary and cardiorespiratory physiology of air-breathing and amphibious fishes. Acta Physiol. 2020, 228, e13406. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D. Air-breathing organs and Nervous control of respiration in Freshwater fishes. Res. J. Sci. Technol. 2020, 12, 143. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, I.S.; Kim, S.Y. Structure and mucous histochemistry of the intestinal respiratory tract of the mud loach, Misgurnus anguillicaudatus (Cantor). J. Appl. Ichthyol. 2003, 19, 215–219. [Google Scholar] [CrossRef]
- Singh, J.P.N.; Yadava, R.Y. Histopathological alterations in labyrinthine organ of an air breathing climbing perch Anabas testudineus exposed to 2,4 dichlorophenoxyacetic acid. J. Exp. Zool. India 2011, 14, 69–76. [Google Scholar]
- Lefevre, S.; Wang, T.; Jensen, A.; Cong, N.V.; Huong, D.T.T.; Phuong, N.T.; Bayley, M. Air-breathing fishes in aquaculture. What can we learn from physiology? J. Fish Biol. 2014, 84, 705–731. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Feng, S.; Xu, J.; Zhang, S.; Li, S.; Sun, X.; Xu, P. Comparative transcriptome analysis between aquatic and aerial breathing organs of Channa argus to reveal the genetic basis underlying bimodal respiration. Mar. Genom. 2016, 29, 89–96. [Google Scholar] [CrossRef]
- Xu, J.; Bian, C.; Chen, K.; Liu, G.; Jiang, Y.; Luo, Q.; You, X.; Peng, W.; Li, J.; Huang, Y. Draft genome of the Northern snake-head, Channa argus. Gigascience 2017, 6, gix011. [Google Scholar] [CrossRef]
- Li, N.; Bao, L.; Zhou, T.; Yuan, Z.; Liu, S.; Dunham, R.; Li, Y.; Wang, K.; Xu, X.; Jin, Y.; et al. Genome sequence of walking catfish (Clarias batrachus) provides insights into terrestrial adaptation. BMC Genom. 2018, 19, 952. [Google Scholar] [CrossRef]
- Luo, W.W.; Cao, X.J.; Xu, X.W.; Huang, S.Q.; Liu, C.S.; Tomljanovic, T. Developmental transcriptome analysis and identifica-tion of genes involved in formation of intestinal air-breathing function of Dojo loach, Misgurnus anguillicaudatus. Sci. Rep. 2016, 6, 31845. [Google Scholar] [CrossRef]
- Huang, S.Q.; Cao, X.J.; Tian, X.C. Transcriptome analysis of compromise between air-breathing and nutrient uptake of posterior intestine in loach (Misgurnus anguillicaudatus), an air-breathing fish. Mar. Biotechnol. 2016, 18, 521–533. [Google Scholar] [CrossRef]
- Huang, S.; Cao, X.; Tian, X.; Wang, W. High-throughput sequencing identifies microRNAs from posterior intestine of loach (Misgurnus anguillicaudatus) and their response to intestinal air-breathing inhibition. PLoS ONE 2016, 11, e0149123. [Google Scholar] [CrossRef]
- Otrock, Z.K.; Mahfouz, R.A.R.; Makarem, J.A.; Shamseddine, A.I. Understanding the biology of angiogenesis: Review of the most important molecular mechanisms. Blood Cells Mol. Dis. 2007, 39, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Patel, D.; Khanna, S.; Gordillo, G.M.; Biswas, S.; Friedman, A.; Sen, C.K. Transcriptome-wide analysis of blood vessels laser captured from human skin and chronic wound-edge tissue. Proc. Natl. Acad. Sci. USA 2007, 104, 14472–14477. [Google Scholar] [CrossRef] [PubMed]
- Helbo, S.; Weber, R.E.; Fago, A. Expression patterns and adaptive functional diversity of vertebrate myoglobins. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2013, 1834, 1832–1839. [Google Scholar] [CrossRef]
- Feng, B.; Yi, S.V.; Li, R.W.; Zhou, X.Y. Comparison of age and growth performance of diploid and tetraploid loach Misgurnus anguillicaudatus in the Yangtze River basin, China. Environ. Biol. Fish. 2017, 100, 815–828. [Google Scholar] [CrossRef]
- Zhang, H.P.; Chen, M.Y.; Xu, Y.X.; Xu, G.Y.; Chen, J.R.; Wang, Y.M.; Kang, Y.H.; Shan, X.F.; Kong, L.C.; Ma, H.X. An effective live attenuated vaccine against Aeromonas veronii infection in the loach (Misgurnus anguillicaudatus). Fish Shellfish. Immunol. 2020, 104, 269–278. [Google Scholar] [CrossRef]
- Gonçalves, A.F.; Castro, L.F.; Pereira-Wilson, C.; Coimbra, J.; Wilson, J. Is there a compromise between nutrient uptake and gas exchange in the gut of Misgurnus anguillicaudatus, an intestinal air-breathing fish? Comp. Biochem. Physiol. Part D Genom. Proteom. 2007, 2, 345–355. [Google Scholar] [CrossRef]
- Seo, E.; Yoon, G.Y.; Kim, H.N.; Lim, J.H.; Kim, S.; Kim, B.; Kim, K.H.; Lee, S.J. Morphological features of mucous secretory organ and mucous secretion of loach Misgurnus anguillicaudatus skin for friction drag reduction. J. Fish Biol. 2020, 96, 83–91. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, I.S. Structure and histochemistry of skin of mud loach, Misgurnus anguillicaudatus (Pisces, Cobitidae), from Korea. Korean L. Ichthyol. 1999, 11, 109–116. [Google Scholar]
- Park, J.; Kim, I.; Kim, S. Morphology and histochemistry of the skin of the mud loach, misgurnus mizolepis, in relation to cutaeneous respiration. Korean J. Biol. Sci. 2001, 5, 303–308. [Google Scholar] [CrossRef][Green Version]
- Yokoya, S.; Tamura, O.S. Fine structure of the skin of the amphibious fishes, Boleophthalmus pectinirostris and Periophthalmus cantonensis, with special reference to the location of blood vessels. J. Morphol. 1992, 214, 287–297. [Google Scholar] [CrossRef]
- Wang, C.; Xie, S.; Zhu, X.; Lei, W.; Yang, Y.; Liu, J. Effects of age and dietary protein level on digestive enzyme activity and gene expression of Pelteobagrus fulvidraco larvae. Aquaculture 2006, 254, 554–562. [Google Scholar] [CrossRef]
- Cao, X.J.; Wang, W.M. Histology and Mucin Histochemistry of The Digestive Tract of Yellow Catfish, Pelteobagrus fulvidraco. Anat. Histol. Embryol. 2009, 38, 254–261. [Google Scholar] [CrossRef]
- Leclair, E.E.; Jacek, T.; Bruce, R. Development and regeneration of the zebrafish maxillary barbel: A novel study system for vertebrate tissue growth and repair. PLoS ONE 2010, 5, e8737. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.; Chen, Y.W.; He, F.C. Integrated nr database in protein annotation system and its localization. Comput. Een. 2006, 32, 71–72. [Google Scholar]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.Z.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, 115–119. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Ali, M.; Kwan, G.; Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome. Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Beon, M.S.; Oh, M.K.; Lee, Y.J.; Kim, C.H.; Park, J.Y. A comparative study on vascularization and the structure of the epidermis of an amphibious mudskipper fish, Scartelaos gigas (Gobiidae, Teleostei), on different parts of the body and the appendages. J. Appl. Ichthyol. 2013, 29, 6. [Google Scholar] [CrossRef]
- Singer, A.J.; Clark, R.A. Cutaneous wound healing. N. Engl. J. Med. 1999, 341, 738–746. [Google Scholar] [CrossRef]
- Sheng, L.; Yang, M.; Liang, Y.; Li, Q. Adipose tissue-derived stem cells (ADSCs) transplantation promotes regeneration of expanded skin using a tissue expansion model. Wound Repair Regen. 2013, 21, 746–754. [Google Scholar] [CrossRef] [PubMed]
- André, L.C.; Prado, T.M.; Laísa, P.R.; Wilfried, K. The potential respiratory surfaces of a fish living in a historically polluted river. Anim. Biol. 2019, 70, 101–108. [Google Scholar]
- Zhang, J.; Taniguchi, T.; Takita, T.; Ali, A.B. A study on the epidermal structure of Periophthalmodon and Periophthalmus mudskippers with reference to their terrestrial adaptation. Ichthyol. Res. 2003, 50, 310–317. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, I.S.; Lee, Y.J. A study on the vascularization and structure of the epidermis of the air-breathing mudskipper, Periophthalmus magnuspinnatus (Gobiidae, Teleostei), along different parts of the body. J. Appl. Ichthyol. 2006, 22, 62–67. [Google Scholar] [CrossRef]
- Silvestre, J.S.; Théry, C.; Hamard, G.; Boddaert, J.; Aguilar, B.; Delcayre, A.; Houbron, C.; Tamarat, R.; Blanc-Brude, O.; Heeneman, S.; et al. Lactadherin promotes VEGF-dependent neovascularization. Nat. Med. 2005, 11, 499–506. [Google Scholar] [CrossRef]
- Wang, J.; Yang, L.; Liang, F.; Chen, Y.; Yang, G. Integrin alpha x stimulates cancer angiogenesis through PI3K/Akt signaling mediated VEGFR2/VEGF-A overexpression in blood vessel endothelial cells. J. Cell. Biochem. 2019, 120, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Herndon, M.E.; Lawler, J. The cell biology of thrombospondin-1. Matrix. Biol. 2000, 19, 597–614. [Google Scholar] [CrossRef]
- Koch, W.; Hoppmann, P.; De Waha, A.; Schömig, A.; Kastrati, A. Polymorphisms in thrombospondin genes and myocardial infarction: A case-control study and a meta-analysis of available evidence. Hum. Mol. Genet. 2008, 17, 1120–1126. [Google Scholar] [CrossRef] [PubMed]
- Hauser, N.; Paulsson, M.; Kale, A.A.; Dicesare, P.E. Tendon extracellular matrix contains pentameric thrombospondin-4 (TSP4). FEBS Lett. 1995, 368, 307–310. [Google Scholar] [CrossRef]
- Zhang, J.; Ito, R.; Oue, N.; Zhu, X.; Kitadai, Y.; Yoshida, K.; Nakayama, H.; Yasui, W. Expression of thrombospondin-1 is correlated with microvessel density in gastric carcinoma. Virchows Arch. 2003, 442, 563–568. [Google Scholar] [CrossRef]
- Hsiao, C.T.; Cheng, H.W.; Huang, C.M.; Li, H.R.; Ou, M.H.; Huang, J.R.; Khoo, K.H.; Yu, H.W.; Chen, Y.; Wang, Y.K.; et al. Fibronectin in cell adhesion and migration via N-glycosylation. Oncotarget 2017, 8, 70653–70668. [Google Scholar] [CrossRef]
- Le, A.T.; Stainier, D. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev. Cell 2004, 6, 371–382. [Google Scholar] [CrossRef]
- Wijelath, E.S.; Murray, J.; Rahman, S.; Patel, Y.; Ishida, A.; Strand, K.; Aziz, S.; Cardona, C.; Hammond, W.P.; Savidge, G.F.; et al. Novel vascular endothelial growth factor binding domains of fibronectin enhance vascular endothelial growth factor biological activity. Circ. Res. 2002, 91, 25–31. [Google Scholar] [CrossRef] [PubMed]
- George, E.L.; Baldwin, H.S.; Hynes, R.O. Fibronectins are essential for heart and blood vessel morphogenesis but are dispensable for initial specification of precursor cells. Blood 1997, 90, 3073–3081. [Google Scholar] [CrossRef]
- de Vinuesa, A.G.; Abdelilah-Seyfried, S.; Knaus, P.; Zwijsen, A.; Bailly, S. BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev. 2016, 27, 65–79. [Google Scholar] [CrossRef]
- Benn, A.; Hiepen, C.; Osterland, M.; Schütte, C.; Zwijsen, A.; Knaus, P. Role of bone morphogenetic proteins in sprouting angiogenesis: Differential BMP receptor-dependent signaling pathways balance stalk vs. tip cell competence. FASEB J. 2017, 31, 4720–4733. [Google Scholar] [CrossRef]
- Garbers, C.; Hermanns, H.M.; Schaper, F.; Müller-Newen, G.; Grötzinger, J.; Rose-John, S.; Scheller, J. Plasticity and cross-talk of Interleukin 6-type cytokines. Cytokine Growth Factor Rev. 2012, 23, 85–97. [Google Scholar] [CrossRef]
- van Loon, K.; Huijbers, E.J.M.; Griffioen, A.W. Secreted frizzled-related protein 2: A key player in noncanonical Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev. 2021, 40, 191–203. [Google Scholar] [CrossRef]
- Wu, L.; Zhu, Y. The function and mechanisms of action of LOXL2 in cancer. Int. J. Mol. Med. 2015, 36, 1200–1204. [Google Scholar] [CrossRef]
- Ferrara, N.; Kerbel, R.S. Angiogenesis as a therapeutic target. Nat. Cell Biol. 2005, 438, 967–974. [Google Scholar] [CrossRef]
- Kerbel, R.S. Tumor angiogenesis. N. Engl. J. Med. 2008, 358, 2039–2049. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.M.; Bird, D.; Welti, J.; Gourlaouen, M.; Lang, G.; Murray, G.I.; Reynolds, A.R.; Cox, T.R.; Erler, J.T. Lysyl oxidase plays a critical role in endothelial cell stimulation to drive tumor angiogenesis. Cancer Res. 2013, 73, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Zaffryar Eilot, S.; Marshall, D.; Voloshin, T.; Bar Zion, A.; Spangler, R.; Kessler, O.; Ghermazien, H.; Brekhman, V.; Suss Toby, E.; Adam, D. Lysyl oxidase-like-2 promotes tumour angiogenesis and is a potential therapeutic target in angiogenic tumours. Carcinogenesis 2013, 34, 2370–2379. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Thijssen, V.L.; Poirier, F.; Baum, L.G.; Griffioen, A.W. Galectins in the tumor endothelium: Opportunities for combined cancer therapy. Blood 2007, 110, 819–2827. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Chen, T.T.; Xia, L.; Guo, M.; Xu, Y.; Yue, F.; Jiang, Y.; Chen, G.Q.; Zhao, K.W. Hypoxia inducible factor-1 mediates expression of galectin-1: The potential role in migration/invasion of colorectal cancer cells. Carcinogenesis 2010, 31, 1367–1375. [Google Scholar] [CrossRef] [PubMed]
- Croci, D.O.; Cerliani, J.P.; Dalotto-Moreno, T.; Méndez-Huergo, S.P.; Mascanfroni, I.D.; Dergan-Dylon, S.; Toscano, M.; Caramelo, J.J.; García-Vallejo, J.J.; Ouyang, J.; et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell 2014, 156, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Kanda, A.; Noda, K.; Saito, W.; Ishida, S. Aflibercept traps Galectin-1, an angiogenic factor associated with diabetic retinopathy. Sci. Rep. 2016, 5, 17946. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, M.; Kufareva, I.; Abagyan, R.; Overduin, M. Membrane and protein interactions of the Pleckstrin homology domain superfamily. Membranes 2015, 5, 646–663. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.; Zhou, X.; Cao, B.; Liao, P.; Liu, H.B.; Chen, Y.; Park, H.W.; Zeng, S.X.; Lu, H. Pleckstrin homology domain containing protein PHLDB3 supports cancer growth via a negative feedback loop involving p53. Nat. Commun. 2016, 7, 13755. [Google Scholar] [CrossRef]
- Mana, G.; Clapero, F.; Panieri, E.; Panero, V.; Böttcher, R.T.; Tseng, H.Y.; Saltarin, F.; Astanina, E.; Wolanska, K.I.; Morgan, M.; et al. PPFIA1 drives active α5β1 integrin recycling and controls fibronectin fibrillogenesis and vascular morphogenesis. Nat. Commun. 2016, 7, 13546. [Google Scholar] [CrossRef] [PubMed]
- Martin, P. Wound Healing—Aiming for Perfect Skin Regeneration. Science 1997, 276, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Kampfer, H.; Pfeilschifter, J.; Frank, S. Expressional regulation of angiopoietin-1 and -2 and the tie-1 and -2 receptor tyrosine kinases during cutaneous wound healing: A comparative study of normal and impaired repair. Lab. Investig. 2001, 81, 361–373. [Google Scholar] [CrossRef]
- Usui, M.L.; Mansbridge, J.N.; Carter, W.G.; Fujita, M.; Olerud, J.E. Keratinocyte Migration, Proliferation, and Differentiation in Chronic Ulcers From Patients With Diabetes and Normal Wounds. J. Histochem. Cytochem. 2008, 56, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Yang, M.; Li, H.; Du, Z.; Yang, Y.; Li, Q. Transplantation of adipose stromal cells promotes neovascularization of random skin flaps. Tohoku J. Exp. Med. 2011, 224, 229–234. [Google Scholar] [CrossRef]

| Items | Loach | Yellow Catfish |
|---|---|---|
| Total length | 36.07 Gb | 44.83 Gb |
| Total number of clean reads | 266,126,010 | 291,478,754 |
| Total number of unigenes | 56,056 | 53,731 |
| Mean length of unigenes (nt) | 991 | 1080 |
| Total number of N50 | 1922 | 2073 |
| ID | Description | p Value | DEGs |
|---|---|---|---|
| ko04512 | ECM-receptor interaction | 6.36 × 10−32 | fn1, itga11, itga5, lama1, lamb1, lamc3, thbs1, thbs2, thbs3a, thbs4b, tnc |
| ko04510 | Focal adhesion | 1.58 × 10−21 | flna, fn1, itga11, itga5, lama1, lamb1, lamc3, mylpf, pdgfc, thbs1, thbs2, thbs3a, thbs4b, tln2, tnc |
| ko04810 | Regulation of actin cytoskeleton | 9.06 × 10−4 | itga5, itga11, fn1, pdgfc, mylpf, bdkrb2, fgfr4, tmsb |
| ko04350 | TGF-beta signaling pathway | 2.42 × 10−2 | bmp2, rps6kb1, thbs1, dcn, nbl1 |
| ko04630 | Jak-STAT signaling pathway | 3.28 × 10−1 | lep-B, il11 |
| ko04310 | Wnt signaling pathway | 6.18 × 10−1 | nfatc1, sfrp1, serpinf1, sfrp2, fzd2 |
| ko04115 | p53 signaling pathway | 7.47 × 10−1 | thbs1, igfbp3 |
| ko04340 | Hedgehog signaling pathway | 8.68 × 10−1 | bmp2, ihhb |
| ko04010 | MAPK signaling pathway | 8.82 × 10−1 | nfatc1, fgfr4, flna, cacna1b, ngf, cacnb4 |
| ko04270 | Vascular smooth muscle contraction | 9.31 × 10−1 | myl6, prkg1 |
| ko04150 | mTOR signaling pathway | 9.60 × 10−1 | rps6kb1 |
| ko04012 | ErbB signaling pathway | 9.60 × 10−1 | rps6kb1 |
| ko04910 | Insulin signaling pathway | 9.70 × 10−1 | rps6kb1, fbp2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Sun, B.; Huang, L.; Yang, L.; Liu, C.; Zhu, J.; Gao, J.; Cao, X. Comparative Transcriptomic Analysis of Regenerated Skins Provides Insights into Cutaneous Air-Breathing Formation in Fish. Biology 2021, 10, 1294. https://doi.org/10.3390/biology10121294
Huang S, Sun B, Huang L, Yang L, Liu C, Zhu J, Gao J, Cao X. Comparative Transcriptomic Analysis of Regenerated Skins Provides Insights into Cutaneous Air-Breathing Formation in Fish. Biology. 2021; 10(12):1294. https://doi.org/10.3390/biology10121294
Chicago/Turabian StyleHuang, Songqian, Bing Sun, Longfei Huang, Lijuan Yang, Chuanshu Liu, Jinli Zhu, Jian Gao, and Xiaojuan Cao. 2021. "Comparative Transcriptomic Analysis of Regenerated Skins Provides Insights into Cutaneous Air-Breathing Formation in Fish" Biology 10, no. 12: 1294. https://doi.org/10.3390/biology10121294
APA StyleHuang, S., Sun, B., Huang, L., Yang, L., Liu, C., Zhu, J., Gao, J., & Cao, X. (2021). Comparative Transcriptomic Analysis of Regenerated Skins Provides Insights into Cutaneous Air-Breathing Formation in Fish. Biology, 10(12), 1294. https://doi.org/10.3390/biology10121294






