Soil Fungal Diversity of the Aguarongo Andean Forest (Ecuador)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Declaration of Ethics
2.2. Sample Collection
2.3. Soil Physicochemical Analysis
2.4. Environmental DNA Extraction from Soil Samples
2.5. PCR Amplification and Next-Generation Sequencing (NGS, Illumina MiSeq)
2.6. Taxonomic Allocation of Sequence Readings and Statistical Analysis
2.7. Access Numbers
3. Results
3.1. Soil Chemical Characteristics
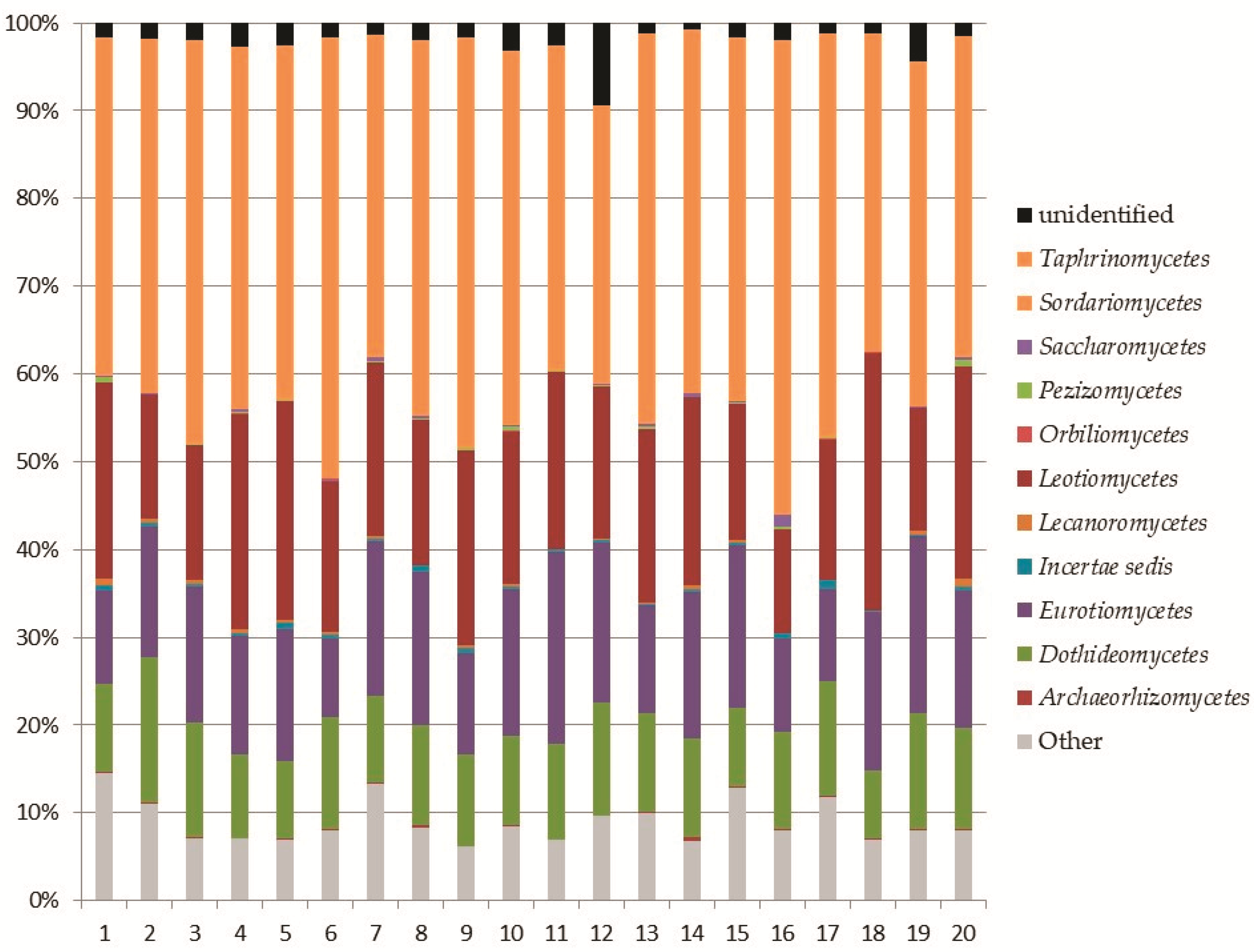
3.2. Soil Fungal Assemblage Composition
3.2.1. Subkingdom Dikarya
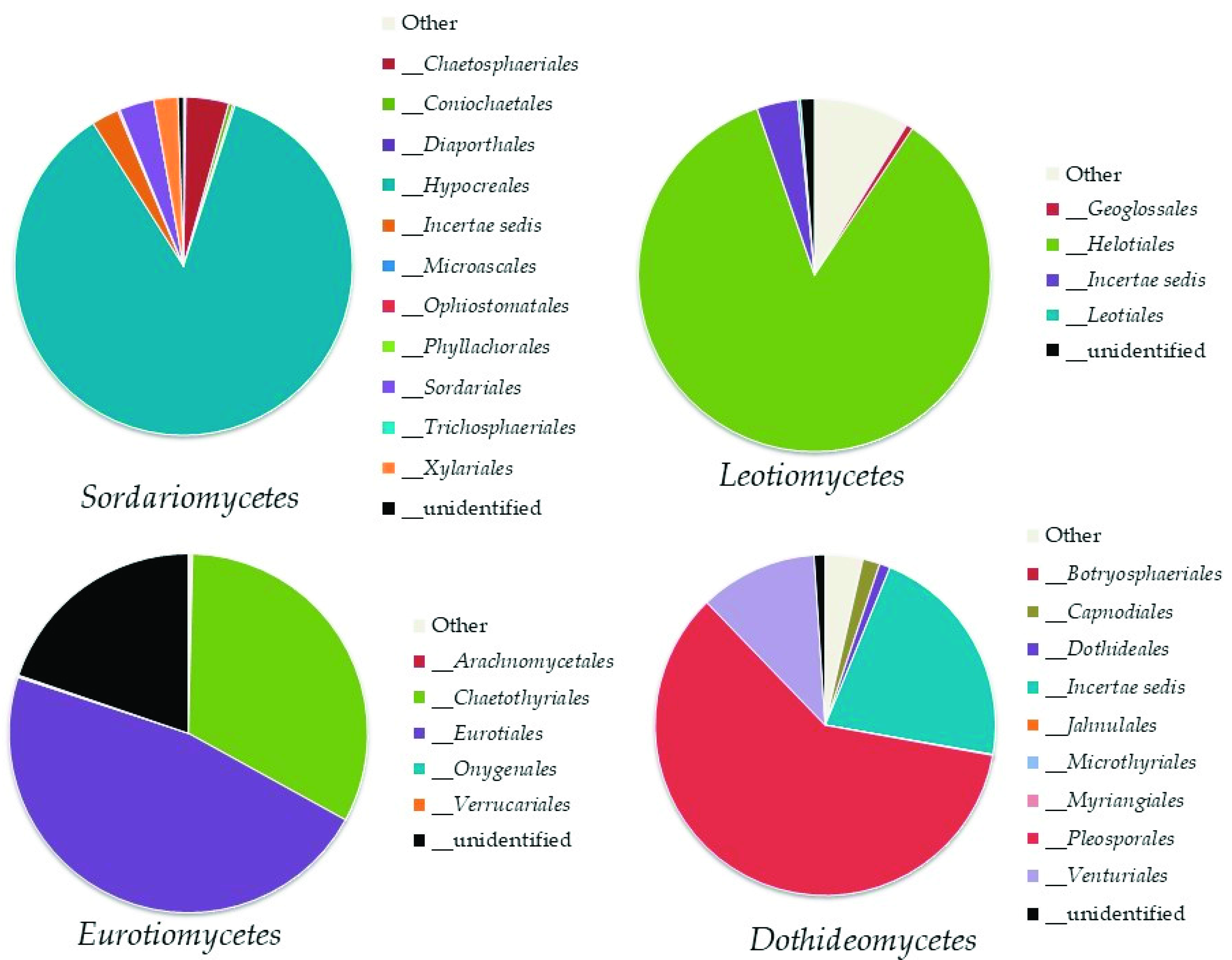
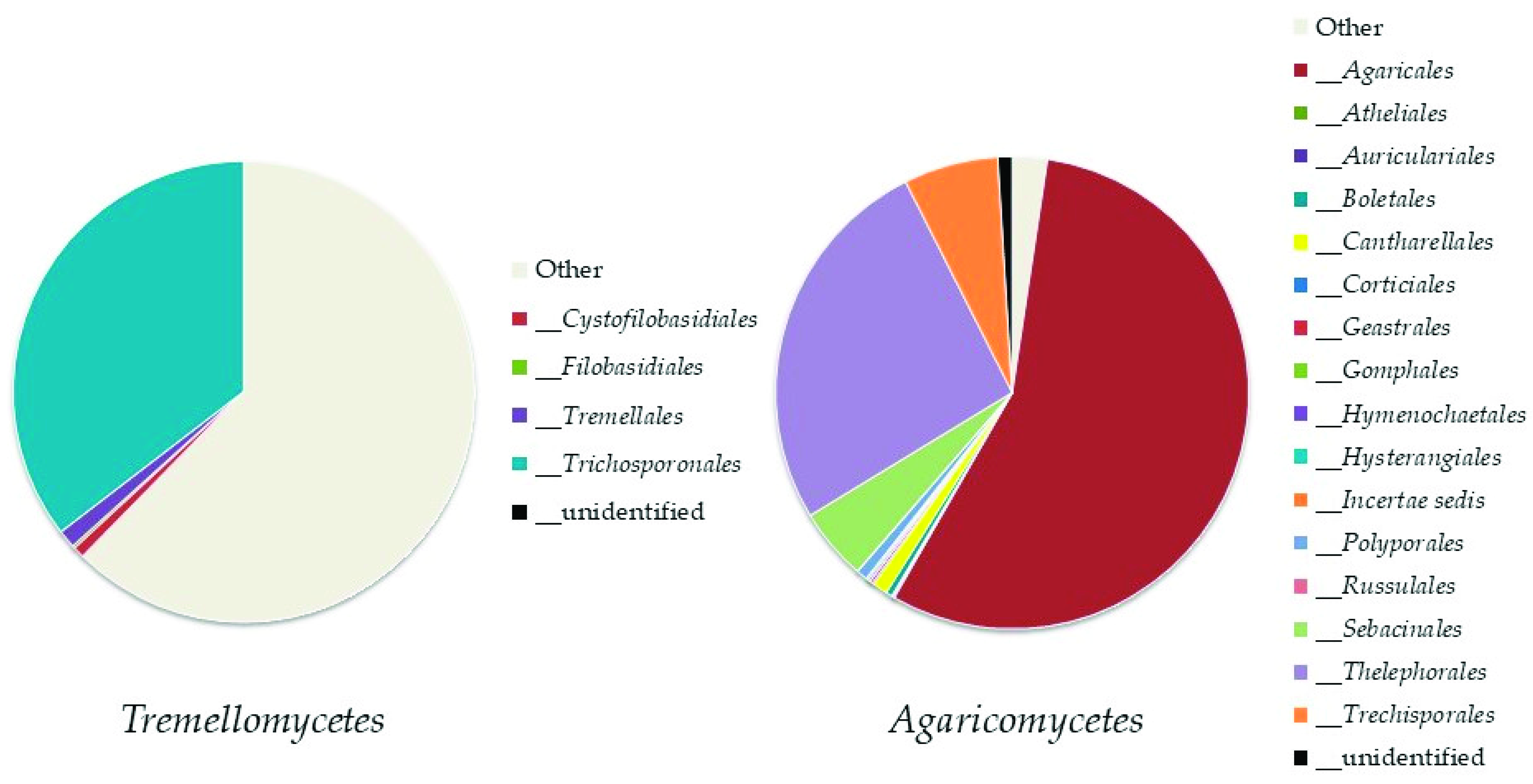
Ascomycota
Top Ten Most Abundant Species of Ascomycota
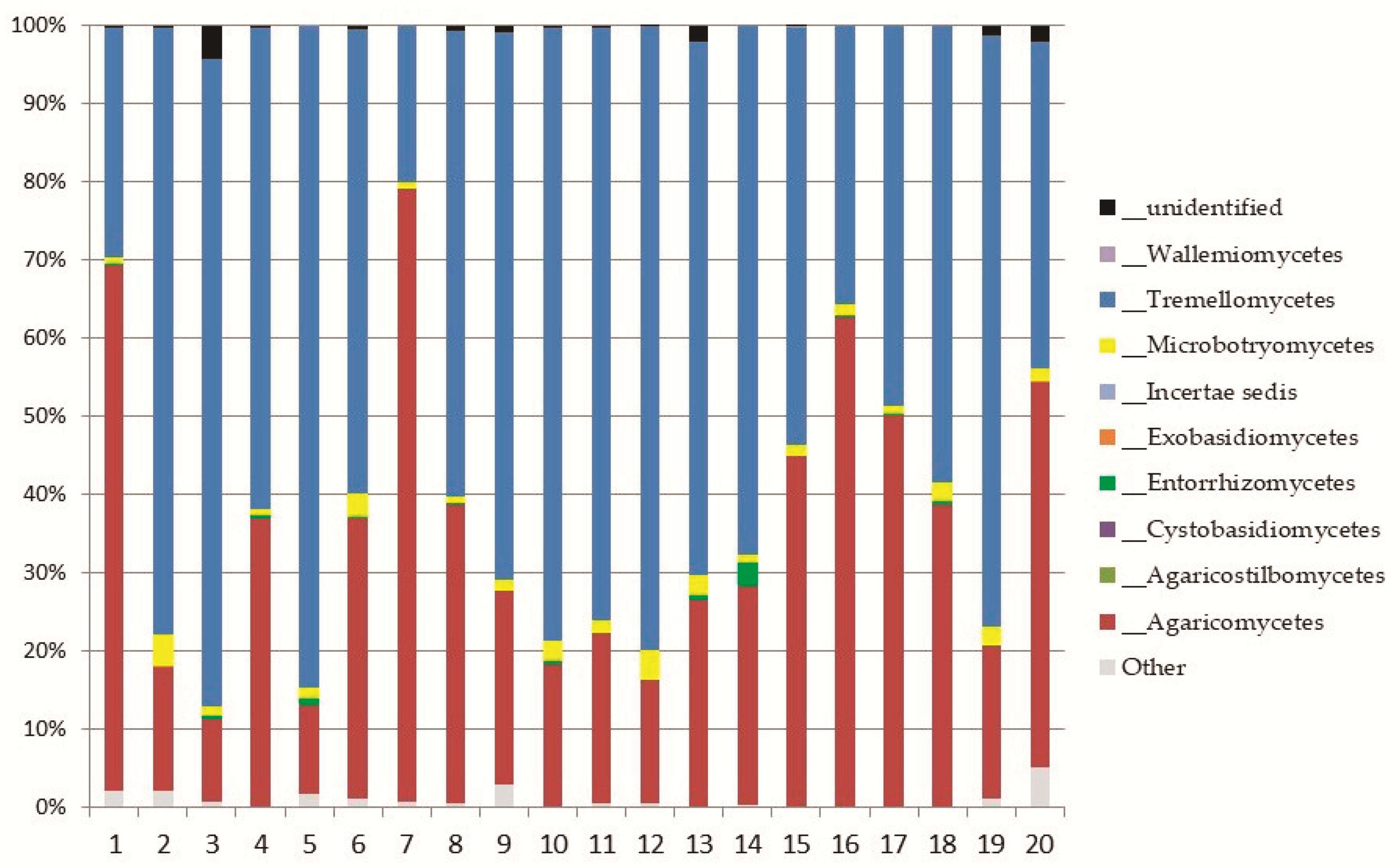
Basidiomycota
Top Ten Abundant Species of Basidiomycota
3.2.2. The Subkingdom Mucoromyceta
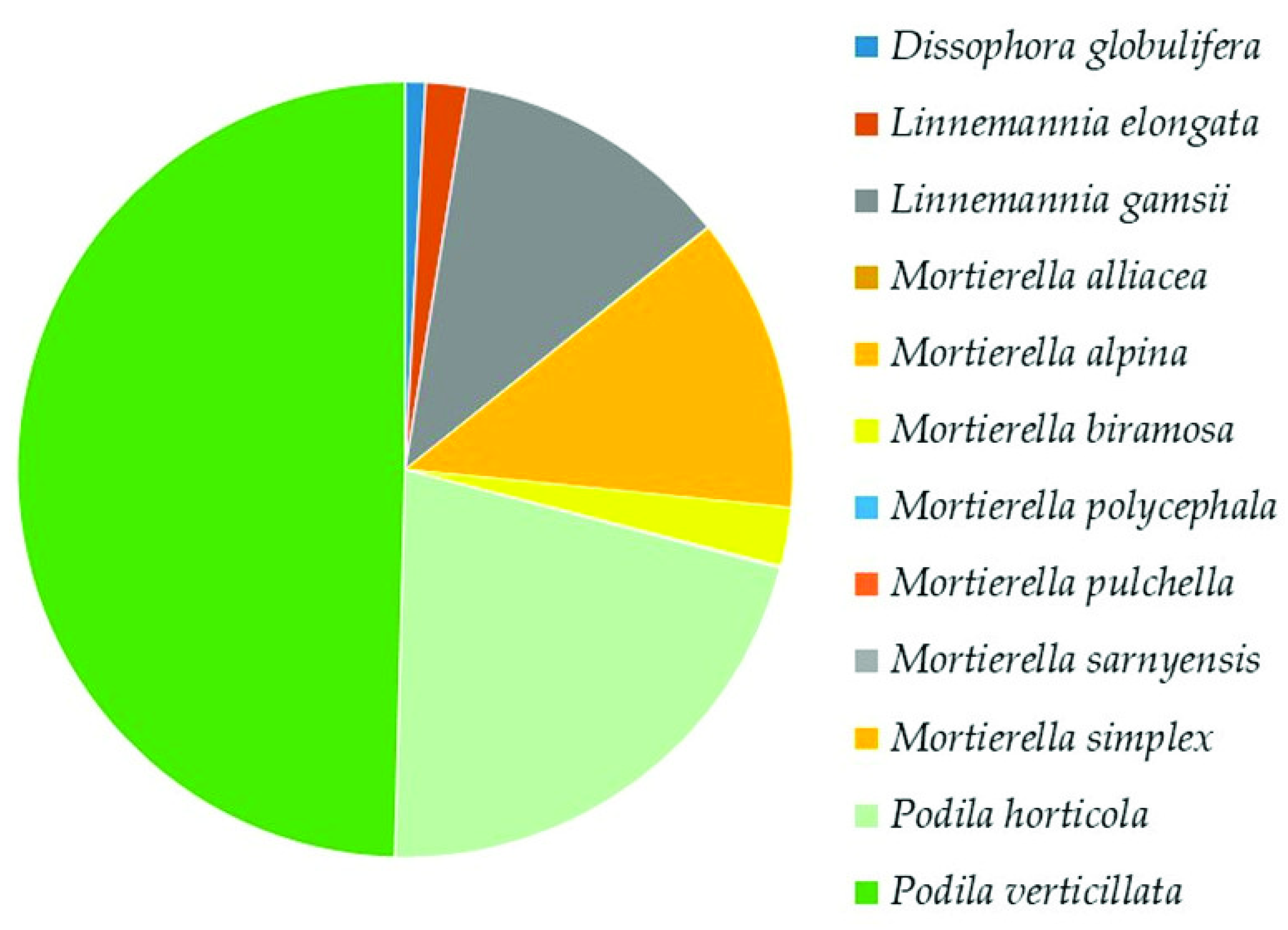
Mortierellomycota
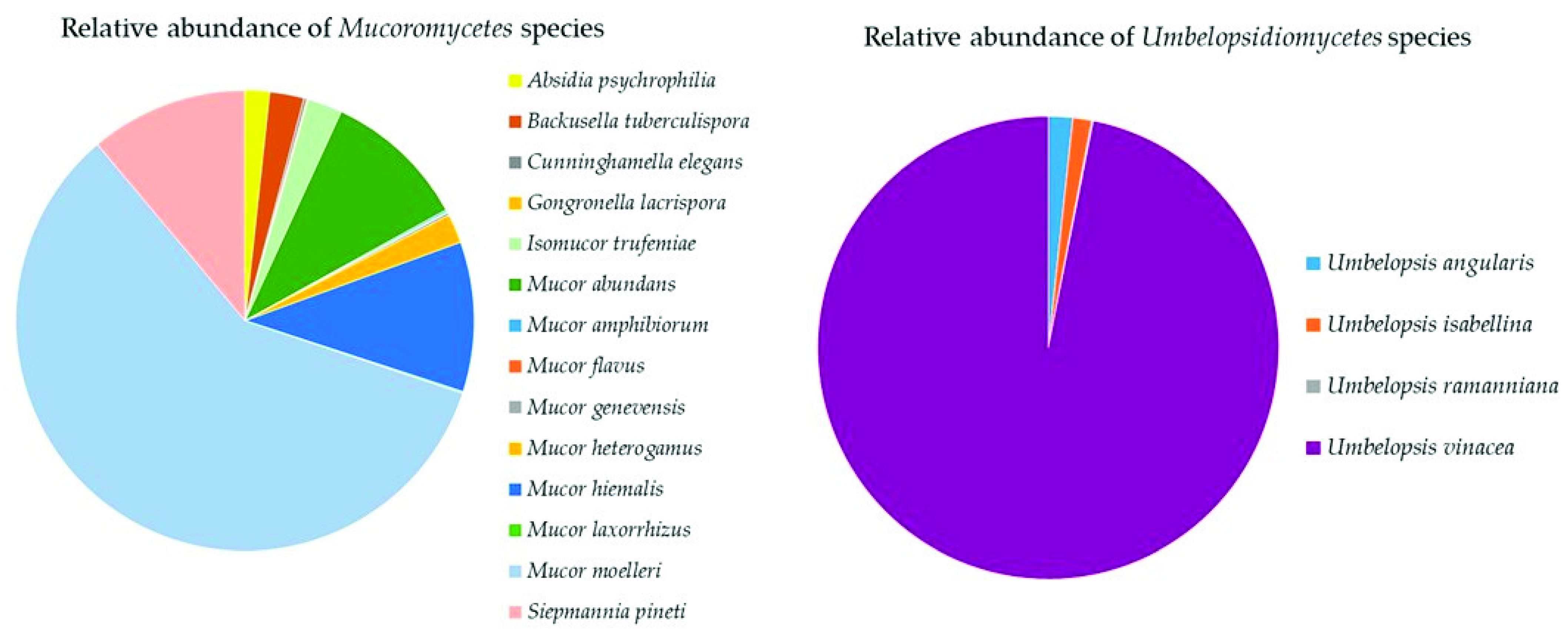
Mucoromycota
Glomeromycota
The Top Mucoromyceta
3.2.3. The Subkingdom Chytridiomyceta
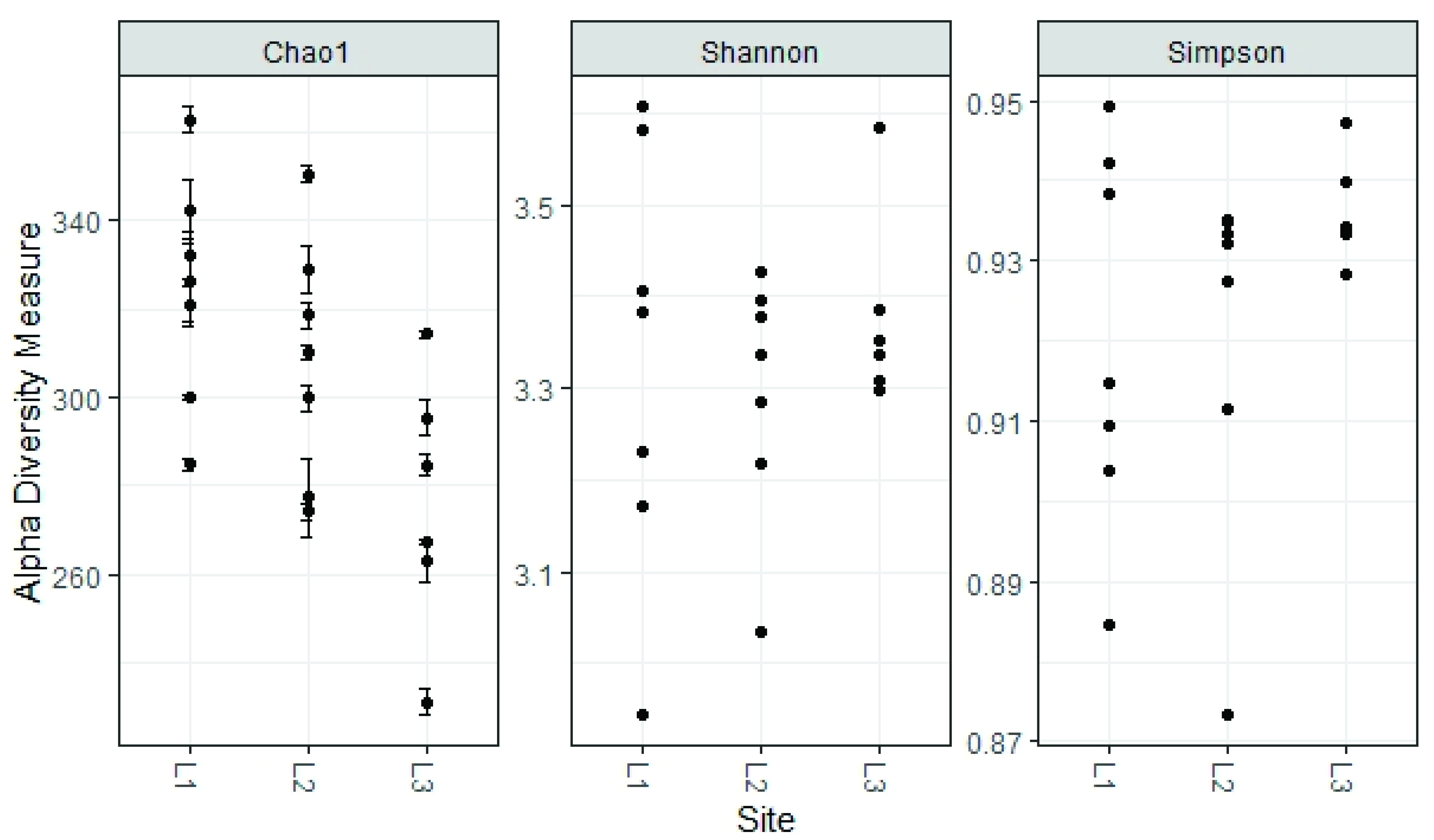
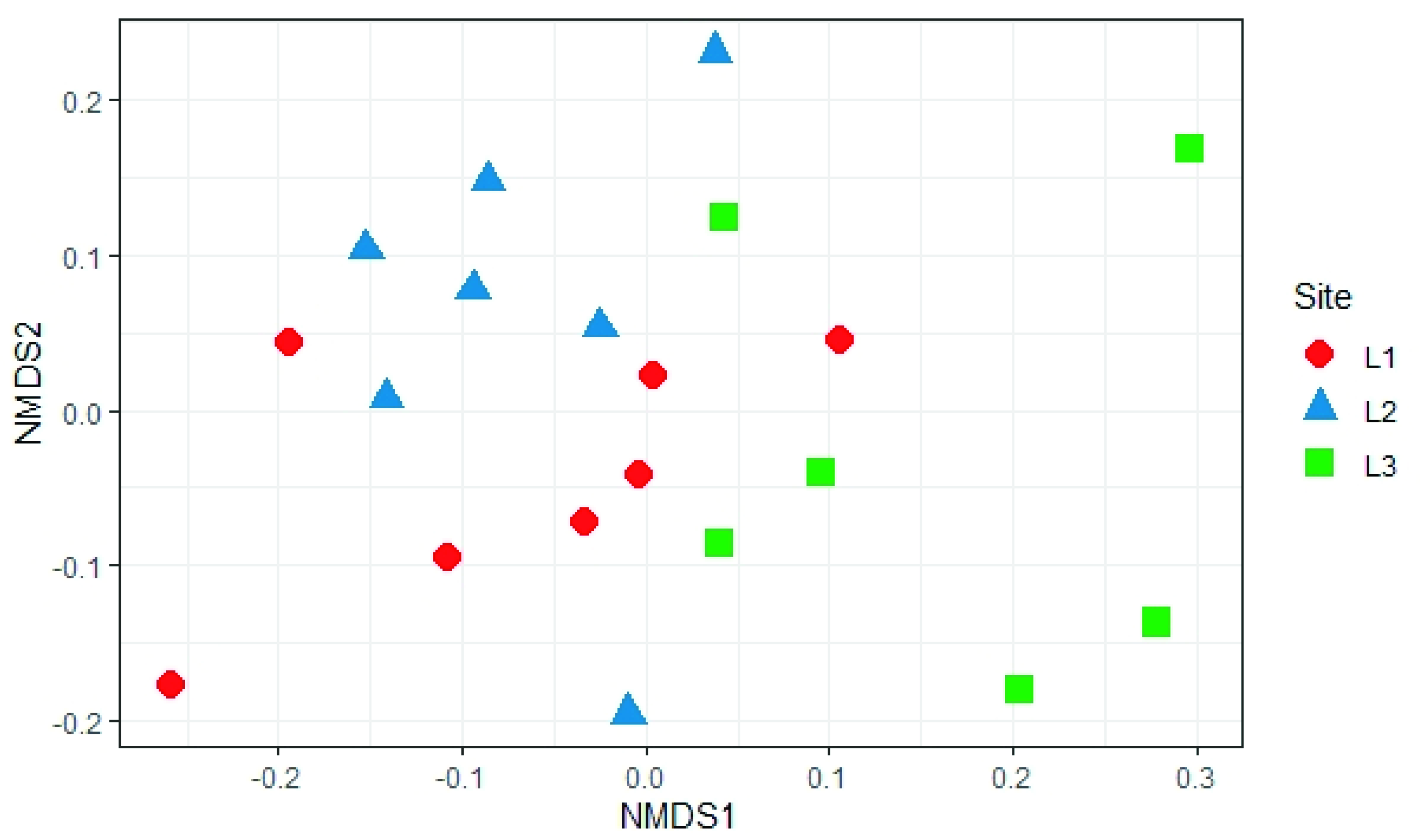
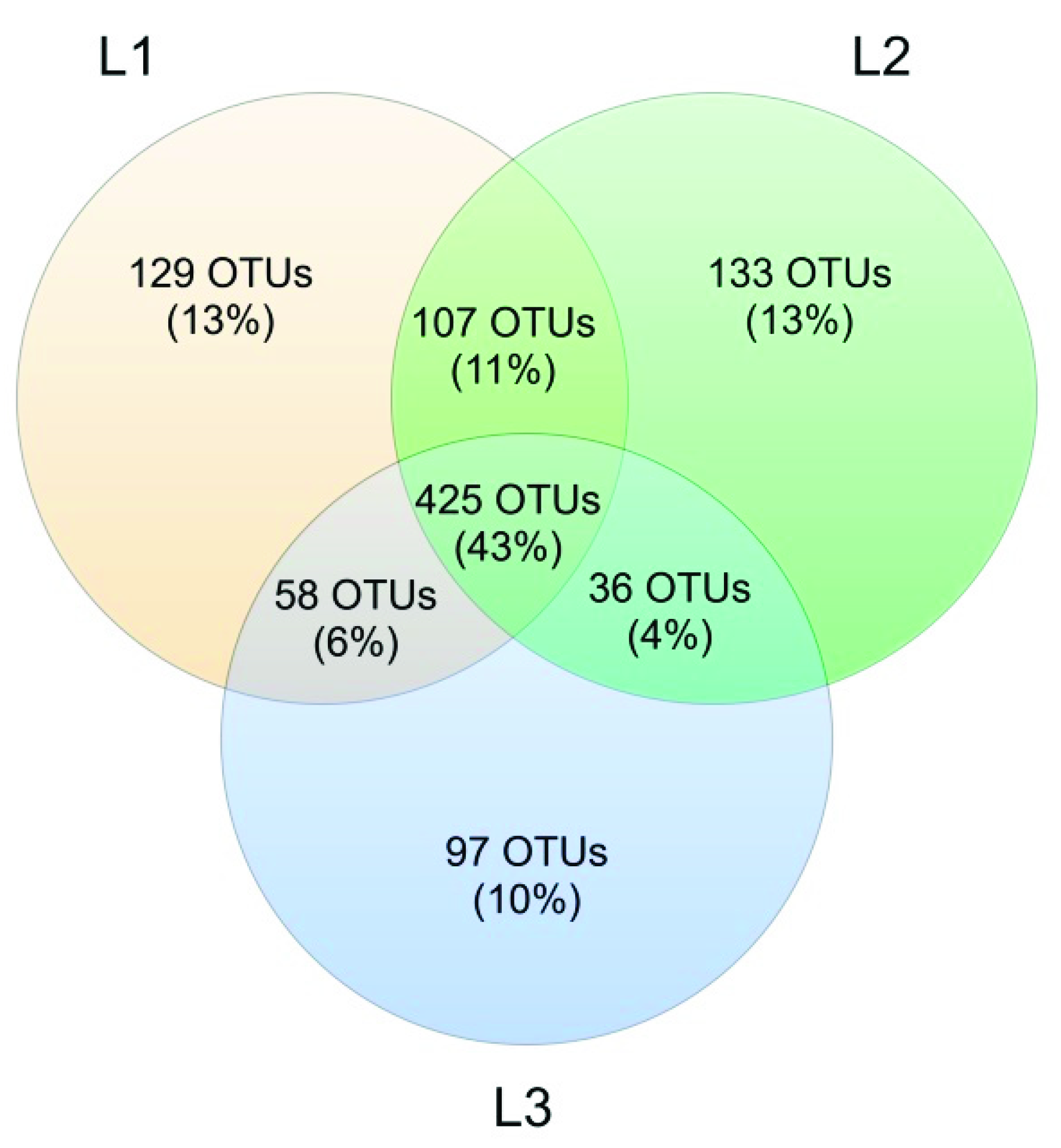
3.3. Soil Mycobiota Diversity in the Sampling Sites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tedersoo, L.; Bahram, M.; Põlme, S.; Koljag, U.; Yorou, N.S.; Wijesundera, R.; Virreal Ruiz, L.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1078. [Google Scholar] [CrossRef] [Green Version]
- Mueller, G.M.; Schmit, J.P.; Leacock, P.R.; Buyck, B.; Cifuentes, J.; Dejardin, D.E.; Halling, R.E.; Hjortstam, K.; Iturriaga, T.; Larsson, K.H.; et al. Global diversity and distribution of macrofungi. Biodivers. Conserv. 2007, 16, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Jairus, T.; Horton, B.M.; Abarenkov, K.; Suvi, T.; Saar, I.; Kõljalg, U. Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol. 2008, 180, 479–490. [Google Scholar] [CrossRef]
- Thakur, M.P.; Geisen, S. Trophic Regulations of the Soil Microbiome. Trends Microbiol. 2019, 27, 771–780. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Baldauf, S.L.; Leyval, C.; Straczek, J.; Young, J.P.W. Extensive fungal diversity in plant roots. Science 2002, 295, 2051. [Google Scholar] [CrossRef]
- Banchi, E.; Ametrano, C.G.; Tordoni, E.; Stankovi, D.; Ongaro, S.; Tetiach, M.; Pallavicini, A.; Muggia, L.; Verardo, P.; Tassan, F.; et al. Environmental DNA assessment of airborne plant and fungal seasonal diversity. Sci. Total Environ. 2020, 738, 140249. [Google Scholar] [CrossRef] [PubMed]
- Coleine, C.; Selbmann, L.; Pombubpa, N.; Stajich, J.E. Amplicon sequencing of rock inhabiting microbial communities from Joshua Tree National Park, USA. Microbiol. Resour. Announc. 2021, 10, e00494-21. [Google Scholar] [CrossRef]
- Seifert, K.A. Progress towards DNA barcoding of fungi. Mol. Ecol. Resour. 2009, 9, 83–89. [Google Scholar] [CrossRef]
- Peay, K.; Kennedy, P.; Talbot, J. Dimensions of biodiversity in the Earth mycobiome. Nat. Rev. Microbiol. 2016, 14, 434–447. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 83, 4–18. [Google Scholar] [CrossRef]
- Toapanta-Alban, C.E.; Ordoñez, M.E.; Barnes, C.W.; Blanchette, R.A. Taxonomy of the major rhizomorphic species of the “Melanopus group” within Polyporaceae in Yasunı’ National Park, Ecuador. PLoS ONE 2021, 16, e0254567. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Haelewaters, D.; Pfliegler, W.P.; Page, R.A.; Dick, C.W.; Aime, M.C. A new species of Gloeandromyces from Ecuador and Panama revealed by morphology and phylogenetic reconstruction, with a discussion of secondary barcodes in Laboulbeniomycetes taxonomy. Mycologia 2020, 112, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef]
- Galarza, L.; Akagi, Y.; Takao, K.; Kim, C.S.; Maekawa, N.; Itai, A.; Peralta, E.; Santos, E.; Kodama, M. Characterization of Trichoderma species isolated in Ecuador and their antagonistic activities against phytopathogenic fungi from Ecuador and Japan. J. Gen. Plant. Pathol. 2015, 81, 201–210. [Google Scholar] [CrossRef]
- Baroni, T.J.; Halling, R.E. New species of Rhodocybe from South America with a key to species. Mycologia 1992, 84, 411–421. [Google Scholar] [CrossRef]
- Goh, T.K.; Hyde, K.D. A new species of Palmicola from Ecuador. Mycol. Res. 1996, 100, 714–716. [Google Scholar] [CrossRef]
- Berndt, R. New Puccinia species on Baccharis from Ecuador and Costa Rica. Mycol. Res. 1998, 102, 1108–1112. [Google Scholar] [CrossRef]
- Garcés, F.F.; Fiallos, F.F.; Silva, E.; Martinez, F.; Aime, M.C.; Comstock, J.C.; Glynn, N.C.; Castlebury, L.A. First Report of Orange Rust of Sugarcane Caused by Puccinia kuehnii in Ecuador. Plant. Dis. 2014, 98, 842. [Google Scholar] [CrossRef] [PubMed]
- Dueñas, J.F.; Camenzind, T.; Roy, J.; Hempel, S.; Homeier, J.; Suárez, J.P.; Rillig, M.C. Moderate phosphorus additions consistently affect community composition of arbuscular mycorrhizal fungi in tropical montane forests in southern Ecuador. New Phytol. 2020, 227, 1505–1518. [Google Scholar] [CrossRef]
- Garcés-Ruiz, M.; Senés-Guerrero, C.; Declerck, S.; Cranenbrouck, S. Community composition of arbuscular mycorrhizal fungi associated with native plants growing in a petroleum-polluted soil of the Amazon region of Ecuador. Microbiologyopen 2019, 8, e00703. [Google Scholar] [CrossRef]
- Jaswal, R.; Pathak, A.; Edwards, B., III; Lewis, R., III; Seaman, J.C.; Stothard, P.; Krivushin, K.; Blom, J.; Rupp, O.; Chauhan, A. Metagenomics-Guided Survey, Isolation, and Characterization of Uranium Resistant Microbiota from the Savannah River Site, USA. Genes 2019, 10, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rundell, S.M.; Spakowicz, D.J.; Narváez-Trujillo, A.; Strobel, S.A. The Biological Diversity and Production of Volatile Organic Compounds by Stem-Inhabiting Endophytic Fungi of Ecuador. J. Fungi 2015, 1, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Novotná, A.; Ángel Benítez, A.; Herrera, P.; Cruz, D.; Filipczyková, E.; Suárez, J.P. High diversity of root-associated fungi isolated from three epiphytic orchids in southern Ecuador. Mycoscience 2018, 59, 24–32. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sadam, A.; Zambrano, M.; Valencia, R.; Bahram, M. Low diversity and high host preference of ectomycorrhizal fungi in Western Amazonia, a neotropical biodiversity hotspot. ISME J. 2010, 4, 465–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haug, I.; Setaro, S.; Suárez, J.P. Global AM fungi are dominating mycorrhizal communities in a tropical premontane dry forest in Laipuna, South Ecuador. Mycol. Progress 2021, 20, 837–845. [Google Scholar] [CrossRef]
- Setaro, S.; Suárez, J.P. Species composition of arbuscular mycorrhizal communities changes with elevation in the Andes of South Ecuador. PLoS ONE 2019, 14, e0221091. [Google Scholar]
- Minga, N.O. Experience to Protection and Management of Native Andean Forests in the South of Ecuador. Lyonia 2003, 4, 157–164. [Google Scholar]
- Dunque-Sarango, P.; Cajamarca-Rivadeneira, R.; Wemple, B.C.; Delgado- Fernández, M.E. Estimation of the water balance of for a small tropical andean catchment. LA GRANJA Rev. Cienc. Vida 2019, 29, 56–69. [Google Scholar]
- Uroz, S.; Ioannidis, P.; Lengelle, J.; Cébron, A.; Morin, E.; Buée, M.; Martin, F. Functional assays and metagenomic analyses reveals differences between the microbial communities inhabiting the soil horizons of a Norway spruce plantation. PLoS ONE 2013, 8, e55929. [Google Scholar] [CrossRef] [PubMed]
- Bloem, J.; Hopkins, D.W.; Benedetti, A. Microbiological Methods for Assessing Soil Quality; CABI Publishing: London, UK, 2006; p. 307. [Google Scholar]
- Euherabide, M.; Saínz Rozas, H.; Barbieri, P.; Echeverría, H.E. Comparación de métodos para determinar carbono orgánico en suelo. Cienc. Suelo 2014, 32, 13–19. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomial RNA genes for phylogenetics. In PCR Protocols a Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Caporaso, J.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nillson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org (accessed on 7 February 2018).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.0–10. 2013. Available online: http://CRAN.R-project.org/package=vegan (accessed on 7 February 2018).
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Doring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef] [Green Version]
- Vandepol, N.; Liber, J.; Desirò, A.; Na, H.; Kennedy, M.; Barry, K.; Grigoriev, I.V.; Miller, A.N.; O’Donnell, K.; Stajich, J.E.; et al. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 2020, 104, 267–289. [Google Scholar] [CrossRef] [PubMed]
- Fredi, F.; Parra, R.P.; Zumba, D.A. Bromeliads of the Aguarongo Protective Forest-Ecuador and Adaptation to Climate Change. J. Eng. Appl. Sci. 2017, 12, 1619–1622. [Google Scholar]
- Vaz, A.B.M.; Fonseca, P.L.C.; Leite, L.R.; Badotti, F.; Salim, A.C.M.; Flavio, M.G.; Araujo, F.M.G.; Cuadros-Orellana, S.C.; Duarte, Â.A.; Rosa, C.A.; et al. Using next-generation sequencing (NGS) to uncover diversity of wood-decaying fungi in neotropical atlantic forests. Phytotaxa 2017, 295, 001–021. [Google Scholar] [CrossRef] [Green Version]
- Landínez-Torres, A.Y.; Panelli, S.; Picco, A.M.; Comandatore, F.; Tosi, S.; Capelli, E. A meta-barcoding analysis of soil myco-biota of the upper Andean Colombian agro-environment. Sci. Rep. 2019, 9, 10085. [Google Scholar] [CrossRef] [Green Version]
- Landínez-Torres, A.Y.; Abril, J.L.B.; Tosi, S.; Nicola, L. Soil Microfungi of the Colombian Natural Regions. Int. J. Environ. Res. Public Heal. 2020, 17, 8311. [Google Scholar] [CrossRef]
- Nicola, L.; Landínez-Torres, A.Y.; Zambuto, F.; Capelli, E.; Tosi, S. The Mycobiota of High Altitude Pear Orchards Soil in Colombia. Biology 2021, 10, 1002. [Google Scholar] [CrossRef]
- Senés-Guerrero, C.; Schüßler, A. A conserved arbuscular mycorrhizal fungal core- species community colonizes potato roots in the Andes. Fungal Divers. 2016, 77, 317–333. [Google Scholar] [CrossRef] [Green Version]
- Loján, P.; Senés-Guerrero, C.; Suárez, J.P.; Kromann, P.; Schüßler, A.; Declerck, S. Potato field-inoculation in Ecuador with Rhizophagus irregularis: No impact on growth performance and associated arbuscular mycorrhizal fungal communities. Symbiosis 2017, 73, 45–56. [Google Scholar] [CrossRef]
- Rodolfi, M.; Longa, C.M.O.; Pertot, I.; Tosi, S.; Savino, E.; Guglielminetti, M.; Altobelli, E.; Del Frate, G.; Picco, A.M. Fungal biodiversity in the periglacial soil of Dosde Glacier (Valtellina, Northern Italy). J. Basic Microbiol. 2016, 56, 263–274. [Google Scholar] [CrossRef]
- Tosi, S.; Casado, B.; Gerdol, R.; Caretta, G. Fungi isolated from antarctic mosses. Polar Biol. 2002, 25, 262–268. [Google Scholar] [CrossRef]
- Santos, J.A.D.; Meyer, E.; Sette, L.D. Fungal Community in Antarctic Soil Along the Retreating Collins Glacier (Fildes Peninsula, King George Island). Microorganisms 2020, 8, 1145. [Google Scholar] [CrossRef]
- Canini, F.; Geml, J.; D’Acqui, L.P.; Selbmann, L.; Onofri, S.; Ventura, S.; Zucconi, L. Exchangeable cations and pH drive diversity and functionality of fungal communities in biological soil crusts from coastal sites of Victoria Land, Antarctica. Fungal Ecol. 2020, 45, 100923. [Google Scholar] [CrossRef]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi; Academic Press: London, UK, 1980. [Google Scholar]
- Tamayo-Vélez, Á.; Osorio, N.W. Soil Fertility Improvement by Litter Decomposition and Inoculation with the Fungus Mortierella sp. in Avocado Plantations of Colombia. Commun. Soil Sci. Plan. 2018, 49, 139–147. [Google Scholar] [CrossRef]
- Gonçalves, C.M.; Oliveira, R.J.V.; Silva, R.M.F.; Souza, C.A.F.; Lima, D.X.; Silva, G.A. Mortierella verticillata Linnem (Mortierellomycota, Mortierellales) isolated from mountainous environments: A first report from South America. Check List 2020, 16, 907–910. [Google Scholar] [CrossRef]
- Smith, S.N. An Overview of Ecological and Habitat Aspects in the Genus Fusarium with Special Emphasis on the Soil Borne Pathogenic Forms. Plant Pathol. Bull. 2007, 16, 97–120. [Google Scholar]
- Chang, J.; Liu, S.; Shi, J.; Guo, N.; Zhang, H.; Chen, J. A new Curvularia lunata variety discovered in Huanghuaihai Region in China. J. Integr. Agr. 2020, 19, 551–560. [Google Scholar] [CrossRef]
- Watanabe, S.; Kato, H.; Kumakura, K.; Ishibashi, E.; Nagayama, K. Properties and biological control activities of aerial and submerged spores in Trichoderma asperellum SKT-1. J. Pestic. Sci. 2006, 31, 375–379. [Google Scholar] [CrossRef] [Green Version]
- Chagas Junior, A.F.; Chagas, L.F.B.; Miller, L.D.O.; de Oliveira, J.C. Efficiency of Trichoderma asperellum UFT 201 as plant growth promoter in soybean. Afr. J. Agr. Res. 2019, 14, 263–271. [Google Scholar]
- Nicola, L.; Tosi, S.; Savini, D. In vitro evaluation of nematophagous activity of fungal isolates. J. Basic Microbiol. 2014, 54, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Hernández, S.; Torres-Hernández, G.; Alonso-Díaz, M.A.; von Son-de-Fernex, E.; Aguilar-Marcelino, L.; González-Garduño, R.; Becerril-Pérez, C.M.; Alcántara-Carbajal, J.L.; Vargas-López, S.; Olmedo-Juárez, A.; et al. Effect of an Arthrobotrys musiformis (Fungi: Orbiliales) culture filtrate on the population of gastrointestinal parasitic nematode eggs in faeces of grazing lambs. Vet. Parasitol. Reg. Stud. Rep. 2021, 24, 100565. [Google Scholar] [CrossRef] [PubMed]
- Iwanicki, N.S.; Pereira, A.A.; Botelho, A.B.R.Z.; Rezende, J.M.; Moral, R.D.; Zucchi, M.I.; Delalibera, I. Monitoring of the field application of Metarhizium anisopliae in Brazil revealed high molecular diversity of Metarhizium spp in insects, soil and sugarcane roots. Sci. Rep. 2019, 9, 4443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amaresan, N.; Senthil Kumar, M.; Sankaranarayanan, M. Beneficial Microbes in Agro-Ecology: Bacteria and Fungi; Academic Press: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Mašínová, T.; Bahnmann, B.D.; Větrovský, T.; Tomšovský, M.; Merunková, K.; Baldrian, P. Drivers of yeast community composition in the litter and soil of a temperate forest. FEMS Microbiol. Ecol. 2017, 93, fiw223. [Google Scholar] [CrossRef]
- Middelhoven, W.J. Trichosporon wieringae sp. nov., an anamorphic basidiomycetous yeast from soil, and assimilation of some phenolic compounds, polysaccharides and other non-conventional carbon sources by saprophytic Trichosporon species. Antonie van Leeuwenhoek 2004, 86, 329–337. [Google Scholar] [CrossRef]
- Larsen, M.J.A. Contribution to the Taxonomy of the Genus Tomentella; New York Botanical Garden: New York, NY, USA, 1974; p. 145. [Google Scholar]
- Gáper, J.; Gáperová, S.; Pristas, P.; Naplavova, K. Medicinal Value and Taxonomy of the Tinder Polypore, Fomes fomentarius (Agaricomycetes): A Review. Int. J. Med. Mushrooms 2016, 18, 851–859. [Google Scholar] [CrossRef]
- Pérez, R.; Yasna Tapia, Y.; Antilén, M.; Manuel Casanova, M.; Vidal, C.; Santander, C.; Aponte, H.; Cornejo, P. Interactive effect of compost application and inoculation with the fungus Claroideoglomus claroideum in Oenothera picensis plants growing in mine tailings. Ecotoxicol. Environ. Safe 2021, 208, 111495. [Google Scholar] [CrossRef] [PubMed]
- Gleason, F.H.; Pilgaard, B.; Henderson, L.; Lange, L. The key ecological role and biology of Rhizophlyctis rosea, a zoosporic, early lineage fungus in soil ecosystems. Curr. Trends Microbiol. 2019, 60, 67–80. [Google Scholar]
- Steciow, M.M.; Arambarri, A.M. Southernmost occurrence of a tropical fungus: Monoblepharella mexicana (Gonapodyaceae, Chytridiomycota). Nova Hedwig. 2000, 70, 107–112. [Google Scholar] [CrossRef]
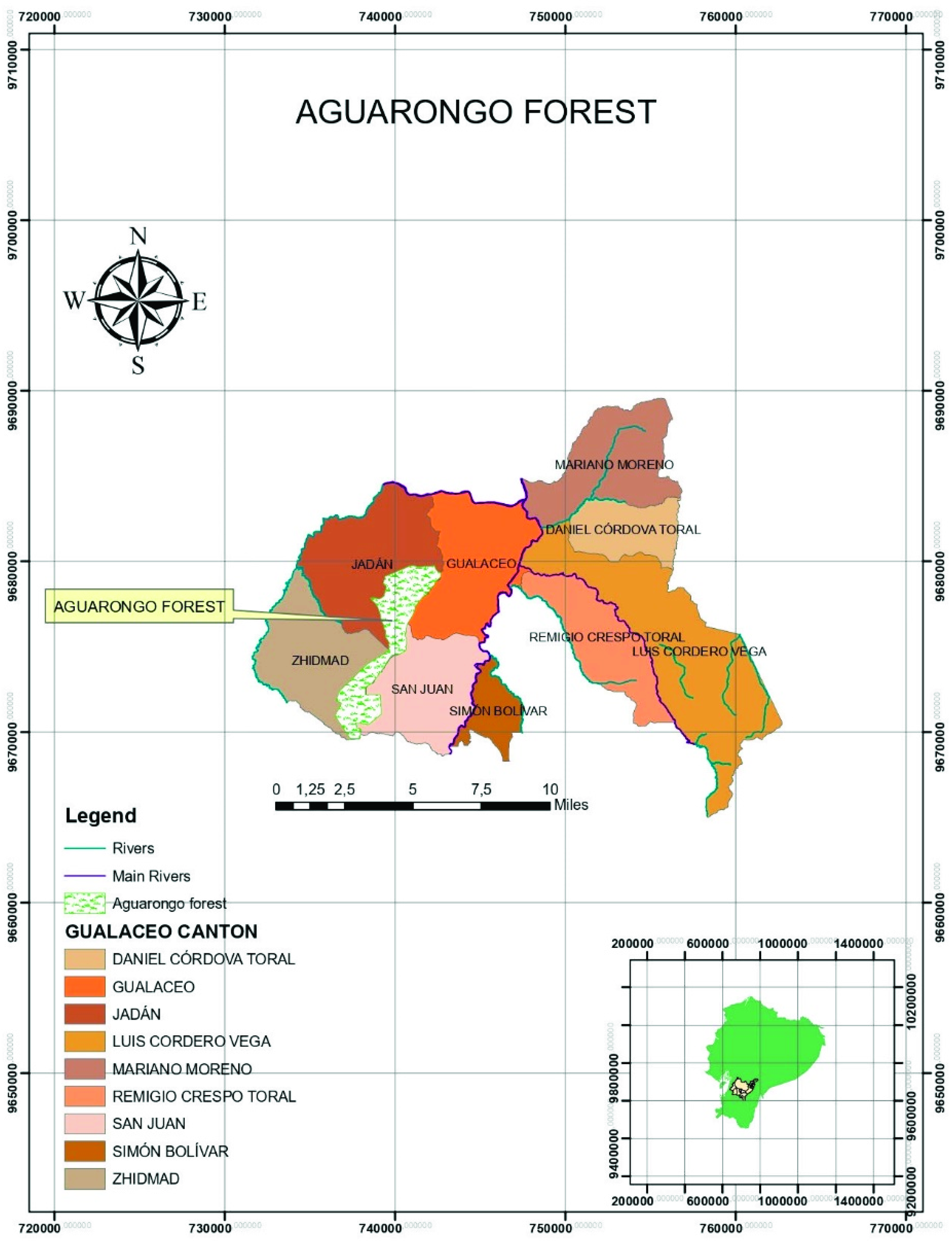
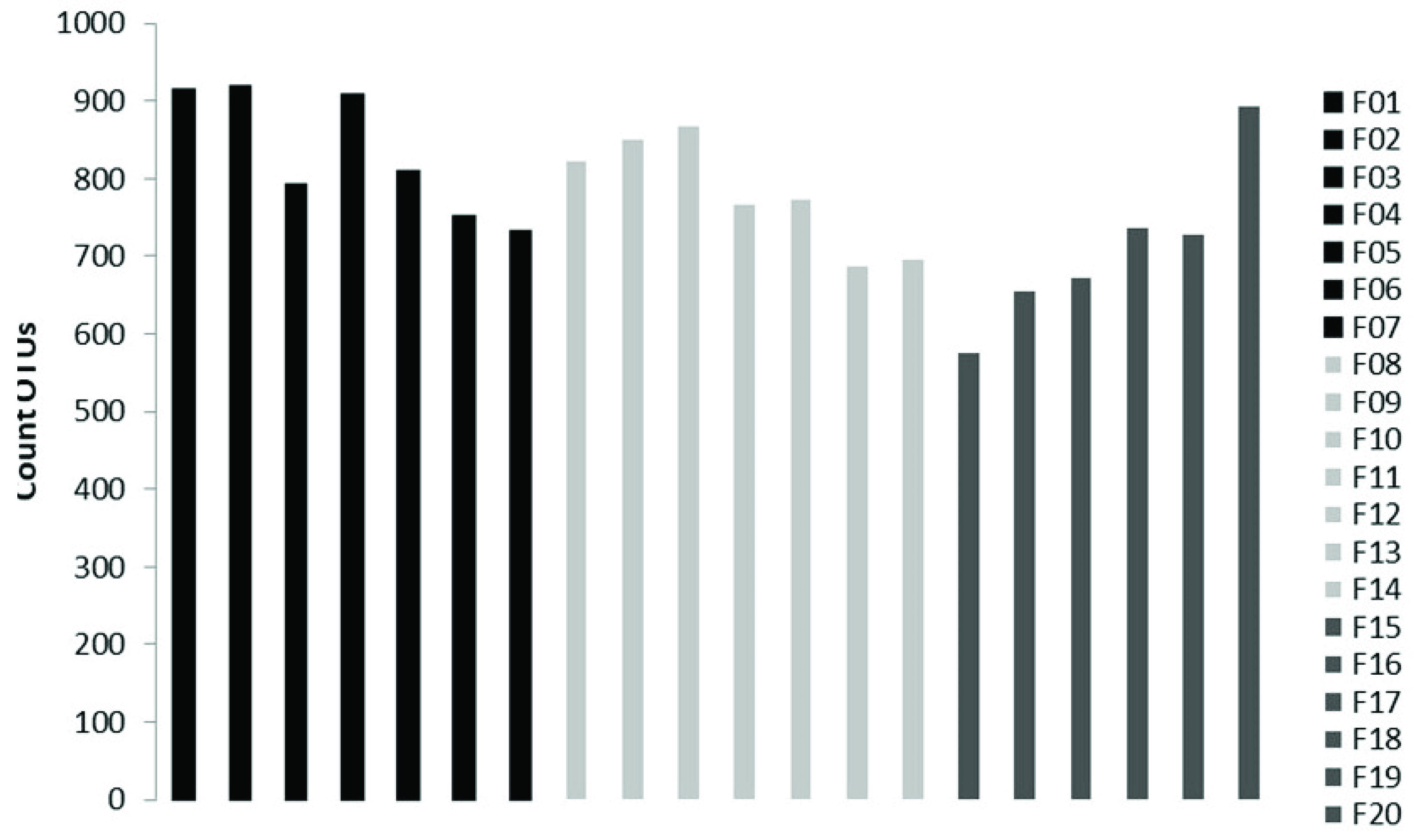
| Sampling Site | Number of Collected Samples | Altitude | Coordinates |
|---|---|---|---|
| L1 | 35 | 3101–3146 | 2.936° S; 78.8339° W |
| L2 | 35 | 3159–3173 | 2.936° S; 78.842° W |
| L3 | 30 | 3175–3221 | 2.959° S; 78.855° W |
| Sampling Sites | pH | Organic Matter | N | P | K | Mg | Ca | S |
| L1 | 4.9 b | 10.9 a | 0.68 b | 3.6 b | 203.3 b | 425.7 b | 1776.3 b | 846.4 b |
| L2 | 4.8 a | 15.5 a | 0.82 a | 5.9 a | 128.0 a | 162.7 a | 564.7 a | 1012.6 a |
| L3 | 4.6 a | 12.3 a | 0.71 a | 5.3 a | 126.7 a | 177.7 a | 885.3 a | 790.4 a |
| Sampling Sites | Cu | Mn | Zn | Fe | Na | Cl | Al | |
| L1 | 10.5 b | 118.7 b | 16.0 a | 834.3 a | 28.7 a | 11.8 a | 6.4 a | |
| L2 | 8.7 a | 72.7 a | 15.7 a | 926.0 a | 31.3 a | 15.1 a | 6.9 a | |
| L3 | 12.7 a | 107.7 a | 16.0 a | 1197.3 b | 24.3 a | 371.8 b | 7.1 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delgado, E.F.; Valdez, A.T.; Covarrubias, S.A.; Tosi, S.; Nicola, L. Soil Fungal Diversity of the Aguarongo Andean Forest (Ecuador). Biology 2021, 10, 1289. https://doi.org/10.3390/biology10121289
Delgado EF, Valdez AT, Covarrubias SA, Tosi S, Nicola L. Soil Fungal Diversity of the Aguarongo Andean Forest (Ecuador). Biology. 2021; 10(12):1289. https://doi.org/10.3390/biology10121289
Chicago/Turabian StyleDelgado, Ernesto F., Adrián T. Valdez, Sergio A. Covarrubias, Solveig Tosi, and Lidia Nicola. 2021. "Soil Fungal Diversity of the Aguarongo Andean Forest (Ecuador)" Biology 10, no. 12: 1289. https://doi.org/10.3390/biology10121289
APA StyleDelgado, E. F., Valdez, A. T., Covarrubias, S. A., Tosi, S., & Nicola, L. (2021). Soil Fungal Diversity of the Aguarongo Andean Forest (Ecuador). Biology, 10(12), 1289. https://doi.org/10.3390/biology10121289








