SSR-Sequencing Reveals the Inter- and Intraspecific Genetic Variation and Phylogenetic Relationships among an Extensive Collection of Radish (Raphanus) Germplasm Resources
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Material Planting
2.2. Phenotypic Observations
2.3. DNA Extraction
2.4. SSR Primer Design and Genotyping
2.5. Analysis of Genetic Diversity and Phylogenetic Relationships among Different Populations
3. Results
3.1. Classical Classification of the Radish Germplasm Collection Based on Basic Phenotypic Characteristics
3.2. Screening of Core Primers for SSR-Seq
3.3. Genetic Diversity of Radish Germplasm Resources
3.4. Genetic Structure of Radish Germplasm Resources
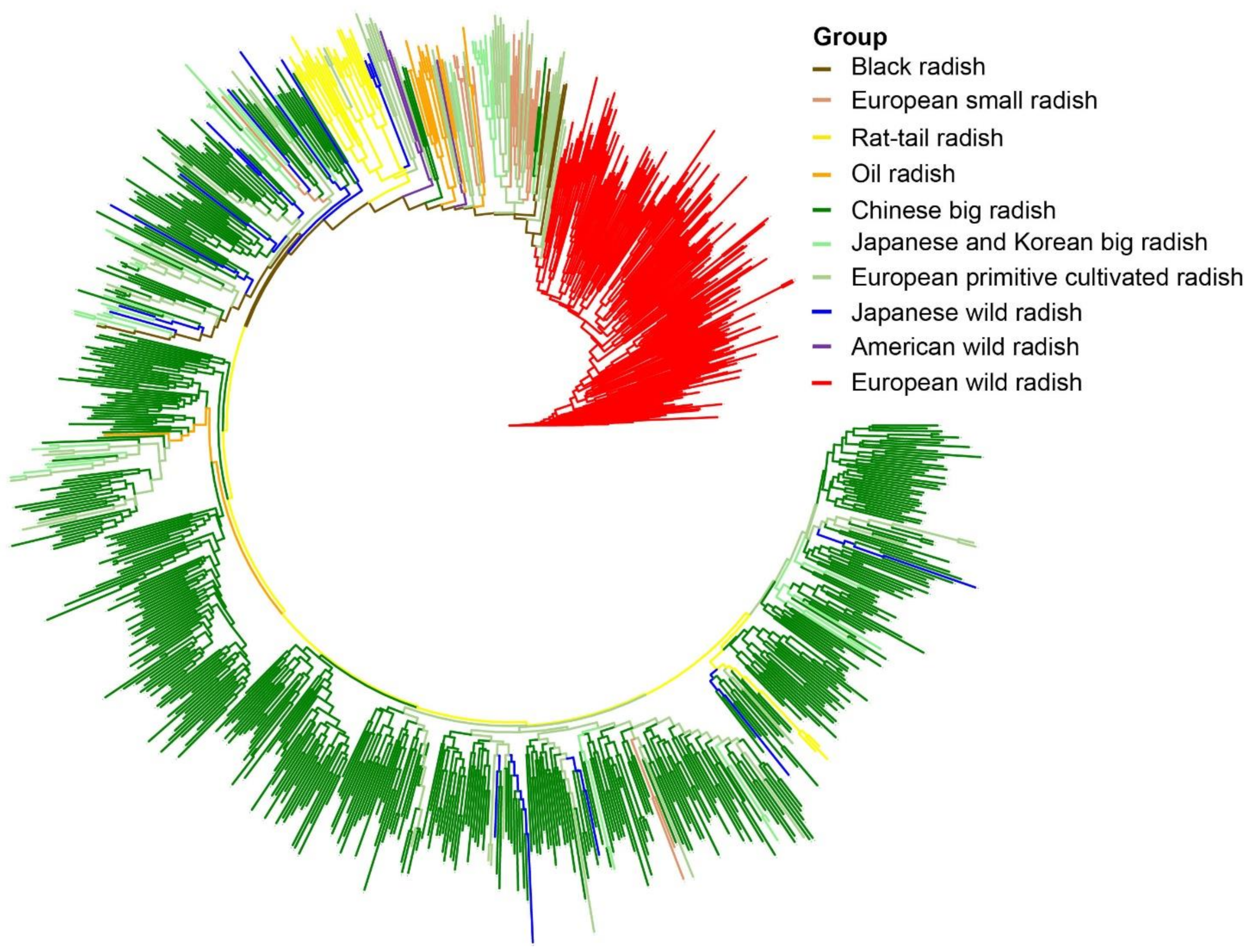
3.5. Phylogenetic Relationships among Radish Germplasm Resources
3.6. Population Differentiation and Gene Flow of Radish Germplasm Resources
4. Discussion
4.1. Development and Application of SSR-Seq Technology Suitable for Genotyping Radish Germplasm Resources
4.2. Radish Origin and Domestication Centers
4.3. Phylogenetic Relationships among Radish Germplasm Resources and Possible Evolutionary Paths
4.4. Taxonomy of R. Sativus var. Raphanistroides, a Semi-Cultivated Type
4.5. The Evolutionary Role of American Wild Radish as a Weed in Competition with Cultivated Crops
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kang, E.S.; Sun, M.H.; Ko, H.C.; Yu, H.J.; Chae, W.B. Reproductive traits and molecular evidence related to the global distribution of cultivated radish (Raphanus sativus L.). Plant. Syst. Evol. 2016, 302, 1367–1380. [Google Scholar] [CrossRef]
- Kitamura, S. Cultivars of Radish and their Change. In Japanese Radish; Japan Society for the Promotion of Science: Tokyo, Japan, 1958; pp. 1–19. [Google Scholar]
- Yamane, K.; Na, L.; Ohnishi, O. Chloroplast DNA variations of cultivated radish and its wild relatives. Plant. Sci. 2005, 168, 627–634. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, L.; Zheng, P. Genetic diversity and evolutionary relationship analyses within and among Raphanus species using EST-SSR markers. Mol. Breed. 2015, 35, 62. [Google Scholar] [CrossRef]
- Lü, N.; Yamane, K.; Ohnishi, O. Genetic diversity of cultivated and wild radish and phylogenetic relations among Raphanus and Brassica species revealed by the analysis of trnK/ matK sequence. Breed. Sci. 2008, 58, 15–22. [Google Scholar] [CrossRef]
- Yamane, K.; Lü, N.; Ohnishi, O. Multiple origins and high genetic diversity of cultivated radish inferred from polymorphism in chloroplast simple sequence repeats. Breed. Sci. 2009, 59, 55–65. [Google Scholar] [CrossRef]
- Muuoz, M.C. La Corola en la Tribu Brassiceae. Anal. Inst. Bot. Cavanilles 1978, 85, 297–334. [Google Scholar]
- Georgescu, M.; Luchian, V.; Groza, O.; Ionescu, N.; Săvulescu, E. Raphanus Raphanistrum Subsp. Landra (Moretti ex DC.) Bonnier & Layens—Adventitious Species of Mediterranean Origin Adapted as Weed in Crops-Some Considerations on Morphological and Anatomical Peculiarities. Agric. Agric. Sci. Procedia 2016, 10, 123–128. [Google Scholar] [CrossRef]
- Warwick, S.I.; Francis, A. The biology of Canadian weeds. 132. Raphanus raphanistrum, L. Can. J. Plant. Sci. 2005, 85, 709–733. [Google Scholar] [CrossRef]
- Ridley, C.E.; Ellstrand, N.C. Rapid evolution of morphology and adaptive life history in the invasive California wild radish (Raphanus sativus) and the implications for management. Evol. Appl. 2010, 3, 64–76. [Google Scholar] [CrossRef]
- Shahal, A.; Ruth, P.O.; Avi, G.; Yehoshua, S.; Itai, O.; Zvi, P. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant. Sci. 2014, 19, 351–360. [Google Scholar] [CrossRef]
- Candolle, A. Origin of Cultivated Plants; Hafner Publisher Company: New York, NY, USA, 1959. [Google Scholar]
- Kole, C. Genome Mapping and Molecular Breeding in Plants. In Vegetables; Springer: Berlin, Germany, 2007; Volume 5. [Google Scholar]
- Kobayashi, H.; Shirasawa, K.; Fukino, N.; Hirakawa, H.; Akanuma, T.; Kitashiba, H. Identification of genome-wide single-nucleotide polymorphisms among geographically diverse radish accessions. DNA Res. 2020, 27, p.dsaa001. [Google Scholar] [CrossRef] [PubMed]
- Schulz, O.E. Part. I: Brassicinae and Raphaninae. In Cruciferae-Brassiceae; Engler, A., Ed.; Verlag von Wilhelm Engelmann: Leipzig, Germany, 1919; pp. 194–210. [Google Scholar]
- Zohary, D.; Hopf, M.; Weiss, E. Domestication of Plants in The Old World: The Origin and Spread of Domesticated Plants in Southwest Asia, Europe, and The Medi-terranean Basin; Oxford University Press on Demand: New York, NY, USA, 2012. [Google Scholar]
- Editorial Committee of Chinese Flora; Chinese Academy of Sciences. Flora Reipublioae Population Sinicae; Science Press: Beijing, China, 1987. [Google Scholar]
- Makino, T. Observation on the flora of Japan. Bot. Mag. 1909, 23, 59–75. [Google Scholar] [CrossRef][Green Version]
- Yamagishi, H.; Terachi, T. Intra- and inter-specific variations in the mitochondrial gene orf138 of Ogura-type male-sterile cytoplasm from Raphanus sativus and Raphanus raphanistrum. Theor. Appl. Genet. 2001, 103, 725–732. [Google Scholar] [CrossRef]
- Yamagishi, H. Assessment of cytoplasmic polymorphisms by PCR-RFLP of mitochondrial orfB region in wild and cultivated radishes (Raphanus). Plant. Breed. 2004, 123, 141–144. [Google Scholar] [CrossRef]
- Kim, N.; Jeong, Y.M.; Jeong, S.; Kim, G.B.; Baek, S.; Kwon, Y.E.; Cho, A.; Choi, S.B.; Kim, J.; Lim, W.J.; et al. Identification of candidate domestication regions in the radish genome based on high-depth resequencing analysis of 17 genotypes. Theor. Appl. Genet. 2016, 129, 1797–1814. [Google Scholar] [CrossRef] [PubMed]
- Ridley, C.E.; Kim, S.C.; Ellstrand, N.C. Bidirectional history of hybridization in California wild radish, Raphanus sativus (Brassicaceae), as revealed by chloroplast DNA. Am. J. Bot. 2008, 95, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Hegde, S.G.; Nason, J.D.; Clegg, J.M.; Ellstrand, N.C. The evolution of California’s wild radish has resulted in the extinction of its progenitors. Evolution 2006, 60, 1187–1197. [Google Scholar] [CrossRef]
- Wang, N.; Kitamoto, N.; Ohsawa, R.; Fujimura, T. Genetic diversity of radish (Raphanus sativus) germplasms and relationships among worldwide accessions analyzed with AFLP markers. Breed. Sci. 2008, 58, 107–112. [Google Scholar] [CrossRef]
- Kong, Q.; Li, X.; Xiang, C.; Qiu, Y.; Shen, D. Phylogenetic Analysis of Cultivated Radish (Raphanus sativus L.) Germplasm with AFLP. Sci. Agric. Sin. 2005, 38, 1017–1023. [Google Scholar] [CrossRef]
- Toth, G. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000, 10, 967. [Google Scholar] [CrossRef]
- Moumeni, A.; Samadi, B.; Wu, J.; Leung, H. Genetic diversity and relatedness of selected Iranian rice cultivars and disease donors assayed by SSR and Candidate defense markers. Euphytica 2003, 131, 275–284. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ma, Q.; Chen, Y.; Wang, B.; Pei, D. Identification of major walnut cultivars grown in China based on nut phenotypes and SSR marker. Sci. Hortic. 2014, 168, 240–248. [Google Scholar] [CrossRef]
- Song, Y.; Fan, L.; Chen, H.; Zhang, M.; Ma, Q.; Zhang, S.; Wu, J. Identifying genetic diversity and preliminary core collection of Pyrus pyrifolia cultivars by a genome-wide set of SSR markers. Sci. Hortic. 2014, 167, 5–16. [Google Scholar] [CrossRef]
- Mekonnen, F.; Mekbib, F.; Kumar, S.; Ahmed, S.; Sharma, T.R. Molecular diversity and population structure of the Ethiopian Lentil (Lens Culinaris Medikus) genotype assessment using SSR Markers. J. Crop. Sci. Biotech. 2015, 19, 1–15. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, T.; Wang, J.; Chen, Q.; Luo, Y.; Zhang, Y.; Tang, H.-R.; Wang, X.-R. Genetic diversity and population structure in cherry (Cerasus pseudocerasus (Lindl). G. Don) along Longmenshan Fault Zones in China with newly developed SSR markers. Sci. Hortic. 2016, 212, 11–19. [Google Scholar] [CrossRef]
- Mashilo, J.; Shimelis, H.; Odindo, A.; Amelework, B. Genetic diversity and differentiation in citron watermelon [Citrullus lanatus var. citroides] landraces assessed by simple sequence repeat markers. Sci. Hortic. 2017, 214, 99–106. [Google Scholar] [CrossRef]
- Brown, J.; Schnell, R.; Motamayor, J.; Lopes, U.; Kuhn, D. Resistance Gene Mapping for Witches’ Broom Disease in Theobroma cacao L. in an F2 Population using SSR Markers and Candidate Genes. J. Am. Soc. Hortic. Sci. 2005, 130, 519–524. [Google Scholar] [CrossRef]
- Babu, B.K.; Mathur, R.K.; Venu, M.; Shil, S.; Ravichandran, G.; Anita, P.; Bhagya, H.P. Genome-wide association study (GWAS) of major QTLs for bunch and oil yield related traits in Elaeis guineensis L. Plant. Sci. 2021, 305, 110810. [Google Scholar] [CrossRef]
- Sarhanova, P.; Pfanzelt, S.; Brandt, R.; Himmelbach, A.; Blattner, F.R. SSR-seq: Genotyping of microsatellites using next-generation sequencing reveals higher level of polymorphism as compared to traditional fragment size scoring. Ecol. Evol. 2018, 8, 10817–10833. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; de Lorenzis, G.; Velasco, D.; Koehmstedt, A.; Maghradze, D.; Bobokashvili, Z.; Musayev, M.; Zdunic, G.; Laucou, V.; Andrew Walker, M.; et al. Genetic diversity analysis of cultivated and wild grapevine (Vitis vinifera L.) accessions around the Mediterranean basin and Central Asia. BMC Plant. Biol. 2018, 18, 137. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, J.; Han, R.; Zhang, F.; Mao, A.; Luo, J.; Dong, B.; Liu, H.; Tang, H.; Wen, C.; et al. Target SSR-Seq: A Novel SSR Genotyping Technology Associate with Perfect SSRs in Genetic Analysis of Cucumber Varieties. Front. Plant. Sci. 2019, 10, 531. [Google Scholar] [CrossRef]
- Lepais, O.; Chancerel, E.; Boury, C.; Salin, F.; Manicki, A.; Taillebois, L.; Dutech, C.; Aissi, A.; Bacles, C.F.E.; Daverat, F.; et al. Fast sequence-based microsatellite genotyping development workflow. PeerJ 2020, 8, e9085. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Li, C.; Qin, S.; Huang, Z.; Gan, B.; Jiang, Z.; Huang, X.; Yang, X.; Li, Q.; Xiang, X.; et al. High-throughput sequencing-based microsatellite genotyping for polyploids to resolve allele dosage uncertainty and improve analyses of genetic diversity, structure and differentiation: A case study of the hexaploid Camellia oleifera. Mol. Ecol. Resources 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shen, D. Descriptors and Data Standard for Radish (Raphanus Sativus L.); China Agriculture Press: Beijing, China, 2008. [Google Scholar]
- Doyle, J. DNA protocols for plants-CTAB total DNA isolation. In Molecular Techniques in Taxonomy; Springer Nature: Basingstoke, UK, 1991. [Google Scholar]
- Zhang, X.; Yu, Z.; Mei, S.; Qiu, Y.; Yang, X.; Chen, X.; Cheng, F.; Wu, Z.; Sun, Y.; Jing, Y.; et al. A de novo Genome of a Chinese Radish Cultivar. Hortic. Plant. J. 2015, 1, 155–164. [Google Scholar] [CrossRef]
- Bruvo, R.; Michiels, N.K.; D’Souza, T.G.; Schulenburg, H. A simple method for the calculation of microsatellite genotype distance irrespective of ploidy level. Mol. Ecol. 2004, 13, 2101–2106. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, S.; Lucas, L.K.; Nice, C.C.; Fordyce, J.A.; Forister, M.L.; Gompert, Z. The predictability of genomic changes underlying a recent host shift in Melissa blue butterflies. Mol. Ecol. 2018, 27, 2651–2666. [Google Scholar] [CrossRef]
- Yamagishi, H.; Tateishi, M.; Terachi, T.; Murayama, S. Genetic Relationships among Japanese Wild Radishes (Raphanus sativus f. raphanistroides Makino), Cultivated Radishes and R. raphanistrum Revealed by RAPD Analysis. J. Jpn. Soc. Hortic. Sci. 1998, 67, 526–531. [Google Scholar] [CrossRef]
- Fukushima, K.; Kanomata, T.; Kon, A.; Masuko-Suzuki, H.; Ito, K.; Ogata, S.; Takada, Y.; Komatsubara, Y.; Nakamura, T.; Watanabe, T.; et al. Spatiogenetic characterization of S receptor kinase (SRK) alleles in naturalized populations of Raphanus sativus L. var. raphanistroides on Yakushima island. Genes Genet. Syst. 2021, 96, 20–00066. [Google Scholar] [CrossRef] [PubMed]
- Chamberlin, K. Molecular Cloning of an SSR Marker Associated with Resistance to Sclerotinia Blight in Peanut and Sequence Variation among Resistant and Susceptible Plant Lines. In Proceeding of the American Peanut Research and Education Society, Young Harris, GA, USA, 8–11 July 2013; Wheat Peanut and other Field Crops Research Unit: Stillwater, OK, USA, 2013. [Google Scholar]
- Tanksley, S.D.; Mccouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef]
- Neale, D.B.; Saghai-Maroof, M.A.; Allard, R.W.; Zhang, Q.; Jorgensen, R.A. Chloroplast DNA diversity in populations of wild and cultivated barley. Genetics 1988, 120, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Abe, J.; Gai, J.; Shimamoto, Y. Diversity of chloroplast DNA SSRs in wild and cultivated soybeans: Evidence for multiple origins of cultivated soybean. Theor. Appl. Genet. 2002, 105, 645–653. [Google Scholar] [CrossRef]
- Dane, F.; Lang, P.; Bakhtiyarova, R. Comparative analysis of chloroplast DNA variability in wild and cultivated Citrullus species. Theor. Appl. Genet. 2004, 108, 958–966. [Google Scholar] [CrossRef]
- Vavilov, N.I. The theory of the origin of cultivated plants according to Darwin. Z. landw. VersWes. Bulg. 1940, 10, 1–31. [Google Scholar]
- Perrino, P. The diversity in Vavilov’s Mediterranean Gene Center. Die Kult. 1988, 36, 85–105. [Google Scholar] [CrossRef]
- Hummer, K.; Hancock, J. Vavilovian Centers of Plant Diversity: Implications and Impacts. HortScience 2015, 50, 780–783. [Google Scholar] [CrossRef]
- Parsons, W.T.; Cuthbertson, E.G. Noxious Weeds of Australia; CSIRO Publishing: Collingwood, Australia, 1992. [Google Scholar]
- Huh, M.K.; Ohnishi, O. Genetic diversity and genetic relationships of East Asian natural populations of wild radish revealed by AFLP. Breed. Sci. 2002, 52, 79–88. [Google Scholar] [CrossRef]
- Iriondo, J.M.; Milla, R.; Volis, S.; Rubio de Casas, R. Reproductive traits and evolutionary divergence between Mediterranean crops and their wild relatives. Plant. Biol. 2018, 20, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Takanori, O.; Masashi, H.; Makoto, Y. Spatial structure of microsatellite variability within and among populations of wild radish Raphanus sativus L. var. hortensis Backer f. raphanistroides Makino (Brassicaceae) in Japan. Breed. Sci. 2010, 60, 195–202. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, T.; Wang, J.; Wang, P.; Qiu, Y.; Zhao, W.; Pang, S.; Li, X.; Wang, H.; Song, J.; et al. Pan-genome of Raphanus highlights genetic variation and introgression among domesticated, wild and weedy radishes. Mol. Plant. 2021. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, H.; Terachi, T. Multiple origins of cultivated radishes as evidenced by a comparison of the structural variations in mitochondrial DNA of Raphanus. Genome 2003, 46, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Higashi, H.; Mitsui, Y.; Setoguchi, H. Distinct Phylogeographic Structures of Wild Radish (Raphanus sativus L. var. raphanistroides Makino) in Japan. PLoS ONE 2015, 10, e0135132. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, A.; Tack, D.; Lale, A.; Goldston Josh Caple, M.; Conner, E. Weed evolution: Genetic differentiation among wild, weedy, and crop radish. Evol. Appl. 2018, 11, 1964–1974. [Google Scholar] [CrossRef] [PubMed]



| Index | EWR | AWR | ESR | OR | RTR | BR | EPCR | JKBR | JWR | CBR | ACR | ECR | AR | Max | Min | Average |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accessions | 201 | 3 | 18 | 16 | 26 | 5 | 60 | 47 | 17 | 546 | 618 | 100 | 939 | |||
| Na | 9.34 | 1.92 | 4.05 | 4.32 | 4.08 | 2.58 | 6.63 | 5.32 | 3.84 | 8.08 | 8.66 | 7.53 | 11.16 | 11.16 | 1.92 | 5.95 |
| Ne | 4.40 | 1.75 | 2.67 | 2.81 | 2.39 | 2.22 | 3.58 | 2.81 | 2.58 | 2.47 | 2.56 | 3.63 | 3.33 | 4.4 | 1.75 | 2.86 |
| Ho | 0.28 | 0.14 | 0.17 | 0.43 | 0.19 | 0.12 | 0.24 | 0.17 | 0.22 | 0.33 | 0.32 | 0.24 | 0.30 | 0.43 | 0.12 | 0.24 |
| He | 0.71 | 0.42 | 0.59 | 0.61 | 0.53 | 0.53 | 0.68 | 0.61 | 0.57 | 0.55 | 0.57 | 0.69 | 0.67 | 0.71 | 0.42 | 0.59 |
| Nei | 0.70 | 0.34 | 0.57 | 0.58 | 0.51 | 0.46 | 0.67 | 0.6 | 0.54 | 0.55 | 0.57 | 0.69 | 0.67 | 0.7 | 0.34 | 0.57 |
| I | 1.59 | 0.52 | 1.05 | 1.10 | 0.96 | 0.77 | 1.39 | 1.16 | 1.00 | 1.08 | 1.13 | 1.43 | 1.45 | 1.59 | 0.52 | 1.12 |
| Indexes | OP | WCR | ACR | ECR |
|---|---|---|---|---|
| Fis | 0.60 | 0.58 | 0.46 | 0.56 |
| Fit | 0.68 | 0.65 | 0.57 | 0.68 |
| Fst | 0.21 | 0.18 | 0.19 | 0.28 |
| Nm | 1.07 | 1.17 | 1.04 | 0.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, J.; Qiu, Y.; Wang, H.; Wang, P.; Zhang, X.; Li, C.; Song, J.; Gui, W.; Shen, D.; et al. SSR-Sequencing Reveals the Inter- and Intraspecific Genetic Variation and Phylogenetic Relationships among an Extensive Collection of Radish (Raphanus) Germplasm Resources. Biology 2021, 10, 1250. https://doi.org/10.3390/biology10121250
Li X, Wang J, Qiu Y, Wang H, Wang P, Zhang X, Li C, Song J, Gui W, Shen D, et al. SSR-Sequencing Reveals the Inter- and Intraspecific Genetic Variation and Phylogenetic Relationships among an Extensive Collection of Radish (Raphanus) Germplasm Resources. Biology. 2021; 10(12):1250. https://doi.org/10.3390/biology10121250
Chicago/Turabian StyleLi, Xiaoman, Jinglei Wang, Yang Qiu, Haiping Wang, Peng Wang, Xiaohui Zhang, Caihua Li, Jiangping Song, Wenting Gui, Di Shen, and et al. 2021. "SSR-Sequencing Reveals the Inter- and Intraspecific Genetic Variation and Phylogenetic Relationships among an Extensive Collection of Radish (Raphanus) Germplasm Resources" Biology 10, no. 12: 1250. https://doi.org/10.3390/biology10121250
APA StyleLi, X., Wang, J., Qiu, Y., Wang, H., Wang, P., Zhang, X., Li, C., Song, J., Gui, W., Shen, D., Yang, W., Cai, B., Liu, L., & Li, X. (2021). SSR-Sequencing Reveals the Inter- and Intraspecific Genetic Variation and Phylogenetic Relationships among an Extensive Collection of Radish (Raphanus) Germplasm Resources. Biology, 10(12), 1250. https://doi.org/10.3390/biology10121250









