Systematic Studies of the Circadian Clock Genes Impact on Temperature Compensation and Cell Proliferation Using CRISPR Tools
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Single Guide RNA Design
2.3. Construction of Vectors and Cell Lines Screening
2.4. Identification of Knock-Out Cell Lines
2.5. Off-Target Analysis
2.6. Bioluminescence Recording
2.7. Immuno-Blotting Assays
2.8. Cell Proliferation Assay
3. Results
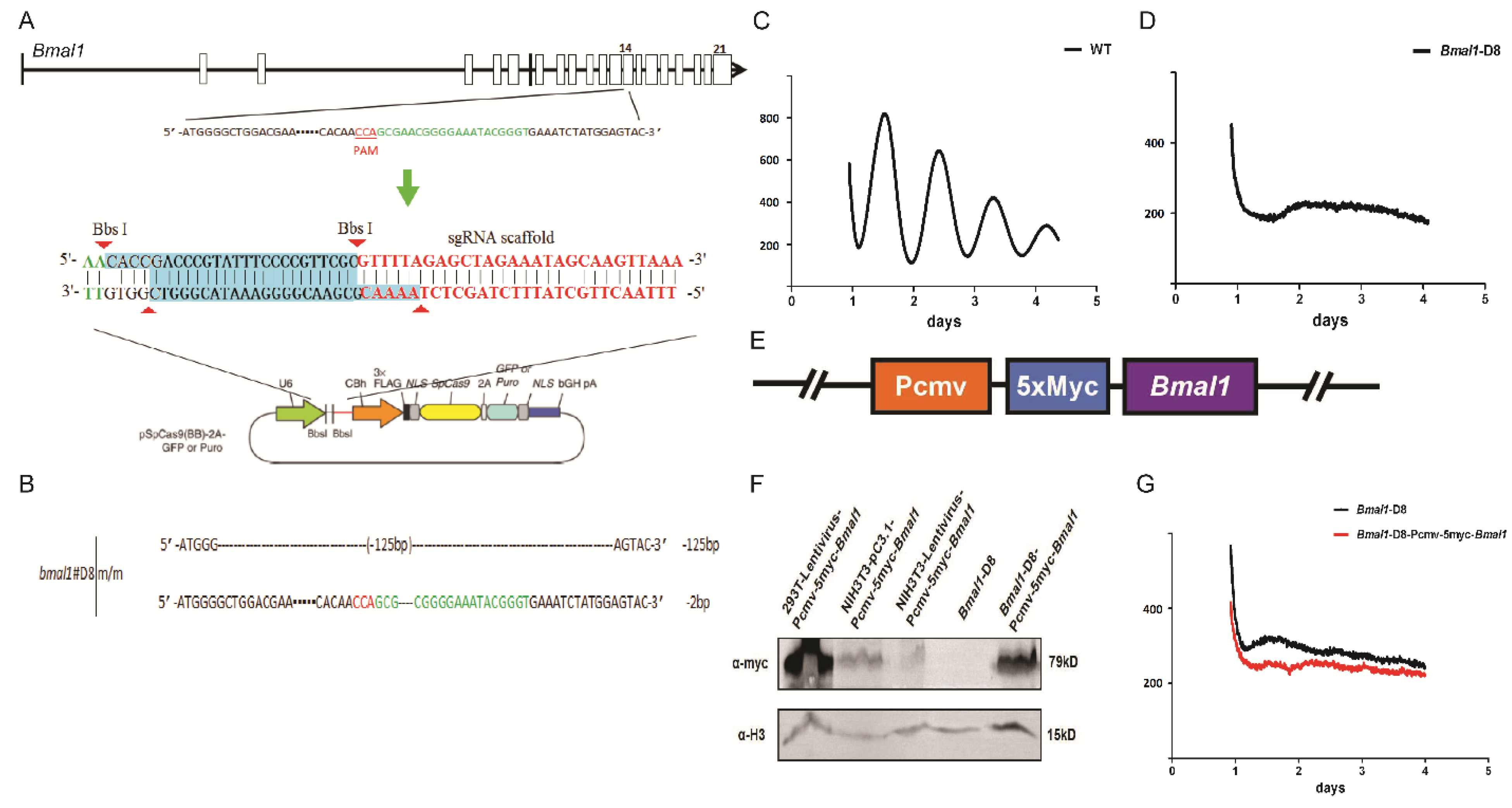
3.1. Cellular Circadian Gene Bmal1 Deletion by the CRISPR System Resulted in Loss of Circadian Rhythms
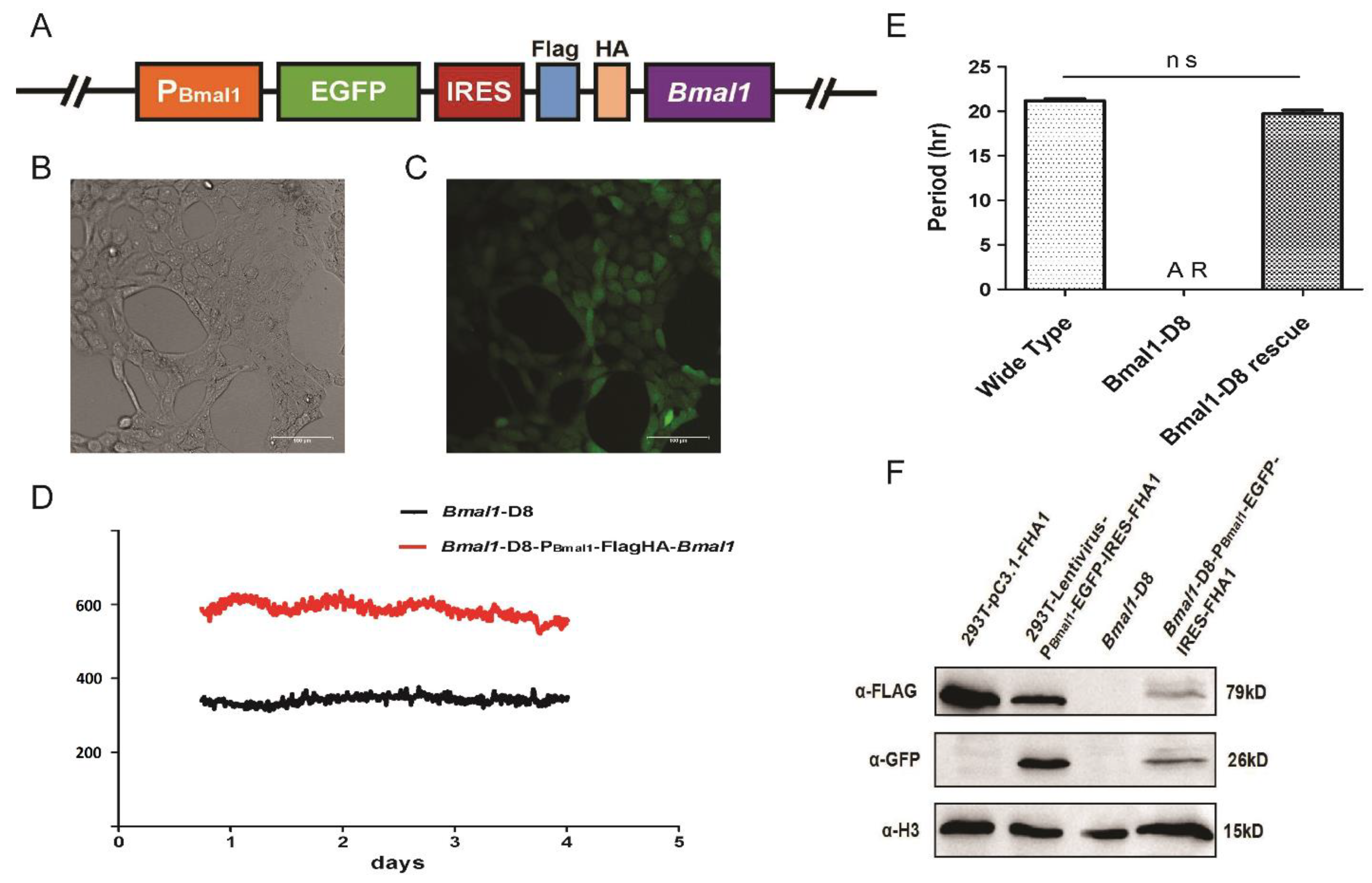
3.2. The Expression of Exogenous Bmal1 under a Bmal1 Promoter Can Rescue the Rhythm in Bmal-D8 Cells
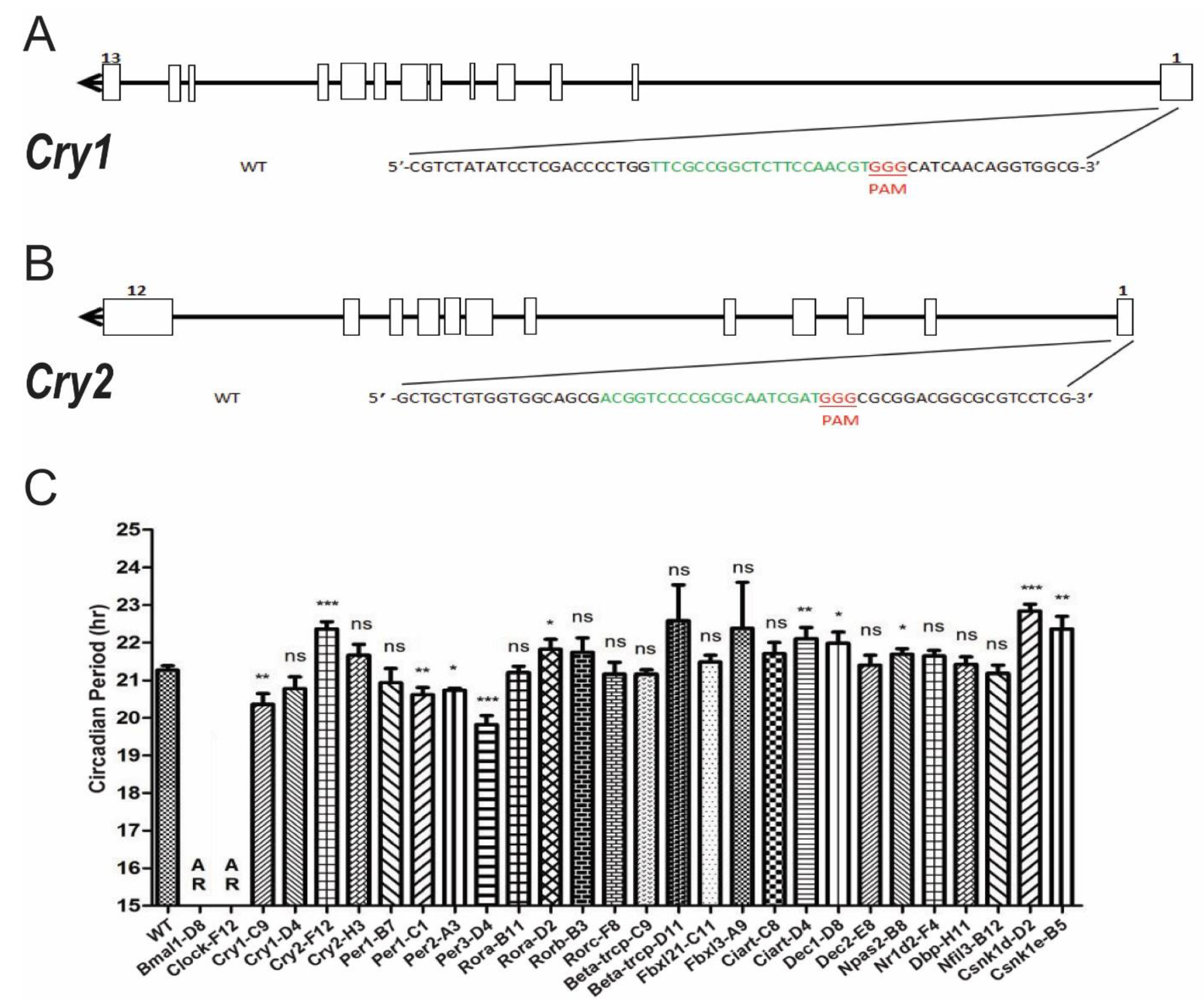
3.3. Systematic Knocking out Clock Genes Using CRISPR/Cas9
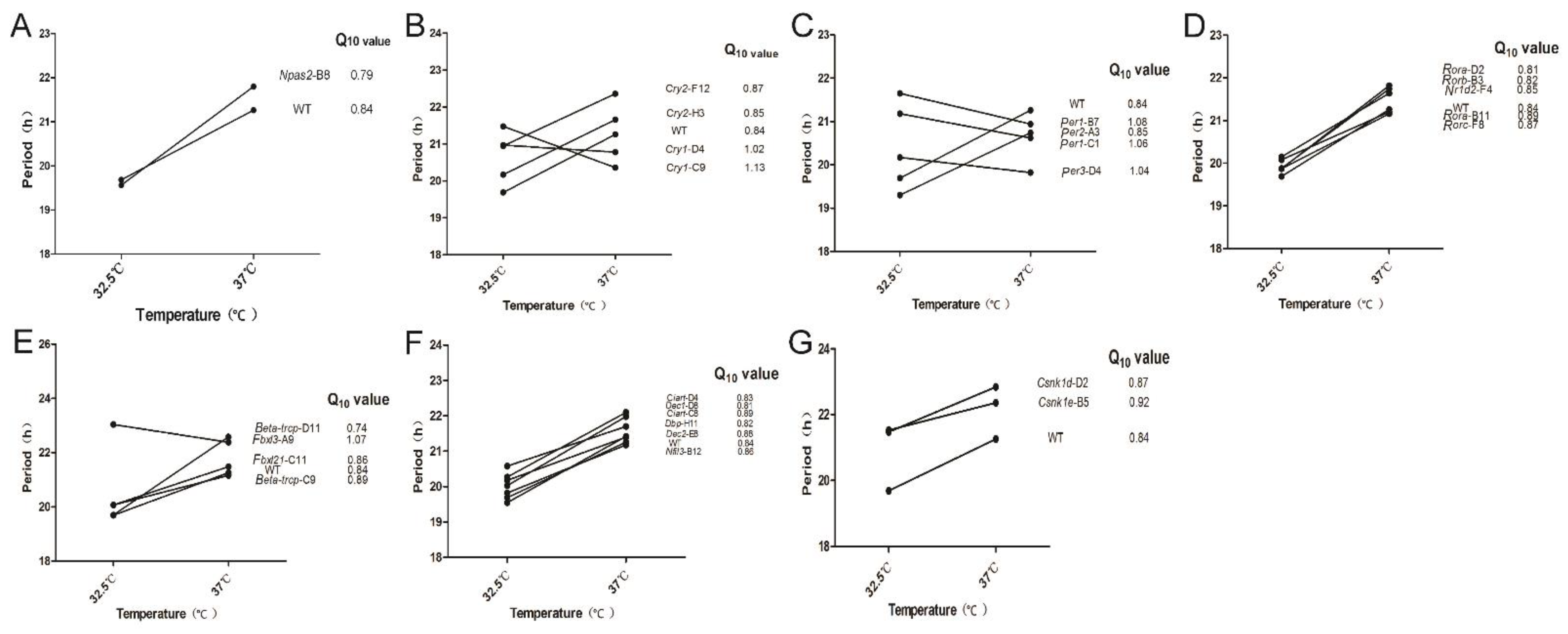
3.4. Temperature Compensation Evaluated in Cells Which Have Systematically Knocked out Clock Genes
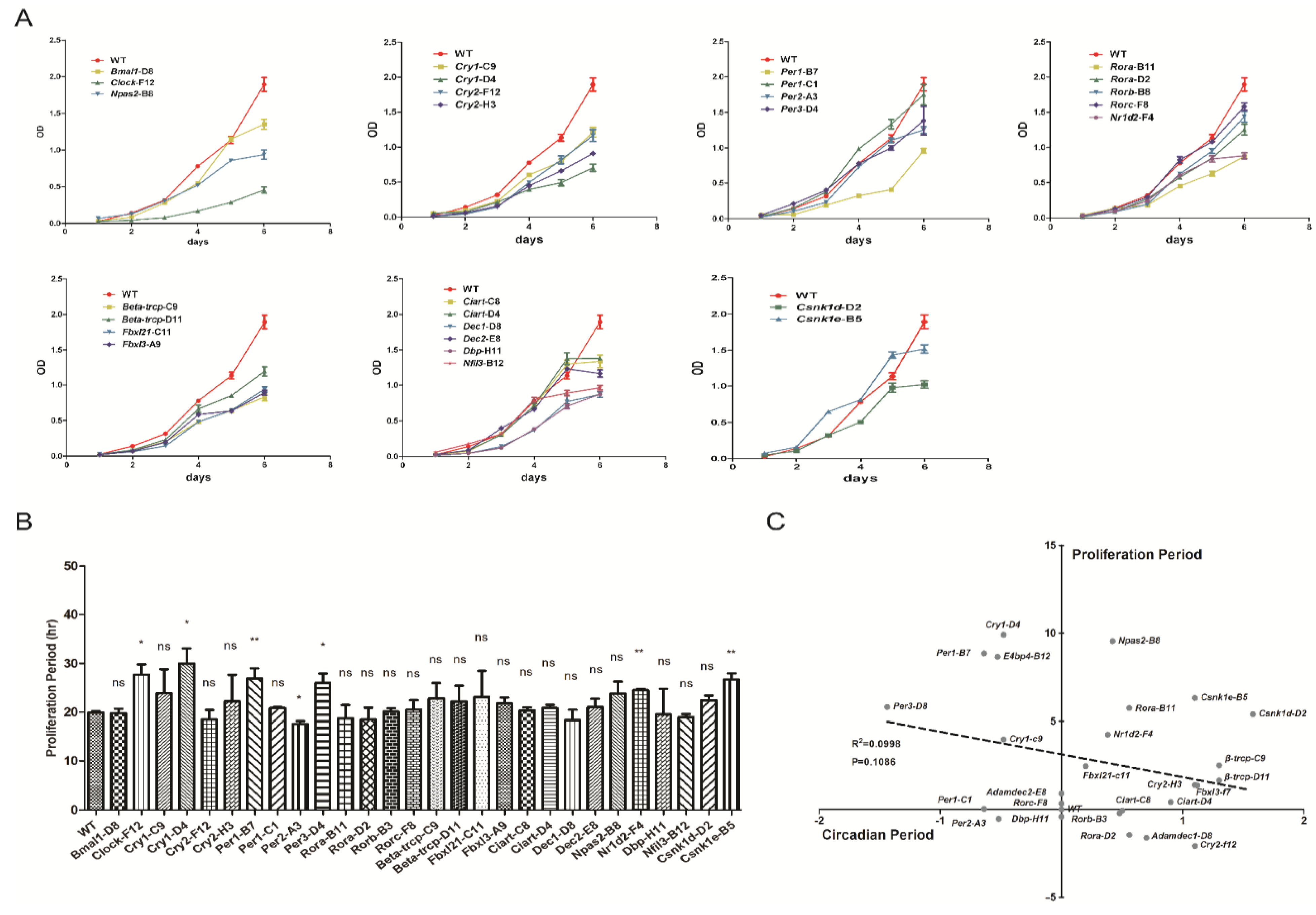
3.5. Proliferation Rates Evaluated in Cells Which Have Systematically Knocked out Clock Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Albrecht, U. Timing to perfection: The biology of central and peripheral circadian clocks. Neuron 2012, 74, 246–260. [Google Scholar] [CrossRef] [PubMed]
- Allada, R.; Chung, B.Y. Circadian organization of behavior and physiology in Drosophila. Annu. Rev. Physiol. 2010, 72, 605–624. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Brown, S.A.; Marcacci, L.; Tronche, F.; Kellendonk, C.; Reichardt, H.M.; Schütz, G.; Schibler, U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 2000, 289, 2344–2347. [Google Scholar] [CrossRef]
- McNamara, P.; Seo, S.B.; Rudic, R.D.; Sehgal, A.; Chakravarti, D.; FitzGerald, G.A. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: A humoral mechanism to reset a peripheral clock. Cell 2001, 105, 877–889. [Google Scholar] [CrossRef]
- Qin, X.; Mori, T.; Zhang, Y.; Johnson, C.H. PER2 Differentially Regulates Clock Phosphorylation versus Transcription by Reciprocal Switching of CK1ε Activity. J. Biol. Rhythm. 2015, 30, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.W.; Laval-Martin, D.L.; Edmunds, L.N., Jr. Cell cycle oscillators. Temperature compensation of the circadian rhythm of cell division in Euglena. Exp. Cell Res. 1985, 157, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Ruby, N.F.; Heller, H.C. Temperature sensitivity of the suprachiasmatic nucleus of ground squirrels and rats in vitro. J. Biol. Rhythm. 1996, 11, 126–136. [Google Scholar] [CrossRef]
- Lakin-Thomas, P.L. Choline depletion, frq mutations, and temperature compensation of the circadian rhythm in Neurospora crassa. J. Biol. Rhythm. 1998, 13, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.J.; Curtin, K.D.; Rosbash, M. PER protein interactions and temperature compensation of a circadian clock in Drosophila. Science 1995, 267, 1169–1172. [Google Scholar] [CrossRef]
- Tosini, G.; Menaker, M. The tau mutation affects temperature compensation of hamster retinal circadian oscillators. Neuroreport 1998, 9, 1001–1005. [Google Scholar] [CrossRef]
- Isojima, Y.; Nakajima, M.; Ukai, H.; Fujishima, H.; Yamada, R.G.; Masumoto, K.H.; Kiuchi, R.; Ishida, M.; Ukai-Tadenuma, M.; Minami, Y.; et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2009, 106, 15744–15749. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, Y.; Koyama, Y.M.; Ukai-Tadenuma, M.; Hirokawa, T.; Kikuchi, M.; Yamada, R.G.; Ukai, H.; Fujishima, H.; Umehara, T.; Tainaka, K.; et al. Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock. Mol. Cell 2017, 67, 783–798.e20. [Google Scholar] [CrossRef] [PubMed]
- Terauchi, K.; Kitayama, Y.; Nishiwaki, T.; Miwa, K.; Murayama, Y.; Oyama, T.; Kondo, T. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. USA 2007, 104, 16377–16381. [Google Scholar] [CrossRef] [PubMed]
- Ito-Miwa, K.; Furuike, Y.; Akiyama, S.; Kondo, T. Tuning the circadian period of cyanobacteria up to 6.6 days by the single amino acid substitutions in KaiC. Proc. Natl. Acad. Sci. USA 2020, 117, 20926–20931. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M.; Moriya, T.; Shibata, S. Physiological, pharmacological and molecular aspects of mammalian biological clocks. Nihon Yakurigaku Zasshi. Folia Pharmacol. Jpn. 1998, 112, 243–250. [Google Scholar] [CrossRef]
- Kondratov, R.V.; Shamanna, R.K.; Kondratova, A.A.; Gorbacheva, V.Y.; Antoch, M.P. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2006, 20, 530–532. [Google Scholar] [CrossRef]
- Edery, I. Circadian rhythms in a nutshell. Physiol. Genom. 2000, 3, 59–74. [Google Scholar] [CrossRef]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef]
- Buhr, E.D.; Takahashi, J.S. Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. 2013, 217, 3–27. [Google Scholar] [CrossRef]
- Gustafson, C.L.; Partch, C.L. Emerging models for the molecular basis of mammalian circadian timing. Biochemistry 2015, 54, 134–149. [Google Scholar] [CrossRef]
- Huang, N.; Chelliah, Y.; Shan, Y.; Taylor, C.A.; Yoo, S.H.; Partch, C.; Green, C.B.; Zhang, H.; Takahashi, J.S. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 2012, 337, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A.; Ripperger, J.; Kadener, S.; Fleury-Olela, F.; Vilbois, F.; Rosbash, M.; Schibler, U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 2005, 308, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Duong, H.A.; Robles, M.S.; Knutti, D.; Weitz, C.J. A molecular mechanism for circadian clock negative feedback. Science 2011, 332, 1436–1439. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Etchegaray, J.P.; Cagampang, F.R.; Loudon, A.S.; Reppert, S.M. Posttranslational mechanisms regulate the mammalian circadian clock. Cell 2001, 107, 855–867. [Google Scholar] [CrossRef]
- Ye, R.; Selby, C.P.; Ozturk, N.; Annayev, Y.; Sancar, A. Biochemical analysis of the canonical model for the mammalian circadian clock. J. Biol. Chem. 2011, 286, 25891–25902. [Google Scholar] [CrossRef] [PubMed]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar] [CrossRef]
- Stratmann, M.; Stadler, F.; Tamanini, F.; van der Horst, G.T.; Ripperger, J.A. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by CRYPTOCHROME1. Genes Dev. 2010, 24, 1317–1328. [Google Scholar] [CrossRef]
- Eide, E.J.; Vielhaber, E.L.; Hinz, W.A.; Virshup, D.M. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J. Biol. Chem. 2002, 277, 17248–17254. [Google Scholar] [CrossRef]
- Keesler, G.A.; Camacho, F.; Guo, Y.; Virshup, D.; Mondadori, C.; Yao, Z. Phosphorylation and destabilization of human period I clock protein by human casein kinase I epsilon. Neuroreport 2000, 11, 951–955. [Google Scholar] [CrossRef]
- Lowrey, P.L.; Shimomura, K.; Antoch, M.P.; Yamazaki, S.; Zemenides, P.D.; Ralph, M.R.; Menaker, M.; Takahashi, J.S. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 2000, 288, 483–492. [Google Scholar] [CrossRef]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Siepka, S.M.; Yoo, S.H.; Park, J.; Song, W.; Kumar, V.; Hu, Y.; Lee, C.; Takahashi, J.S. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 2007, 129, 1011–1023. [Google Scholar] [CrossRef]
- Hirano, A.; Yumimoto, K.; Tsunematsu, R.; Matsumoto, M.; Oyama, M.; Kozuka-Hata, H.; Nakagawa, T.; Lanjakornsiripan, D.; Nakayama, K.I.; Fukada, Y. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 2013, 152, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Shirogane, T.; Jin, J.; Ang, X.L.; Harper, J.W. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J. Biol. Chem. 2005, 280, 26863–26872. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef] [PubMed]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Mitsui, S.; Yan, L.; Yagita, K.; Miyake, S.; Okamura, H. Role of DBP in the circadian oscillatory mechanism. Mol. Cell. Biol. 2000, 20, 4773–4781. [Google Scholar] [CrossRef]
- Oishi, K.; Miyazaki, K.; Kadota, K.; Kikuno, R.; Nagase, T.; Atsumi, G.; Ohkura, N.; Azama, T.; Mesaki, M.; Yukimasa, S.; et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 2003, 278, 41519–41527. [Google Scholar] [CrossRef]
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98, 193–205. [Google Scholar] [CrossRef]
- Fukada, Y.; Okano, T. Circadian clock system in the pineal gland. Mol. Neurobiol. 2002, 25, 19–30. [Google Scholar] [CrossRef]
- Annayev, Y.; Adar, S.; Chiou, Y.Y.; Lieb, J.D.; Sancar, A.; Ye, R. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J. Biol. Chem. 2014, 289, 5013–5024. [Google Scholar] [CrossRef] [PubMed]
- Price, J.L.; Blau, J.; Rothenfluh, A.; Abodeely, M.; Kloss, B.; Young, M.W. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 1998, 94, 83–95. [Google Scholar] [CrossRef]
- Altman, B.J.; Hsieh, A.L.; Sengupta, A.; Krishnanaiah, S.Y.; Stine, Z.E.; Walton, Z.E.; Gouw, A.M.; Venkataraman, A.; Li, B.; Goraksha-Hicks, P.; et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015, 22, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Zhang, F. Dissecting neural function using targeted genome engineering technologies. ACS Chem. Neurosci. 2012, 3, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Haurwitz, R.E.; Doudna, J.A. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 2012, 18, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Traynard, P.; Feillet, C.; Soliman, S.; Delaunay, F.; Fages, F. Model-based investigation of the circadian clock and cell cycle coupling in mouse embryonic fibroblasts: Prediction of RevErb-α up-regulation during mitosis. Bio. Syst. 2016, 149, 59–69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gao, L.; Chen, H.; Li, C.; Xiao, Y.; Yang, D.; Zhang, M.; Zhou, D.; Liu, W.; Wang, A.; Jin, Y. ER stress activation impairs the expression of circadian clock and clock-controlled genes in NIH3T3 cells via an ATF4-dependent mechanism. Cell. Signal. 2019, 57, 89–101. [Google Scholar] [CrossRef]
- Yeom, M.; Pendergast, J.S.; Ohmiya, Y.; Yamazaki, S. Circadian-independent cell mitosis in immortalized fibroblasts. Proc. Natl. Acad. Sci. USA 2010, 107, 9665–9670. [Google Scholar] [CrossRef]
- Bieler, J.; Cannavo, R.; Gustafson, K.; Gobet, C.; Gatfield, D.; Naef, F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 2014, 10, 739. [Google Scholar] [CrossRef] [PubMed]
- van der Horst, G.T.; Muijtjens, M.; Kobayashi, K.; Takano, R.; Kanno, S.; Takao, M.; De Wit, J.; Verkerk, A.; Eker, A.P.; Van Leenen, D.; et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 1999, 398, 627–630. [Google Scholar] [CrossRef]
- Kitanishi, K.; Igarashi, J.; Hayasaka, K.; Hikage, N.; Saiful, I.; Yamauchi, S.; Uchida, T.; Ishimori, K.; Shimizu, T. Heme-binding characteristics of the isolated PAS-A domain of mouse Per2, a transcriptional regulatory factor associated with circadian rhythms. Biochemistry 2008, 47, 6157–6168. [Google Scholar] [CrossRef] [PubMed]
- Cermakian, N.; Monaco, L.; Pando, M.P.; Dierich, A.; Sassone-Corsi, P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001, 20, 3967–3974. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Larkin, D.W.; Albrecht, U.; Sun, Z.S.; Sage, M.; Eichele, G.; Lee, C.C.; Bradley, A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 1999, 400, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Shearman, L.P.; Jin, X.; Lee, C.; Reppert, S.M.; Weaver, D.R. Targeted disruption of the mPer3 gene: Subtle effects on circadian clock function. Mol. Cell. Biol. 2000, 20, 6269–6275. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.O.; Pappu, B.P.; Nurieva, R.; Akimzhanov, A.; Kang, H.S.; Chung, Y.; Ma, L.; Shah, B.; Panopoulos, A.D.; Schluns, K.S.; et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 2008, 28, 29–39. [Google Scholar] [CrossRef]
- Liu, A.C.; Tran, H.G.; Zhang, E.E.; Priest, A.A.; Welsh, D.K.; Kay, S.A. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008, 4, e1000023. [Google Scholar] [CrossRef]
- Pourcet, B.; Zecchin, M.; Ferri, L.; Beauchamp, J.; Sitaula, S.; Billon, C.; Delhaye, S.; Vanhoutte, J.; Mayeuf-Louchart, A.; Thorel, Q.; et al. Nuclear Receptor Subfamily 1 Group D Member 1 Regulates Circadian Activity of NLRP3 Inflammasome to Reduce the Severity of Fulminant Hepatitis in Mice. Gastroenterology 2018, 154, 1449–1464.e20. [Google Scholar] [CrossRef]
- Pittendrigh, C.S. On temperature independence in the clock system controlling emergence time in Drosophila. Proc. Natl. Acad. Sci. USA 1954, 40, 1018–1029. [Google Scholar] [CrossRef]
- Goriki, A.; Hatanaka, F.; Myung, J.; Kim, J.K.; Yoritaka, T.; Tanoue, S.; Abe, T.; Kiyonari, H.; Fujimoto, K.; Kato, Y.; et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 2014, 12, e1001839. [Google Scholar] [CrossRef] [PubMed]
- Honma, S.; Kawamoto, T.; Takagi, Y.; Fujimoto, K.; Sato, F.; Noshiro, M.; Kato, Y.; Honma, K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature 2002, 419, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Ohsaki, K.; Oishi, K.; Kozono, Y.; Nakayama, K.; Nakayama, K.I.; Ishida, N. The role of {beta}-TrCP1 and {beta}-TrCP2 in circadian rhythm generation by mediating degradation of clock protein PER2. J. Biochem. 2008, 144, 609–618. [Google Scholar] [CrossRef]
- Barnes, J.W.; Tischkau, S.A.; Barnes, J.A.; Mitchell, J.W.; Burgoon, P.W.; Hickok, J.R.; Gillette, M.U. Requirement of mammalian Timeless for circadian rhythmicity. Science 2003, 302, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Eng, G.W.L.; Virshup, D.M. Site-specific phosphorylation of casein kinase 1 δ (CK1δ) regulates its activity towards the circadian regulator PER2. PLoS ONE 2017, 12, e0177834. [Google Scholar] [CrossRef]
- Meng, Q.J.; Logunova, L.; Maywood, E.S.; Gallego, M.; Lebiecki, J.; Brown, T.M.; Sládek, M.; Semikhodskii, A.S.; Glossop, N.R.J.; Piggins, H.D.; et al. Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 2008, 58, 78–88. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Umemura, Y.; Minami, Y.; Koike, N.; Hosokawa, T.; Hara, M.; Ito, H.; Inokawa, H.; Yagita, K. Effect of Multiple Clock Gene Ablations on the Circadian Period Length and Temperature Compensation in Mammalian Cells. J. Biol. Rhythm. 2016, 31, 48–56. [Google Scholar] [CrossRef]
| Genes | Alleles | Mouse Mutant Phenotypes | Cellular Mutation Phenotypes |
|---|---|---|---|
| Bmal1 | Bmal1−/− | Rhythm disorder [39] | Rhythm disorder |
| Clock | Clock∆19/∆19 | Rhythm disorder [39] | Rhythm disorder |
| Npas2 | Npas2−/− | Shorten 0.2 h [53] | Lengthen 0.42 h |
| Cry1 | Cry1−/− | Shorten 0.5 h [52] | Shorten 0.48 h |
| Cry2 | Cry2−/− | Lengthen 1 h [52] | Lengthen 1.1 h |
| Per1 | Per1−/− | Shorten 0.5 h [54] | Shorten 0.64 h |
| Per2 | Per2brdml | Shorten 1.5 h [55] | Shorten 0.52 h |
| Per3 | Per3−/− | Shorten 0–0.5 h [56] | Shorten 1.44 h |
| Rora | Staggerer | Shorten 0.5 h [35] | Lengthen 0.56 h |
| Rorb | Rorb−/− | Lengthen 0.5 h [57] | Lengthen 0.48 h |
| Rorc | Rorc−/− | WT [58] | WT |
| Nr1d1 | Nr1d1−/− | Shorten 0.5 h [59] | No cell lines |
| Nr1d2 | Nr1d2−/− | N/A | Lengthen 0.38 h |
| Dbp | Dbp−/− | Shorten 0.5 h [60] | WT |
| Nfil3 | Nfil3−/−Nfile3 | N/A | WT |
| Ciart | Ciart−/− | Lengthen [61] | Lengthen 0.5–0.9 h |
| Dec1 | Dec1−/− | Lengthen 0–0.15 h [62] | Lengthen 0.7 h |
| Dec2 | Dec2−/− | WT [62] | WT |
| Beta-trcp | Beta-trcp−/− | WT [63] | WT/Lengthen 1.3 h |
| Fbxl21 | Fbxl21−/− | WT [33] | Lengthen 0.2 h |
| Fbxl3 | Fbxl3−/− | Lengthen 2–3 h [32] | Lengthen 1.12 h |
| Myc | Myc−/− | Lengthen [44] | No cell lines |
| Timeless | Timeless−/− | Embryo death [64] | No cell lines |
| Csnk1d | Csnk1d−/+ | Shorten 0.5 h [65] | Lengthen 1.58 h |
| Csnk1e | Csnk1e−/− | Lengthen 0.3 h [66] | Lengthen 1.1 h |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Tian, T.; Wu, Y.; Yang, Y.; Zhang, Y.; Qin, X. Systematic Studies of the Circadian Clock Genes Impact on Temperature Compensation and Cell Proliferation Using CRISPR Tools. Biology 2021, 10, 1204. https://doi.org/10.3390/biology10111204
Wu Y, Tian T, Wu Y, Yang Y, Zhang Y, Qin X. Systematic Studies of the Circadian Clock Genes Impact on Temperature Compensation and Cell Proliferation Using CRISPR Tools. Biology. 2021; 10(11):1204. https://doi.org/10.3390/biology10111204
Chicago/Turabian StyleWu, Yue, Tian Tian, Yin Wu, Yu Yang, Yunfei Zhang, and Ximing Qin. 2021. "Systematic Studies of the Circadian Clock Genes Impact on Temperature Compensation and Cell Proliferation Using CRISPR Tools" Biology 10, no. 11: 1204. https://doi.org/10.3390/biology10111204
APA StyleWu, Y., Tian, T., Wu, Y., Yang, Y., Zhang, Y., & Qin, X. (2021). Systematic Studies of the Circadian Clock Genes Impact on Temperature Compensation and Cell Proliferation Using CRISPR Tools. Biology, 10(11), 1204. https://doi.org/10.3390/biology10111204





