Simple Summary
Non-small cell lung cancer (NSCLC) is a major contributor to cancer related deaths worldwide. The progression of NSCLC is linked to epithelial-mesenchymal transition (EMT), a biologic process that enables tumor cells to acquire an invasive phenotype and resistance to therapies. Discovery of novel biomarkers in NSCLC progression is essential for improved prognosis and pharmacological interventions. We performed an integrated bioinformatics analysis on available gene expression datasets of transforming growth factor β (TGF-β) induced EMT in NSCLC cell lines aiming to establish new prognostic biomarkers in the disease. The retrieved candidate genes were involved in protein modifications, regulation of cell death and cell adhesions, oxidation-reduction reactions of aerobic respiration and mitochondrial translation. Out of these genes, we identified ten prognostic gene biomarkers, mostly involved in protein modifications, whose expressions correlated with patient survival in NSCLC. This ten-gene prognostic signature will be useful to improve risk prediction and guide treatment strategies in NSCLC. Deciphering the exact functions of the biomarker genes previously not linked with NSCLC will also lead to a better understanding of the pathomechanism of NSCLC progression, revealing novel therapeutic targets in the disease.
Abstract
The progression of non-small cell lung cancer (NSCLC) is linked to epithelial-mesenchymal transition (EMT), a biologic process that enables tumor cells to acquire a migratory phenotype and resistance to chemo- and immunotherapies. Discovery of novel biomarkers in NSCLC progression is essential for improved prognosis and pharmacological interventions. In the current study, we performed an integrated bioinformatics analysis on gene expression datasets of TGF-β-induced EMT in NSCLC cells to identify novel gene biomarkers and elucidate their regulation in NSCLC progression. The gene expression datasets were extracted from the NCBI Gene Expression Omnibus repository, and differentially expressed genes (DEGs) between TGF-β-treated and untreated NSCLC cells were retrieved. A protein-protein interaction network was constructed and hub genes were identified. Functional and pathway enrichment analyses were conducted on module DEGs, and a correlation between the expression levels of module genes and survival of NSCLC patients was evaluated. Prediction of interactions of the biomarker genes with transcription factors and miRNAs was also carried out. We described four protein clusters in which DEGs were associated with ubiquitination (Module 1), regulation of cell death and cell adhesions (Module 2), oxidation-reduction reactions of aerobic respiration (Module 3) and mitochondrial translation (Module 4). From the module genes, we identified ten prognostic gene biomarkers in NSCLC. Low expression levels of KCTD6, KBTBD7, LMO7, SPSB2, RNF19A, FOXA2, DHTKD1, CDH1 and PDHB and high expression level of KLHL25 were associated with reduced overall survival of NSCLC patients. Most of these biomarker genes were involved in protein ubiquitination. The regulatory network of the gene biomarkers revealed their interaction with tumor suppressor miRNAs and transcription factors involved in the mechanisms of cancer progression. This ten-gene prognostic signature can be useful to improve risk prediction and therapeutic strategies in NSCLC. Our analysis also highlights the importance of deregulation of ubiquitination in EMT-associated NSCLC progression.
1. Introduction
Lung cancer is the leading cause of cancer-related deaths across the globe. Its high mortality rate is due to advanced stages of the disease at the time of diagnosis [1,2]. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer cases and includes the histological subtypes of adenocarcinoma (LUAD), squamous cell carcinoma (LUSC) and large cell carcinoma [3]. Long-term survival of patients diagnosed with either subtype is poor due to local recurrence of the tumor and the development of metastatic lesions after complete resection [4,5].
In NSCLC progression, epithelial-mesenchymal transition (EMT) has been described as a key process, endowing cancer cells with enhanced motility, invasiveness, resistance to apoptosis and acquisition of stem cell-like properties which further enhance tumor survival [5,6,7]. Accumulating evidence has highlighted an association between EMT and resistance to anti-cancer therapies [8,9]. Sustained by hypoxia and cellular stress, EMT is induced by a plethora of signaling molecules, including epidermal growth factor (EGF), hepatocyte growth factor (HGF) and fibroblast growth factor (FGF) [10,11,12].
Transforming growth factor beta (TGF-β) is considered one of the most potent inducers of EMT, both in vitro and in vivo, exerting a critical tumor-promoting function in advanced stages of NSCLC [13,14]. Secreted by both the cancer cells and the cellular components of the tumor microenvironment, TGF-β also acts as a regulator of multiple biological processes essential in NSCLC progression, including angiogenesis, immunoevasion and immunosuppression [15,16]. While the effect of TGF-β is context dependent, acting as a tumor suppressor at the early stages, its expression level correlates with tumor progression and metastasis [17,18]. Genetic variations in the TGF-β1 signaling pathway can improve prediction of overall survival of patients with NSCLC [19]. Moreover, a monoclonal antibody against TGF-β1, 2 and 3 (fresolimumab), vaccines targeting TGF-β signaling (LucanixTM, FANGTM), and a small molecule inhibitor of the TGF-β receptor I (galunisertib) are in clinical trials for the treatment of NSCLC [20].
Attributed to the biological complexity and poor prognosis of the disease, not all patients who are positive for acknowledged biomarkers of NSCLC (e.g., mutated EGFR, ALK, or ROS1) benefit from existing molecular therapies [21]. Therefore, integrated bioinformatics analysis of available gene expression datasets on NSCLC meets the current clinical needs for novel prognostic biomarkers that can inform therapeutic decision-making.
In a previous study, a 16-gene EMT signature was found inversely associated with T-cell infiltration in NSCLC [22]. A TGF-β-induced EMT gene signature was also reported to predict significantly worse metastasis-free survival of NSCLC patients [23]. High expression of EGFR- and EMT-related proteins was shown in the peripheral leading edge of NSCLC samples and found associated with poor prognosis [24]. In our study, we conducted an integrated bioinformatics analysis on available microarray datasets of TGF-β-induced EMT in NSCLC cells, which increases statistical power and robustness of the results retrieved. Our aim was to identify potential gene biomarkers strongly correlated with the progression of NSCLC, informing overall survival, survival until first progression and pots (first) progression survival of patients.
2. Methods
2.1. Data Sources and Search Strategy
The National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database was searched using the keyword “epithelial-mesenchymal transition”. Two authors (P.G. and K.S.K.) searched the database independently, and no language restrictions were included for article retrieval. Studies were selected according to organism type (Homo sapiens), gene expression profiling (microarray), cancer type (NSCLC) and in vitro treatment modality of EMT initiation (TGF-β supplementation).
2.2. Identification of Differentially Expressed Genes
Identification of differentially expressed genes (DEGs) from the pre-normalized pooled microarray profiles of NSCLC cells with TGF-β treatment vs. untreated NSCLC cells was performed using ImaGEO [25]. Integration of DEGs was conducted via the random effect model which combines effect sizes across all datasets into a meta-effect size. Whilst incorporating cross-study heterogeneity, DEGs with the strongest average effect across all studies were identified. Genes with p < 0.05 corrected by the Benjamini-Hochberg (BH) false discovery rate (FDR) were considered significant. DEGs were regarded upregulated based on z > 1.96, while considered downregulated with z < −1.96 (both corresponding to a 5% significance level). Across the gene expression datasets, the homogeneity and heterogeneity magnitudes of each significant DEG were quantified using Cochran’s Q test and Tau squared (τ2). DEGs based on PCochran’s Q > 0.05 and τ2 = 0 were considered highly homogeneous.
2.3. GO Functional and KEGG Pathway Enrichment
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of total and module DEGs were conducted using the ToppGene Suite [26]. Functional enrichment was categorized into three groups of GO terms: biological process (BP), molecular function (MF) and cellular component (CC). Enriched GO terms and KEGG pathways with a p < 0.05, corrected by BH FDR, were considered significant.
2.4. Protein-Protein Interaction Network Construction and Module Analysis
Network construction of proteins encoded by the total DEGs with a probabilistic confidence score > 0.4 was attained using the Search Tool for the Retrieval of Interacting Genes and Proteins (STRING) database followed by visualization with Cytoscape [27,28]. Protein nodes lacking a connection in the network were excluded.
Protein-protein interaction (PPI) network-based clustered modules were retrieved using the Molecular Complex Detection (MCODE) tool [29]. Modules with MCODE score ≥ 5.5 and nodes ≥ 5 were considered significant. Protein nodes with a higher number of incident edges were regarded hubs according to a degree centrality index ≥ 11.0 using the CentiScaPe plugin [30].
2.5. Construction of Gene Regulatory Network
Prediction of transcription factor and miRNA-gene interactions was obtained using MSigDB and TargetScan [26,31,32]. A gene regulatory network (GRN) of the biomarkers, depicting their interactions with transcription factors and miRNAs, was constructed using Cytoscape [27]. Enriched transcription factors and miRNAs with a p < 0.01, corrected by BH FDR, were considered significant.
2.6. Survival Analysis
Correlation between the expression levels of module genes and survival of patients with NSCLC was evaluated using the Kaplan-Meier-plotter [33]. Gene expression profiles of NSCLC tumor samples of 3251 patients from GEO, The Cancer Genome Atlas (TCGA) and caArray databases were sourced to obtain and verify the prognostic values of the module genes. Patients were divided into groups with high or low gene expression based on auto-selected best cut-off whereby each possible cut-off between the lower and upper quartiles was examined and the most robust threshold was selected. Module DEGs associated with reduced survival until first progression, post (first) progression survival and overall survival upon a log-rank test p < 0.001 and corrected p < 0.05 by BH FDR for the patient cut-off selection method were considered significant and presented as potential gene biomarkers in NSCLC progression.
3. Results
3.1. Overview of the Datasets Included in the Analysis
From the GEO database, 430 EMT studies were retrieved, of which 185 microarray studies were obtained having excluded other types of gene expression datasets. Further exclusion based on cancer types resulted in 15 NSCLC studies. Exclusion based on treatment modality yielded three independent gene expression studies of TGF-β-induced EMT in NSCLC, incorporating the microarray datasets GSE17708, GSE42373 and GSE49644 (Figure 1) [34,35,36,37].
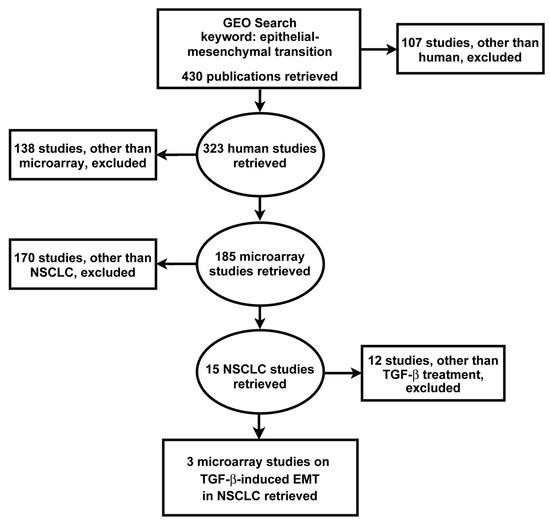
Figure 1.
Search strategy for the identification, screening and selection of eligible gene expression studies. Searching the GEO yielded 430 studies which were screened by applying the indicated exclusion criteria. Three microarray datasets met the search criteria for subsequent integrated bioinformatics analysis of TGF-β-induced EMT in NSCLC cells.
All three gene expression studies used the A549 cell line, and one also included the HCC827 and NCI-H358 cell lines, all LUAD, the most common subtype of NSCLC, and responsive to TGF-β (Table 1). In one study, TGF-β treatment was preceded by TNF-α treatment to initiate EMT. The duration of TGF-β treatment varied across the studies, ranging from 0.5 h to 3 weeks (Table 1).

Table 1.
Description of the microarray datasets selected for the analysis.
3.2. Identification of Functions and Pathways in NSCLC Progression
The pre-normalized microarray datasets were subjected to a significance analysis using the random effect model to reveal genes with significantly altered expressions between the test (TGF-β-treated) and the control (untreated) NSCLC cell samples. A total of 725 DEGs were obtained, among which 566 were highly homogenous. Of these, 215 genes were upregulated (z > 1.96) and 351 genes were downregulated (z < −1.96) (Figure S1, Tables S1 and S2).
The upregulated genes were associated with regulation of morphogenesis, cellular response and differentiation (BP enrichment), focal adhesion and cell junctions (CC enrichment) and SMAD and cytoskeletal protein binding (MF enrichment). KEGG pathway mapping revealed an association of the upregulated genes with proteoglycans and pathways in cancer (Figure 2A). The downregulated genes were linked with metabolic and oxidation-reduction processes (BP), the mitochondrion (CC) and oxidoreductase activity (MF). Pathway enrichment connected the downregulated genes with metabolic pathways (Figure 2B). The upregulated genes point to the morphological changes NSCLC cells undergo to gain a mesenchymal phenotype, while the downregulated genes indicate that a complex metabolic rewiring is taking place in EMT, consistent with increasing evidence from recent studies [38].
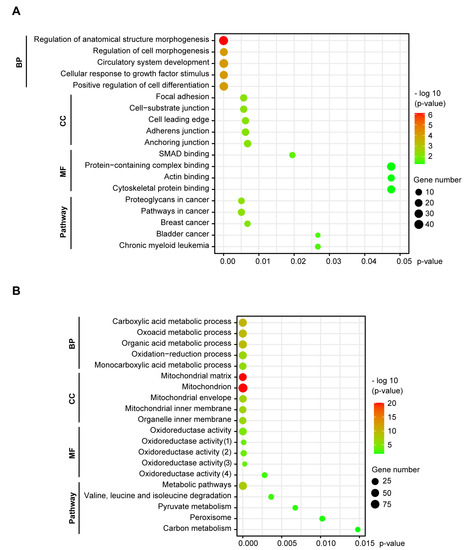
Figure 2.
GO functional and KEGG pathway enrichment of DEGs in NSCLC cells undergoing EMT. Functional enrichment of DEGs was categorized into three groups of GO terms: biological process (BP), cellular component (CC) and molecular function (MF). The significance threshold was set to p < 0.05, corrected by BH FDR. (A) The upregulated genes were involved in morphogenesis, cell adhesions, SMAD and cytoskeleton binding, and associated with cancer pathways. (B) The downregulated genes were linked with metabolic processes, the mitochondrion, oxidoreductase activity and metabolic pathways. (1) Acting on a sulfur group of donors, NAD(P) as acceptor; (2) Acting on NAD(P)H; (3) Acting on the aldehyde or oxo group of donors, disulfide as acceptor; (4) Acting on a sulfur group of donors.
3.3. Protein-Protein Interaction Network and Module Genes in NSCLC Progression
A network was constructed based on predicted interactions between proteins encoded by the DEGs. A total of 541 nodes and 1263 edges were obtained with a combined score > 0.4, including 170 upregulated and 286 downregulated genes (Figure S2). Among these, 60 hub gene candidates with a degree centrality index of ≥11.0 were identified. The complete PPI network was divided into highly dense clustering modules. Among the total 14 modules retrieved, four significant modules with MCODE score ≥ 5.5 and nodes ≥ 5 were identified and the fold changes (z-scores) of DEGs indicated (Figure 3, Table S3).
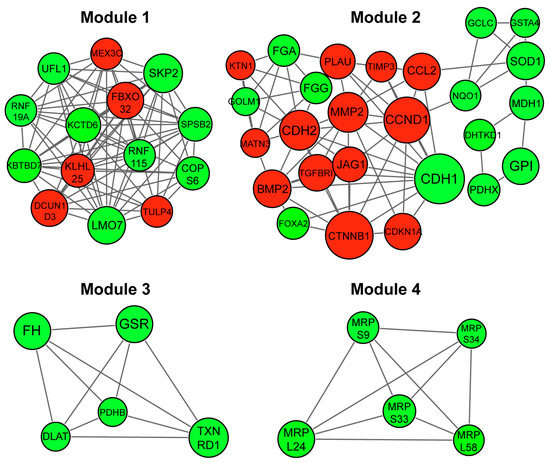
Figure 3.
Module analysis of protein-protein interaction (PPI) network of DEGs in EMT of NSCLC cells. PPI network-based clustered modules were retrieved and clustering modules with a combined node interaction score of > 0.4 and MCODE score ≥ 5.5 and nodes ≥ 5 are displayed. Red indicates upregulated and green shows downregulated node genes. Larger nodes show hub genes with a centrality degree of ≥12.0. Module 1 (score = 12.154), Module 2 (score = 6.000), Module 3 (score = 5.000) and Module 4 (score = 5.000).
Functional GO enrichment of the four modules revealed that Module 1 was involved in protein modification and ubiquitination via, e.g., the Cullin-RING (CLR) and Skp, Cullin, F-box (SCF) ubiquitin ligase complexes (Figure 4A). Module 2 related to regulation of programmed cell death, morphogenesis, cell adhesions, SMAD and phosphatase binding (Figure 4B). Module 3 was associated with generation of energy/aerobic respiration, the mitochondrial matrix and oxidoreductase activity (Figure 4C). Module 4 was connected with mitochondrial translation and the mitochondrial ribosome (Figure 4D). KEGG pathway enrichment of the modules revealed that Module 2 was associated with pathways in cancer, including the Hippo signaling pathway, while Module 3 was involved in glycolysis, pyruvate metabolism and the tricarboxylic acid (TCA) cycle (Figure 4B,C).
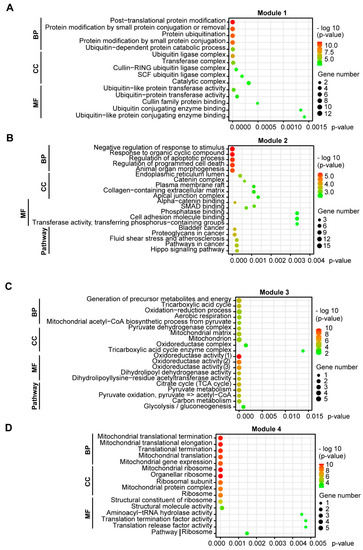
Figure 4.
GO functional and KEGG pathway enrichment of clustering module DEGs in EMT of NSCLC cells. Functional enrichment of DEGs was categorized into three groups of GO terms: biological process (BP), cellular component (CC) and molecular function (MF). The significance threshold was set to p < 0.05, corrected by BH FDR. (A) Module 1 was involved in protein modification and ubiquitination. (B) Module 2 was linked with regulation of programmed cell death and morphogenesis, cell adhesions and pathways in cancer. (C) Module 3 was associated with aerobic respiration, the mitochondrial matrix, and pathways in glycolysis, pyruvate metabolism and the TCA cycle. (D) Module 4 related to mitochondrial translation and the mitochondrial ribosome.
3.4. Prognostic Gene Biomarkers in NSCLC
The prognostic value of potential biomarkers in the module gene sets was determined using gene expression profiles of NSCLC tumor samples from 3251 patients (Table S4). Low expression levels of nine genes, including forkhead box A2 (FOXA2), potassium channel tetramerization domain containing 6 (KCTD6), kelch repeat and BTB domain containing 7 (KBTBD7), dehydrogenase E1 and transketolase domain containing 1 (DHTKD1), LIM domain 7 (LMO7), pyruvate dehydrogenase E1 beta subunit (PDHB), splA/ryanodine receptor domain and SOCS box containing 2 (SPSB2), E-cadherin (CDH1) and ring finger protein 19A (RNF19A), and high expression level of the kelch-like family member 25 (KLHL25) were associated with significantly reduced overall survival (Table 2, Figure 5).

Table 2.
Prognostic gene biomarkers in NSCLC, ranked according to their z-scores.

Figure 5.
Prognostic value of genes associated with EMT in NSCLC. Kaplan-Meier association between the expression levels of module DEGs and overall survival of patients with NSCLC (n = 1144) is displayed. Significance thresholds for potential gene biomarkers include a log-rank test p < 0.001 and a corrected p < 0.05 by BH FDR for the patient cut-off selection method. Low expression levels of CDH1, LMO7, PDHB, KBTBD7, RNF19A, KCTD6, DHTKD1, SPSB2 and FOXA2 while high expression level of KLHL25 were associated with reduced overall survival.
Low expression levels of CDH1, LMO7, PDHB, RNF19A and FOXA2 and high expression level of KLHL25 were associated with reduced survival until first progression (Figure S3). Low expression levels of PDHB, KBTBD7, RNF19A, KCTD6 and FOXA2 were linked to decreased post-progression survival (Figure S4).
Most of the gene biomarkers (KLHL25, KCTD6, KBTBD7, LMO7, SPSB2 and RNF19A) were associated with ubiquitination (Table 2), suggesting an important role of the process in NSCLC progression.
3.5. Gene Regulatory Network of the Proposed Gene Biomarkers
To shed light on the regulation of the gene biomarkers, an interaction network of the biomarker DEGs with transcription factors and miRNAs was constructed. Tumor suppressor miRNAs, including the let-7 and miR-26 family members [39,40,41] were found to be prominent in the regulation of PDHB (Figure 6). Two of the PDHB-interacting miRNAs, miR-200a-3p and miR-141-3p [42,43], were also involved in the regulation of FOXA2. DHTKD1 was found interacting with the tumor suppressive miR-29 family members [44,45,46]. These miR-29 family members appeared to also regulate KLHL25, and one of them, miR-29b-3p, was interacting with RNF19A too (Figure 6).
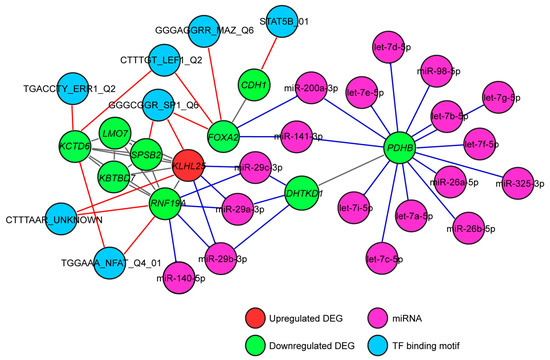
Figure 6.
Gene regulatory network of the proposed gene biomarkers in NSCLC progression. A gene regulatory network for the biomarkers was constructed, and enriched transcription factors and miRNAs with a p < 0.01, corrected by BH FDR, were considered significant. Transcription factors associated with tumor progression and tumor suppressor miRNAs were found involved in the regulation of the gene biomarkers.
Transcription factors associated with various mechanisms of lung cancer progression were also involved in the regulation of the biomarker genes [47,48,49,50]. SP1 was found regulating FOXA2, SPSB2 and KLHL25. NFAT, ERR1, LEF1 and MAZ were linked with the downregulated gene biomarkers only, and STAT5B was the only transcription factor associated with the regulation of CDH1 (Figure 6). No connection of the biomarker genes with EMT transcription factors (EMT-TFs) such as SNAIL, SLUG or TWIST was revealed in our analysis.
4. Discussion
Our integrated bioinformatics analysis on microarray datasets of TGF-β-induced EMT in NSCLC cells revealed four significant protein clusters within the 215 upregulated and 351 downregulated DEGs. These modules were associated with protein ubiquitination (Module 1), morphogenesis and cell adhesions (Module 2), oxidoreductase activity of aerobic respiration (Module 3) and mitochondrial translation (Module 4) confirming the significance of these processes in the regulation of EMT.
We found ten genes correlated with overall survival of patients with NSCLC, and only one of them (CDH1) was a canonical EMT-marker. Low expressions of nine gene biomarkers were associated with decreased survival. Most of them, including KCTD6, KBTBD7, LMO7, SPSB2 and RNF19A were Module 1 genes, involved in protein modifications and ubiquitination [51,52,53,54,55].
In an earlier study, KBTBD7 which regulates extracellular-cytoskeletal signal transduction, appeared to be protective in early-stage NSCLC as one of an 8-gene prognostic signature [55,56]. LMO7 which regulates the actin cytoskeleton and adherens junctions, was described to be downregulated in malignant lung tissue, and LMO7 deficiency was found associated with genetic predisposition to lung cancer [51,57].
The downregulation of FOXA2, DHTKD1 and CDH1, involved in embryonic development, oxidation reactions and intercellular adhesions, respectively, were also linked to worse prognosis of NSCLC patients in our study.
FOXA2 was reported to inhibit EMT and suppress metastasis in human lung cancer cell lines via repressing the SLUG promoter [58]. Loss of FOXA2 expression was found frequent in lung cancer cell lines and NSCLC tumor samples [59].
So far, no correlation of DHTKD1 (component of a mitochondrial 2-oxoglutarate-dehydrogenase-complex) expression with survival of NSCLC patients has been described. In breast carcinoma, differential DNA methylation between tumors and normal tissues was found correlated with the expression level of DHTKD1 [60].
CDH1, a tumor suppressor gene encoding E-cadherin, plays a crucial role in maintaining intercellular junctions in the epithelium [61]. In an earlier study, downregulation of E-cadherin was found to promote EGFR transcription in NSCLC [62]. In NSCLC patients, the mRNA level of the Wilms tumor gene (WT1) was reported to be negatively correlated with that of CDH1 and was associated with pathological stage, metastasis, and survival rate [63]. High E-cadherin and low vimentin expression was linked to better overall survival of NSCLC patients [64].
PDHB, the mitochondrial pyruvate dehydrogenase E1 component subunit beta, a Module 3 gene in our study, catalyzes the decarboxylation of pyruvate to acetyl-CoA, linking the glycolytic pathway to the TCA cycle [65]. PDBH was described as a biomarker within energy metabolism heterogeneity of ovarian cancer cells for the diagnosis and prognosis of ovarian cancer [66]. We found the downregulation of PDHB linked to worse prognosis in NSCLC.
In our integrated bioinformatics analysis, KLHL25 was the only gene whose high expression was associated with a significantly reduced overall survival of NSCLC patients, thus representing an unfavorable prognostic marker in NSCLC progression.
KLHL25, a member of the Kelch-like (KLHL) gene family, a substrate-specific adaptor in the cullin-3 (Cul3)-dependent ubiquitin ligase complex, is required for eukaryotic translational control [67]. Mutations in four of the Kelch-family members have been linked to cancer [68]. However, the exact mechanism how KLHL25 contributes to NSCLC progression warrants further investigations.
Although we found key members of the zinc-finger transcription factor family upregulated in TGF-β-induced EMT in NSCLC cells (ZEB1, z-score = 4.40 and adj. p = 1.09 × 10−5; SNAI2, z-score = 3.07 and adj. p = 4.83 × 10−2), binding motifs of EMT-TFs were not significantly enriched in the gene regulatory network, suggesting no direct interaction of EMT-TFs with CDH1 or the other proposed gene biomarkers in NSCLC progression. Our findings are consistent with recent evidence showing that EMT-TFs also act via indirect mechanisms to downregulate E-cadherin and induce EMT and NSCLC progression [69,70].
Overall, to clarify the role of our biomarker genes and their regulation in NSCLC progression, further validation and functional studies are required. The limitation of our study is the lack of such biological experiments.
Of the gene biomarkers we propose in NSCLC progression, KBTBD7 has already been described as a favorable prognostic indicator in NSCLC [55,56]. An association between the deficiency of LMO7 and genetic predisposition to lung cancer has also been reported [57]. Moreover, CDH1 and FOXA2 are linked to each other in carcinoma progression, evidenced by the loss of silencing FOXA2 leading to E-cadherin downregulation, EMT and metastasis [71,72]. However, most of the gene biomarkers we put forward have not been linked to NSCLC prognosis so far.
Our results also reveals that deregulation of protein ubiquitination has an important function in EMT-associated NSCLC progression. Furthermore, deciphering the function of KLHL25 in the pathomechanism of NSCLC may lead to new therapeutic targets.
5. Conclusions
Genes regulated during EMT of NSCLC cells are closely linked to NSCLC progression. The ten prognostic gene biomarkers we obtained via an integrated bioinformatics analysis of EMT-associated gene expression data can provide improved risk prediction and lead to new therapeutic targets in NSCLC.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/biology10111200/s1, Figure S1: Significant DEGs of the combined microarray datasets of TGF-β-induced EMT in NSCLC cell lines, Figure S2: Protein-protein interaction network of DEGs in EMT of NSCLC cells, Figure S3: Prognostic value of genes associated with EMT in NSCLC–Survival until first progression (FP), Figure S4: Prognostic value of genes associated with EMT in NSCLC–Post (first) progression survival (PPS), Table S1: List of the 20 most highly ranked upregulated genes in EMT of NSCLC cells with their z-scores, listed in order of their FDR values, Table S2: List of the 20 most highly ranked downregulated genes in EMT of NSCLC cells with their z-scores, listed in order of their FDR values, Table S3: Genes with their z-scores from the PPI network-based clustered modules in EMT of NSCLC cells, listed in order of their FDR values, Table S4: List of datasets sourced for patient survival analysis.
Author Contributions
This report is based on the results of P.G.’s undergraduate research project. P.G. contributed with conceptualization, methodology, formal analysis, visualization and writing the original draft of the manuscript. K.S.K. carried out formal analysis, reviewing and editing work, while A.G.’s contribution included writing the original draft, reviewing and editing, visualization and supervision of the project. All authors have read and agreed to the published version of the manuscript.
Funding
No funding was used to conduct this research.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All relevant data generated during this study are included in this published article.
Acknowledgments
We thank the Imperial Open Access Fund by Imperial College London for funding the article processing charges. Author P.G. is supported by the Bodossaki Foundation, Greece.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Walters, S.; Maringe, C.; Coleman, M.P.; Peake, M.D.; Butler, J.; Young, N.; Bergström, S.; Hanna, L.; Jakobsen, E.; Kölbeck, K. Lung cancer survival and stage at diagnosis in Australia, Canada, Denmark, Norway, Sweden and the UK: A population-based study, 2004–2007. Thorax 2013, 68, 551–564. [Google Scholar] [CrossRef]
- Malapelle, U.; Pisapia, P.; Rocco, D.; Smeraglio, R.; di Spirito, M.; Bellevicine, C.; Troncone, G. Next generation sequencing techniques in liquid biopsy: Focus on non-small cell lung cancer patients. Transl. Lung Cancer Res. 2016, 5, 505–510. [Google Scholar] [CrossRef]
- Zappa, C.; Mousa, S.A. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016, 5, 288–300. [Google Scholar] [CrossRef]
- Mahapatra, A. Lung Cancer—Genomics and Personalized Medicine. ACS Chem. Biol. 2010, 5, 529–531. [Google Scholar] [CrossRef]
- Mittal, V. Epithelial mesenchymal transition in aggressive lung cancers. In Lung Cancer and Personalized Medicine: Novel Therapies and Clinical Management; Springer: Cham, Switzerland, 2016; pp. 37–56. [Google Scholar]
- Xiao, D.; He, J. Epithelial mesenchymal transition and lung cancer. J. Thorac. Dis. 2010, 2, 154–159. [Google Scholar]
- O’Leary, K.; Shia, A.; Schmid, P. Epigenetic regulation of EMT in non-small cell lung cancer. Curr. Cancer Drug Targets 2018, 18, 89–96. [Google Scholar] [CrossRef]
- Jakobsen, K.R.; Demuth, C.; Sorensen, B.S.; Nielsen, A.L. The role of epithelial to mesenchymal transition in resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Transl. Lung Cancer Res. 2016, 5, 172–182. [Google Scholar] [CrossRef]
- Weng, C.-H.; Chen, L.-Y.; Lin, Y.-C.; Shih, J.-Y.; Lin, Y.-C.; Tseng, R.-Y.; Chiu, A.-C.; Yeh, Y.-H.; Liu, C.; Lin, Y.-T. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene 2019, 38, 455–468. [Google Scholar] [CrossRef]
- Liu, F.; Song, S.; Yi, Z.; Zhang, M.; Li, J.; Yang, F.; Yin, H.; Yu, X.; Guan, C.; Liu, Y. HGF induces EMT in non-small-cell lung cancer through the hBVR pathway. Eur. J. Pharmacol. 2017, 811, 180–190. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Oeck, S.; Glazer, P.M. Hypoxia promotes resistance to EGFR inhibition in NSCLC cells via the histone demethylases, LSD1 and PLU-1. Mol. Cancer Res. 2018, 16, 1458–1469. [Google Scholar] [CrossRef]
- Schelch, K.; Wagner, C.; Hager, S.; Pirker, C.; Siess, K.; Lang, E.; Lin, R.; Kirschner, M.B.; Mohr, T.; Brcic, L. FGF2 and EGF induce epithelial-mesenchymal transition in malignant pleural mesothelioma cells via a MAPKinase/MMP1 signal. Carcinogenesis 2018, 39, 534–545. [Google Scholar] [CrossRef]
- Ikushima, H.; Miyazono, K. Cellular context-dependent “colors” of transforming growth factor-β signaling. Cancer Sci. 2010, 101, 306–312. [Google Scholar] [CrossRef]
- Hao, Y.; Baker, D.; Ten Dijke, P. TGF-β-mediated epithelial-mesenchymal transition and cancer metastasis. Int. J. Mol. Sci. 2019, 20, 2767. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.-H.; Landström, M.; Moustakas, A. Mechanism of TGF-β signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 2009, 21, 166–176. [Google Scholar] [CrossRef]
- Batlle, E.; Massagué, J. Transforming growth factor-β signaling in immunity and cancer. Immunity 2019, 50, 924–940. [Google Scholar] [CrossRef]
- Tian, M.; Schiemann, W.P. The TGF-β paradox in human cancer: An update. Future Oncol. 2009, 5, 259–271. [Google Scholar] [CrossRef]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, W.; Pi, W.; Bi, N.; DesRosiers, C.; Kong, F.; Cheng, M.; Yang, L.; Lautenschlaeger, T.; Jolly, S. Genetic variations in the transforming growth factor-β1 pathway may improve predictive power for overall survival in non-small cell lung cancer. Front. Oncol. 2021, 11, 599719. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Chung, C.-L.; Hu, T.-H.; Chen, J.-J.; Liu, P.-F.; Chen, C.-L. Recent progress in TGF-β inhibitors for cancer therapy. Biomed. Pharmacother. 2021, 134, 111046. [Google Scholar] [CrossRef] [PubMed]
- Villalobos, P.; Wistuba, I.I. Lung cancer biomarkers. Hematol./Oncol. Clin. 2017, 31, 13–29. [Google Scholar] [CrossRef]
- Chae, Y.K.; Chang, S.; Ko, T.; Anker, J.; Agte, S.; Iams, W.; Choi, W.M.; Lee, K.; Cruz, M. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC). Sci. Rep. 2018, 8, 2918. [Google Scholar] [CrossRef] [PubMed]
- Gordian, E.; Welsh, E.A.; Gimbrone, N.; Siegel, E.M.; Shibata, D.; Creelan, B.C.; Cress, W.D.; Eschrich, S.A.; Haura, E.B.; Muñoz-Antonia, T. Transforming growth factor β-induced epithelial-to-mesenchymal signature predicts metastasis-free survival in non-small cell lung cancer. Oncotarget 2019, 10, 810–824. [Google Scholar] [CrossRef]
- Mahmood, M.Q.; Ward, C.; Muller, H.K.; Sohal, S.S.; Walters, E.H. Epithelial mesenchymal transition (EMT) and non-small cell lung cancer (NSCLC): A mutual association with airway disease. Med. Oncol. 2017, 34, 45. [Google Scholar] [CrossRef] [PubMed]
- Toro-Domínguez, D.; Martorell-Marugán, J.; López-Domínguez, R.; García-Moreno, A.; González-Rumayor, V.; Alarcón-Riquelme, M.E.; Carmona-Sáez, P. ImaGEO: Integrative gene expression meta-analysis from GEO database. Bioinformatics 2019, 35, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Scardoni, G.; Tosadori, G.; Faizan, M.; Spoto, F.; Fabbri, F.; Laudanna, C. Biological network analysis with CentiScaPe: Centralities and experimental dataset integration. F1000Research 2015, 3, 139. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B.; Surowiak, P.; Budczies, J.; Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar]
- Sartor, M.A.; Mahavisno, V.; Keshamouni, V.G.; Cavalcoli, J.; Wright, Z.; Karnovsky, A.; Kuick, R.; Jagadish, H.; Mirel, B.; Weymouth, T. ConceptGen: A gene set enrichment and gene set relation mapping tool. Bioinformatics 2010, 26, 456–463. [Google Scholar] [CrossRef]
- Cieślik, M.; Hoang, S.A.; Baranova, N.; Chodaparambil, S.; Kumar, M.; Allison, D.F.; Xu, X.; Wamsley, J.J.; Gray, L.; Jones, D.R. Epigenetic coordination of signaling pathways during the epithelial-mesenchymal transition. Epigenet. Chromatin 2013, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Wamsley, J.J.; Kumar, M.; Allison, D.F.; Clift, S.H.; Holzknecht, C.M.; Szymura, S.J.; Hoang, S.A.; Xu, X.; Moskaluk, C.A.; Jones, D.R. Activin Upregulation by NF-κB Is Required to Maintain Mesenchymal Features of Cancer Stem-Like Cells in Non-Small Cell Lung Cancer. Cancer Res. 2015, 75, 426–435. [Google Scholar] [CrossRef]
- Sun, Y.; Daemen, A.; Hatzivassiliou, G.; Arnott, D.; Wilson, C.; Zhuang, G.; Gao, M.; Liu, P.; Boudreau, A.; Johnson, L. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014, 2, 20. [Google Scholar] [CrossRef]
- Hua, W.; Ten Dijke, P.; Kostidis, S.; Giera, M.; Hornsveld, M. TGFβ-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell. Mol. Life Sci. 2020, 77, 2103–2123. [Google Scholar] [CrossRef]
- Pop-Bica, C.; Pintea, S.; Magdo, L.; Cojocneanu, R.; Gulei, D.; Ferracin, M.; Berindan-Neagoe, I. The Clinical Utility of miR-21 and let-7 in Non-small Cell Lung Cancer (NSCLC). A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 516850. [Google Scholar] [CrossRef]
- Dang, X.; Ma, A.; Yang, L.; Hu, H.; Zhu, B.; Shang, D.; Chen, T.; Luo, Y. MicroRNA-26a regulates tumorigenic properties of EZH2 in human lung carcinoma cells. Cancer Genet. 2012, 205, 113–123. [Google Scholar] [CrossRef]
- Li, D.; Wei, Y.; Wang, D.; Gao, H.; Liu, K. MicroRNA-26b suppresses the metastasis of non-small cell lung cancer by targeting MIEN1 via NF-κB/MMP-9/VEGF pathways. Biochem. Biophys. Res. Commun. 2016, 472, 465–470. [Google Scholar] [CrossRef]
- Huang, Y.; Bao, T.; Li, Z.; Ji, G.; Zhang, L. Function of miR-200a in proliferation and apoptosis of non-small cell lung cancer cells. Oncol. Lett. 2020, 20, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, Y.; Wang, D.; Wang, Y.; Wang, L. MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2019, 509, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, S.; Mei, R.; Kang, Y.; Duan, J.; Wei, R.; Xiang, C.; Wu, Y.; Lu, X.; Cai, Z. miR-29a suppresses IL-13-induced cell invasion by inhibiting YY1 in the AKT pathway in lung adenocarcinoma A549 cells. Oncol. Rep. 2018, 39, 2613–2623. [Google Scholar] [CrossRef]
- Jia, R.; Wang, C. MiR-29b-3p reverses cisplatin resistance by targeting COL1A1 in non-small-cell lung cancer A549/DDP cells. Cancer Manag. Res. 2020, 12, 2559–2566. [Google Scholar] [CrossRef]
- Liu, L.; Bi, N.; Wu, L.; Ding, X.; Men, Y.; Zhou, W.; Li, L.; Zhang, W.; Shi, S.; Song, Y. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol. Cancer 2017, 16, 50. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.M.; Liao, C.G.; Fei, F.; Guo, X.; Xing, J.L.; Chen, Z.N. Transcription factor Sp1 regulates expression of cancer-associated molecule CD147 in human lung cancer. Cancer Sci. 2010, 101, 1463–1470. [Google Scholar] [CrossRef] [PubMed]
- Shou, J.; Jing, J.; Xie, J.; You, L.; Jing, Z.; Yao, J.; Han, W.; Pan, H. Nuclear factor of activated T cells in cancer development and treatment. Cancer Lett. 2015, 361, 174–184. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Chiang, A.C.; Zhang, X.H.-F.; Kim, J.Y.; Kris, M.G.; Ladanyi, M.; Gerald, W.L.; Massagué, J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 2009, 138, 51–62. [Google Scholar] [CrossRef]
- Luo, W.; Zhu, X.; Liu, W.; Ren, Y.; Bei, C.; Qin, L.; Miao, X.; Tang, F.; Tang, G.; Tan, S. MYC associated zinc finger protein promotes the invasion and metastasis of hepatocellular carcinoma by inducing epithelial mesenchymal transition. Oncotarget 2016, 7, 86420–86432. [Google Scholar] [CrossRef]
- Ooshio, T.; Irie, K.; Morimoto, K.; Fukuhara, A.; Imai, T.; Takai, Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and α-actinin in epithelial cells. J. Biol. Chem. 2004, 279, 31365–31373. [Google Scholar] [CrossRef]
- Huang, Y.; Niwa, J.-I.; Sobue, G.; Breitwieser, G.E. Calcium-sensing receptor ubiquitination and degradation mediated by the E3 ubiquitin ligase dorfin. J. Biol. Chem. 2006, 281, 11610–11617. [Google Scholar] [CrossRef]
- De Smaele, E.; Di Marcotullio, L.; Moretti, M.; Pelloni, M.; Occhione, M.A.; Infante, P.; Cucchi, D.; Greco, A.; Pietrosanti, L.; Todorovic, J. Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia 2011, 13, 374–385, IN21–IN23. [Google Scholar] [CrossRef]
- Nishiya, T.; Matsumoto, K.; Maekawa, S.; Kajita, E.; Horinouchi, T.; Fujimuro, M.; Ogasawara, K.; Uehara, T.; Miwa, S. Regulation of inducible nitric-oxide synthase by the SPRY domain-and SOCS box-containing proteins. J. Biol. Chem. 2011, 286, 9009–9019. [Google Scholar] [CrossRef]
- Genau, H.M.; Huber, J.; Baschieri, F.; Akutsu, M.; Dötsch, V.; Farhan, H.; Rogov, V.; Behrends, C. CUL3-KBTBD6/KBTBD7 ubiquitin ligase cooperates with GABARAP proteins to spatially restrict TIAM1-RAC1 signaling. Mol. Cell 2015, 57, 995–1010. [Google Scholar] [CrossRef]
- He, R.; Zuo, S. A robust 8-gene prognostic signature for early-stage non-small cell lung cancer. Front. Oncol. 2019, 9, 693. [Google Scholar] [CrossRef]
- Karlsson, T.; Kvarnbrink, S.; Holmlund, C.; Botling, J.; Micke, P.; Henriksson, R.; Johansson, M.; Hedman, H. LMO7 and LIMCH1 interact with LRIG proteins in lung cancer, with prognostic implications for early-stage disease. Lung Cancer 2018, 125, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Shu, G.; Yuan, X.; Jing, N.; Song, J. FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers. Cell Res. 2011, 21, 316–326. [Google Scholar] [CrossRef]
- Basseres, D.S.; D’Alò, F.; Yeap, B.Y.; Löwenberg, E.C.; Gonzalez, D.A.; Yasuda, H.; Dayaram, T.; Kocher, O.N.; Godleski, J.J.; Richards, W.G. Frequent downregulation of the transcription factor Foxa2 in lung cancer through epigenetic silencing. Lung Cancer 2012, 77, 31–37. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Li, X. Comprehensive analysis of prognosis-related methylated sites in breast carcinoma. Mol. Genet. Genom. Med. 2020, 8, e1161. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.; Melo, S.; Carneiro, P.; Moreira, A.M.; Fernandes, M.S.; Ribeiro, A.S.; Guilford, P.; Paredes, J.; Seruca, R. Clinical spectrum and pleiotropic nature of CDH1 germline mutations. J. Med. Genet. 2019, 56, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Su, L.; Liu, X. Loss of CDH1 up-regulates epidermal growth factor receptor via phosphorylation of YBX1 in non-small cell lung cancer cells. FEBS Lett. 2013, 587, 3995–4000. [Google Scholar] [CrossRef]
- Wu, C.; Zhu, W.; Qian, J.; He, S.; Wu, C.; Chen, Y.; Shu, Y. WT1 promotes invasion of NSCLC via suppression of CDH1. J. Thorac. Oncol. 2013, 8, 1163–1169. [Google Scholar] [CrossRef]
- Tsoukalas, N.; Aravantinou-Fatorou, E.; Tolia, M.; Giaginis, C.; Galanopoulos, M.; Kiakou, M.; Kostakis, I.D.; Dana, E.; Vamvakaris, I.; Korogiannos, A. Epithelial-mesenchymal transition in non small-cell lung cancer. Anticancer Res. 2017, 37, 1773–1778. [Google Scholar]
- Patel, M.S.; Nemeria, N.S.; Furey, W.; Jordan, F. The pyruvate dehydrogenase complexes: Structure-based function and regulation. J. Biol. Chem. 2014, 289, 16615–16623. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhan, X.; Zhan, X. Energy metabolism heterogeneity-based molecular biomarkers for ovarian cancer. In Molecular Medicine; IntechOpen: London, UK, 2018. [Google Scholar]
- Zhang, C.; Liu, J.; Huang, G.; Zhao, Y.; Yue, X.; Wu, H.; Li, J.; Zhu, J.; Shen, Z.; Haffty, B.G. Cullin3-KLHL25 ubiquitin ligase targets ACLY for degradation to inhibit lipid synthesis and tumor progression. Genes Dev. 2016, 30, 1956–1970. [Google Scholar] [CrossRef] [PubMed]
- Dhanoa, B.S.; Cogliati, T.; Satish, A.G.; Bruford, E.A.; Friedman, J.S. Update on the Kelch-like (KLHL) gene family. Hum. Genom. 2013, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-H.; Tsai, M.-F.; Gow, C.-H.; Wu, S.-G.; Liu, Y.-N.; Chang, Y.-L.; Yu, S.-L.; Tsai, H.-C.; Lin, S.-W.; Chen, Y.-W. Upregulation of microRNA-137 expression by Slug promotes tumor invasion and metastasis of non-small cell lung cancer cells through suppression of TFAP2C. Cancer Lett. 2017, 402, 190–202. [Google Scholar] [CrossRef] [PubMed]
- Manshouri, R.; Coyaud, E.; Kundu, S.T.; Peng, D.H.; Stratton, S.A.; Alton, K.; Bajaj, R.; Fradette, J.J.; Minelli, R.; Peoples, M.D. ZEB1/NuRD complex suppresses TBC1D2b to stimulate E-cadherin internalization and promote metastasis in lung cancer. Nat. Commun. 2019, 10, 5125. [Google Scholar] [CrossRef]
- Song, Y.; Washington, M.K.; Crawford, H.C. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010, 70, 2115–2125. [Google Scholar] [CrossRef] [PubMed]
- Bow, Y.D.; Wang, Y.Y.; Chen, Y.K.; Su, C.W.; Hsu, C.W.; Xiao, L.Y.; Yuan, S.S.; Li, R.N. Silencing of FOXA2 decreases E-cadherin expression and is associated with lymph node metastasis in oral cancer. Oral Dis. 2020, 26, 756–765. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).