Protective Roles of Folic Acid in the Responses of Bovine Mammary Epithelial Cells to Different Virulent Staphylococcus aureus Strains
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
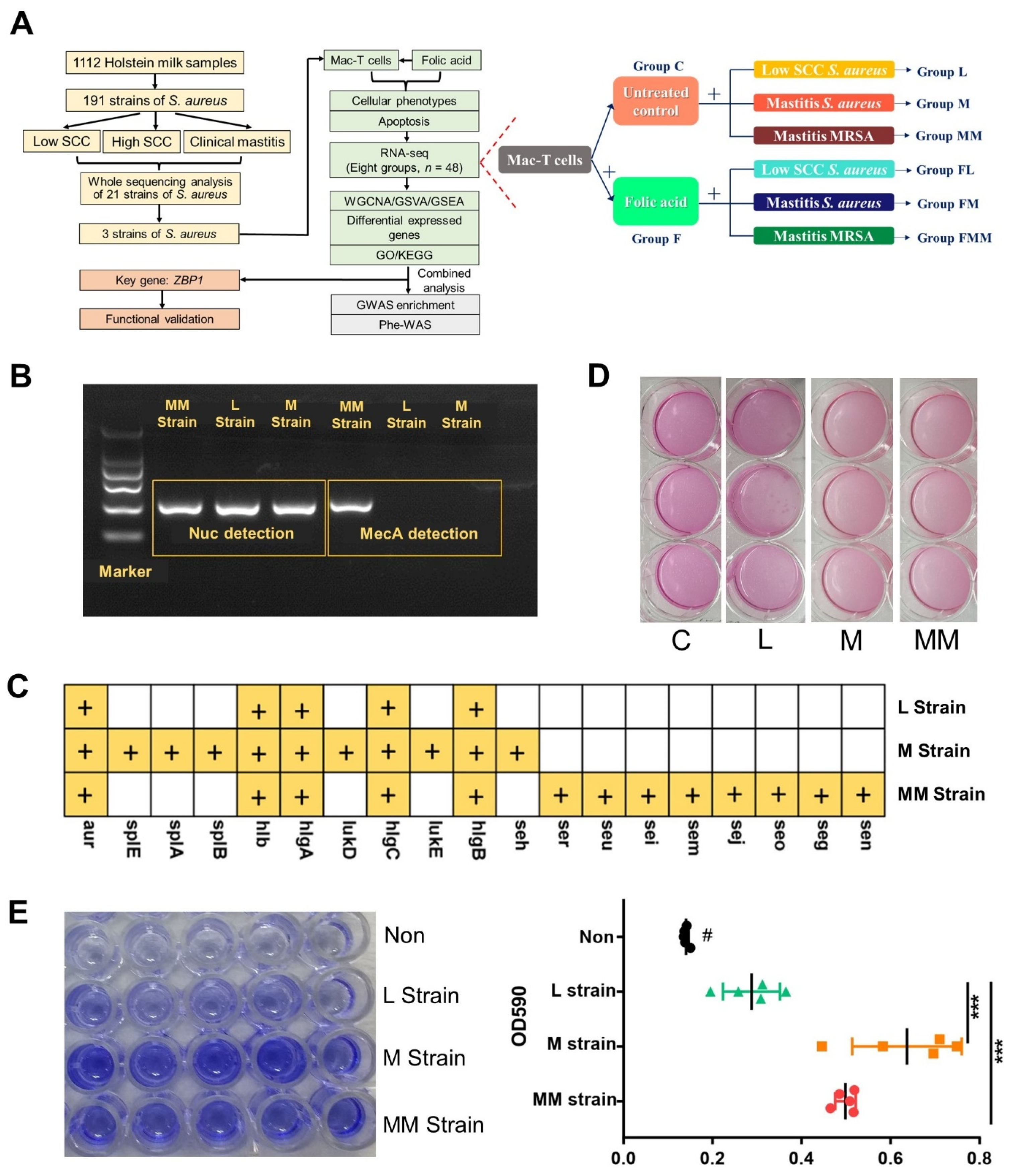
2.1. Bacterial Strains
2.2. DNA Extraction, Whole Sequencing and Biofilm Assay of S. aureus Strains
2.3. Cell Culture, S. aureus Challenge and Folic Acid Treatment of Mac-T Cells
2.4. RNA Extraction and RNA-Seq
2.5. RNA-Seq Data Analysis
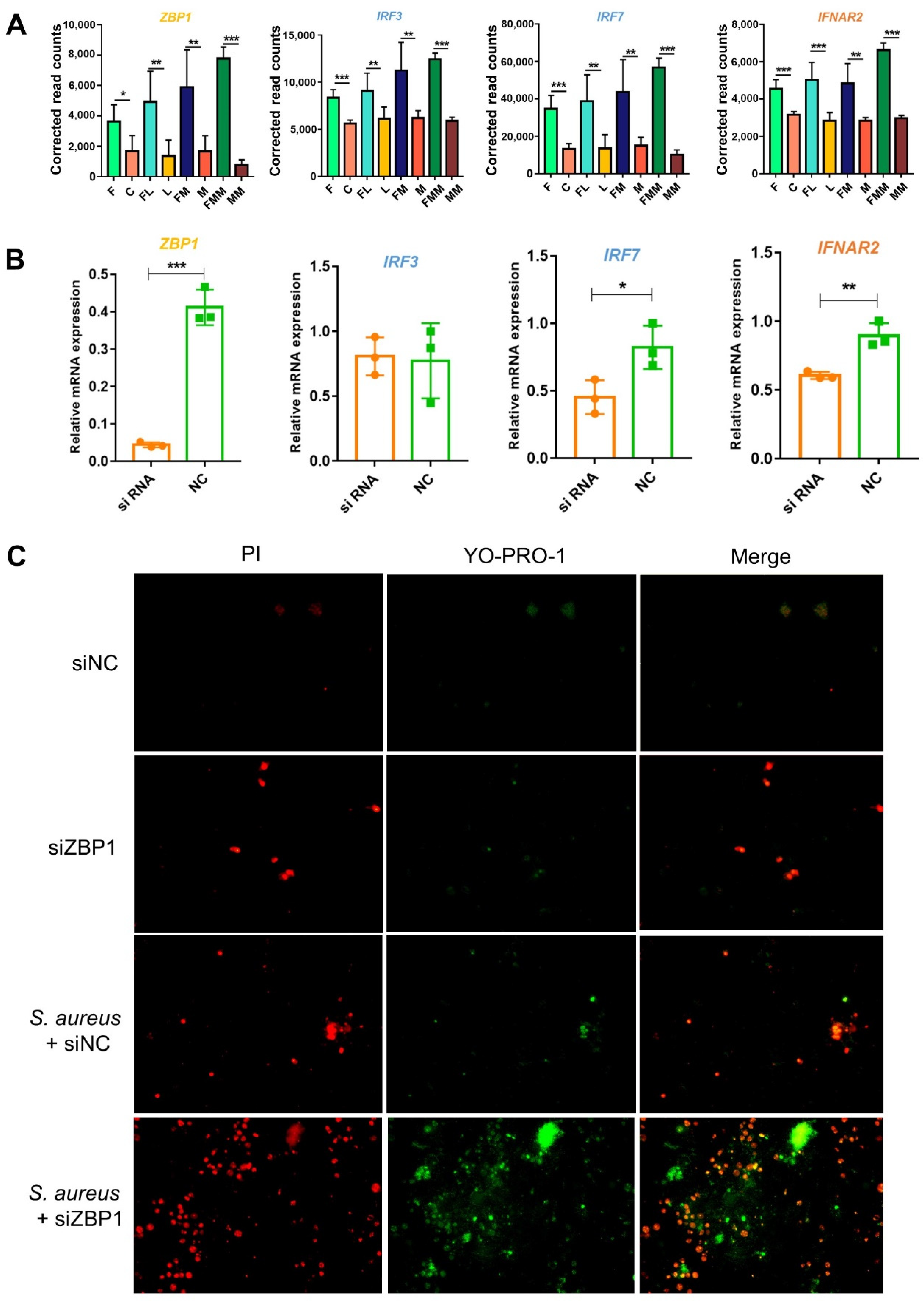
2.6. RNA Interference
2.7. PCR and Fluorescence Quantitative PCR
2.8. Cell Cytotoxicity-Related Assays
2.9. GWAS Enrichment Analysis and Phenome-Wide Association Analysis (Phe-WAS)
3. Results
3.1. Phenotypic Discrepancies of Three Bovine-Originated S. aureus Strains
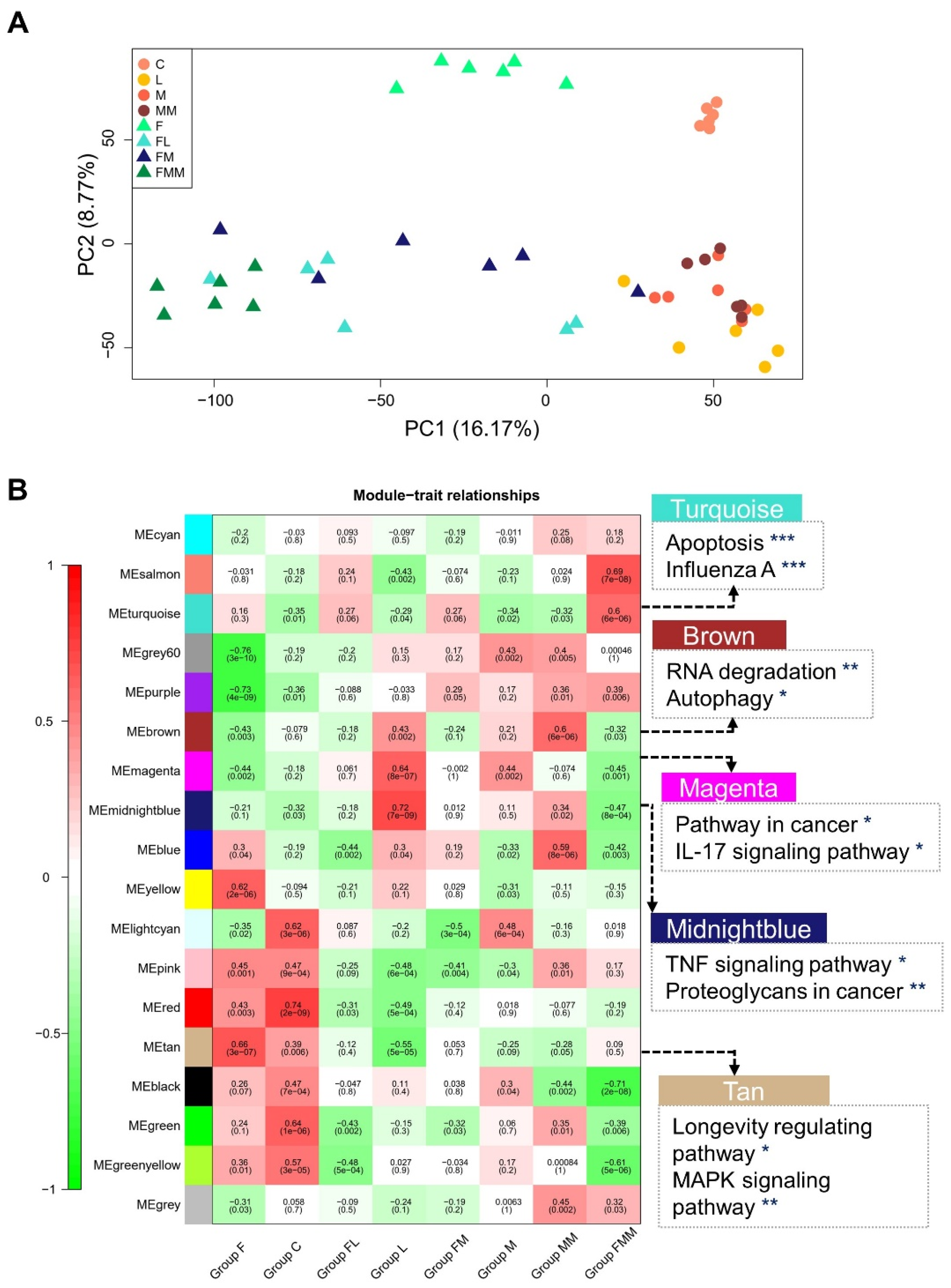
3.2. Transcriptomic Features of Host Cellular Responses to Three S. aureus Strains Challenged with or without FA Supplementation
3.3. Key Gene Co-Expression Modules and Gene Sets Enriched in Immune-Related Pathways
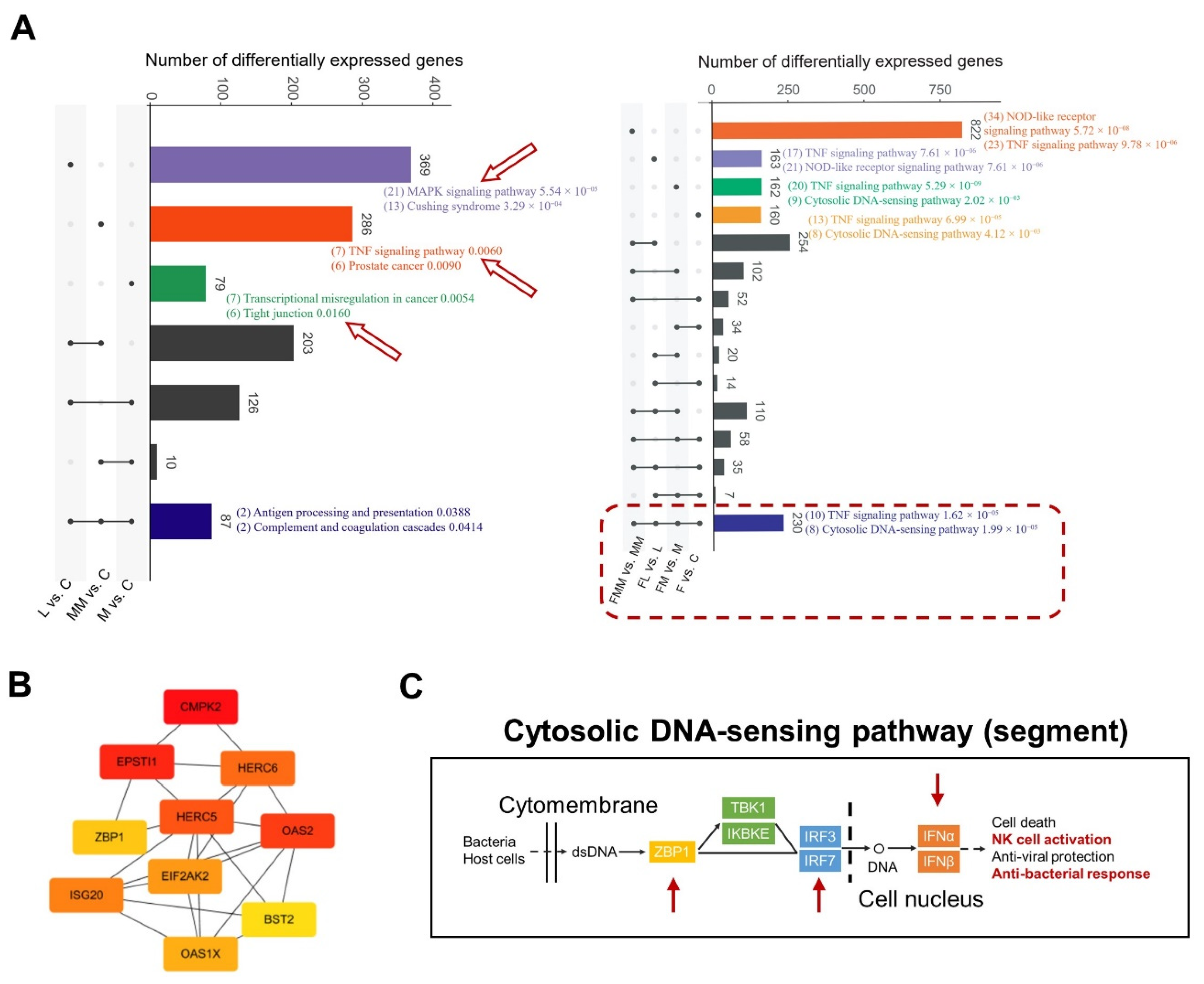
3.4. Transcriptional Changes of Mac-T Cells Induced by FA Supplementation and S. aureus Challenge
3.5. FA Activates the Anti-Bacterial Responses by Elevating the Abilities of Cytosolic DNA Sensing and Tight Junction Assembly
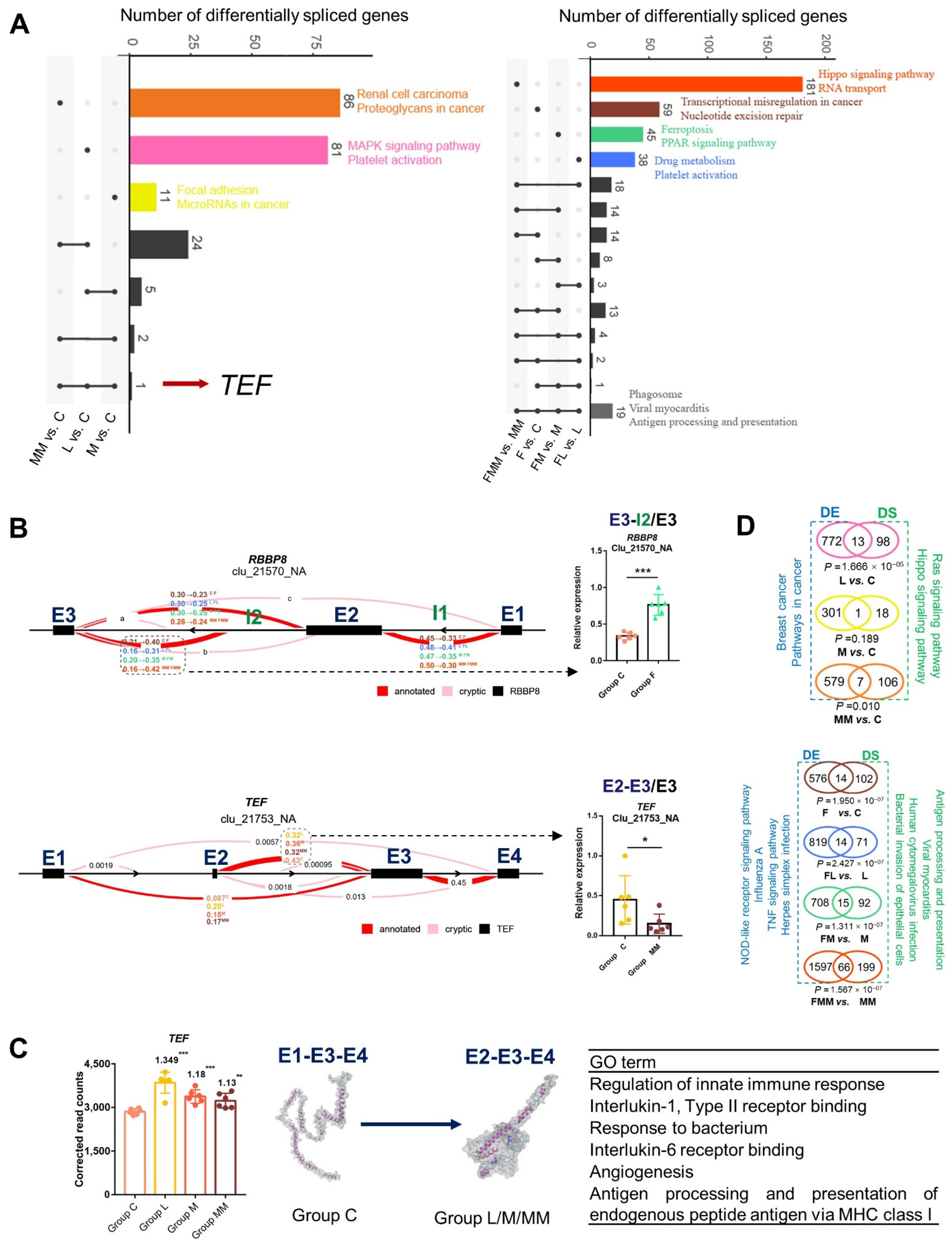
3.6. Differentially Spliced Transcripts Are Enriched in Immune or Inflammatory Pathways
3.7. Association of Key Modules and Gene Sets with Complex Traits
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFC | Age at first calving |
| pbMECs | Primary bovine mammary epithelial cells |
| BMI | Body mass index |
| CFU | Colony-forming unit |
| CALC | Hypocalcemia |
| Cow.con.rate | Cow conception rate |
| DASB | Displaced abomasum |
| DEG | Differentially expressed gene |
| DFB | Days to first breeding |
| DMEM | Dulbecco’s modified Eagle’s medium |
| DSG | Differentially spliced gene |
| Dtr.cal.ease | Daughter calving ease |
| Dtr.preg.rate | Daughter pregnancy rate |
| Dtr.still.birth | Daughter stillbirth |
| FA | Folic acid |
| Fat | Fat yield |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| GSVA | Gene Set variation Analysis |
| GWAS | Genome-wide association studies |
| Heifer.con.rate | Heifer conception rate |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KETO | Ketosis |
| LDH | Lactate dehydrogenase |
| Liva | Livability |
| Mac-T | Bovine mammary alveolar cells |
| MAST | Mastitis |
| METR | Metritis |
| Milk | Milk yield |
| MLST | Multilocus sequence typing |
| MOI | Multiplicity of infection |
| MRSA | Methicillin-resistant S. aureus |
| OD | Optical density |
| PCA | Plate count agar |
| PCA | Principal component analysis |
| PCR | Polymerase chain reaction |
| Phe-WAS | Phenome-wide association study |
| Pro.life | Productive life |
| Pro.percent | Protein percentage |
| Protein | Protein yield |
| RETP | Retained placenta |
| RIN | RNA integrity number |
| RT | Reaction time |
| S. aureus | Staphylococcus aureus |
| SCC | Somatic cell count |
| SCS | Somatic cell score |
| Sire.calv.ease | Sire calving ease |
| Sire.still.ease | Sire stillbirth |
| TEF | TEF transcription factor, PAR bZIP family member |
| WGCNA | Weighted Correlation Network Analysis |
| ZBP1 | Z-DNA binding protein 1 |
References
- Rainard, P.; Foucras, G.; Fitzgerald, J.R.; Watts, J.L.; Koop, G.; Middleton, J.R. Knowledge gaps and research priorities in Staphylococcus aureus mastitis control. Transbound. Emerg. Dis. 2018, 65, 149–165. [Google Scholar] [CrossRef]
- Le Maréchal, C.; Jardin, J.; Jan, G.; Even, S.; Pulido, C.; Guibert, J.-M.; Hernandez, D.; François, P.; Schrenzel, J.; Demon, D.; et al. Staphylococcus aureus seroproteomes discriminate ruminant isolates causing mild or severe mastitis. Vet. Res. 2011, 42, 35. [Google Scholar] [CrossRef] [PubMed]
- de Jong, N.; van Kessel, K.; van Strijp, J. Immune evasion by Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 20. [Google Scholar] [CrossRef]
- Röhrig, C.; Huemer, M.; Lorgé, D.; Luterbacher, S.; Phothaworn, P.; Schefer, C.; Sobieraj, A.M.; Zinsli, L.V.; Shambat, S.M.; Leimer, N.; et al. Targeting Hidden Pathogens: Cell-Penetrating Enzybiotics Eradicate Intracellular Drug-Resistant Staphylococcus aureus. mBio 2020, 11, e00209-20. [Google Scholar] [CrossRef] [PubMed]
- Aqib, A.I.; Ijaz, M.; Farooqi, S.H.; Ahmed, R.; Shoaib, M.; Ali, M.M.; Mehmood, K.; Zhang, H. Emerging discrepancies in conventional and molecular epidemiology of methicillin resistant Staphylococcus aureus isolated from bovine milk. Microb. Pathog. 2018, 116, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef]
- Murphy, M.P.; Niedziela, D.A.; Leonard, F.C.; Keane, O.M. The in vitro host cell immune response to bovine-adapted Staphylococcus aureus varies according to bacterial lineage. Sci. Rep. 2019, 9, 6134. [Google Scholar] [CrossRef]
- Hoekstra, J.; Rutten, V.P.M.G.; Lam, T.J.G.M.; Van Kessel, K.P.M.; Spaninks, M.P.; Stegeman, J.A.; Benedictus, L.; Koop, G. Activation of a bovine mammary epithelial cell line by ruminant-associated Staphylococcus aureus is lineage dependent. Microorganisms 2019, 7, 688. [Google Scholar] [CrossRef]
- Gombart, A.F.; Pierre, A.; Maggini, S. A review of micronutrients and the immune system—Working in harmony to reduce the risk of infection. Nutrients 2020, 12, 236. [Google Scholar] [CrossRef]
- Wang, X.; Yu, Y. Infectious status of mastitis in dairy cattle induced by Staphylococcus aureus and its advances on epidemio-logical patterns and antimicrobial resistance in northern China. Acta Vet. Zootech. Sin. 2015, 46, 1477–1488. [Google Scholar]
- Wang, X.; Wang, Y.; Guo, G.; Usman, T.; Hao, D.; Tang, X.; Zhang, Y.; Yu, Y. Antimicrobial resistance and toxin gene profiles of Staphylococcus aureus strains from Holstein milk. Lett. Appl. Microbiol. 2014, 58, 527–534. [Google Scholar] [CrossRef]
- Biesalski, H.K. Nutrition meets the microbiome: Micronutrients and the microbiota. Ann. N. Y. Acad. Sci. 2016, 1372, 53–64. [Google Scholar] [CrossRef]
- Miszewski, S.G.; Trott, J.F.; E Berryhill, G.; Tat, L.; Green, R.; Borowsky, A.D.; Miller, J.W.; Hovey, R.C. Folate deficiency inhibits development of the mammary gland and its associated lymphatics in FVB mice. J. Nutr. 2020, 150, 2120–2130. [Google Scholar] [CrossRef]
- Lin, P.-H.; Sermersheim, M.; Li, H.; Lee, P.H.U.; Steinberg, S.M.; Matthew, S. Zinc in wound healing modulation. Nutrients 2017, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Khan, A.; Xiao, J.; Dou, J.; Liu, L.; Yu, Y. Overview of folic acid supplementation alone or in combination with vitamin B12 in dairy cattle during periparturient period. Metabolites 2020, 10, 263. [Google Scholar] [CrossRef] [PubMed]
- Munyaka, P.M.; Tactacan, G.; Jing, M.; Karmin, O.; House, J.D.; Paul, M.S.; Sharif, S.; Rodriguez-Lecompte, J.C. Response of older laying hens to an Escherichia coli lipopolysaccharide challenge when fed diets with or without supplemental folic acid. Poult. Sci. 2013, 92, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Gonda, T.A.; Kim, Y.; Salas, M.C.; Gamble, M.V.; Shibata, W.; Muthupalani, S.; Sohn, K.; Abrams, J.A.; Fox, J.G.; Wang, T.C.; et al. Folic acid increases global DNA methylation and reduces inflammation to prevent helicobacter-associated gastric cancer in mice. Gastroenterology 2012, 142, 824–833. [Google Scholar] [CrossRef]
- Feng, D.; Zhou, Y.; Xia, M.; Ma, J. Folic acid inhibits lipopolysaccharide-induced inflammatory response in RAW264.7 macrophages by suppressing MAPKs and NF-κB activation. Inflamm. Res. 2011, 60, 817–822. [Google Scholar] [CrossRef]
- Chen, P.; Li, C.; Li, X.; Li, J.; Chu, R.; Wang, H. Higher dietary folate intake reduces the breast cancer risk: A systematic review and meta-analysis. Br. J. Cancer 2014, 110, 2327–2338. [Google Scholar] [CrossRef]
- Khan, M.Z.; Liu, L.; Zhang, Z.; Khan, A.; Wang, D.; Mi, S.; Usman, T.; Liu, G.; Guo, G.; Li, X.; et al. Folic acid supplementation regulates milk production variables, metabolic associated genes and pathways in perinatal Holsteins. J. Anim. Physiol. Anim. Nutr. 2019, 104, 483–492. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Zhang, Z.; Liu, L.; Wang, D.; Mi, S.; Liu, X.; Liu, G.; Guo, G.; Li, X.; Wang, Y.; et al. Folic acid supplementation regulates key immunity-associated genes and pathways during the periparturient period in dairy cows. Asian-Australas. J. Anim. Sci. 2020, 33, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Tango, C.N.; Akkermans, S.; Hussain, M.S.; Khan, I.; Van Impe, J.; Jin, Y.-G.; Oh, D.H. Modeling the effect of pH, water activity, and ethanol concentration on biofilm formation of Staphylococcus aureus. Food Microbiol. 2018, 76, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Jiang, J.; Cole, J.B.; Freebern, E.; Da, Y.; VanRaden, P.M.; Ma, L. Functional annotation and Bayesian fine-mapping reveals candidate genes for important agronomic traits in Holstein bulls. Commun. Biol. 2019, 2, 212. [Google Scholar] [CrossRef]
- Freebern, E.; Santos, D.; Fang, L.; Jiang, J.; Parker, G.K.; Liu, G.E.; Vanraden, P.M.; Maltecca, C.; Cole, J.B.; Ma, L. GWAS and fine-mapping of livability and six disease traits in Holstein cattle. BMC Genom. 2020, 21, 41. [Google Scholar] [CrossRef]
- Fang, L.; Cai, W.; Liu, S.; Canela-Xandri, O.; Gao, Y.; Jiang, J.; Rawlik, K.; Li, B.; Schroeder, S.G.; Rosen, B.D.; et al. Compre-hensive analyses of 723 transcriptomes enhance genetic and biological interpretations for complex traits in cattle. Genome Res. 2020, 30, 790–801. [Google Scholar] [CrossRef]
- Watanabe, K.; Stringer, S.; Frei, O.; Mirkov, M.U.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Juhász-Kaszanyitzky, É.; Jánosi, S.; Somogyi, P.; Dán, Á.; Bloois, L.V.D.G.-V.; Van Duijkeren, E.; Wagenaar, J.A. MRSA Transmission between Cows and Humans. Emerg. Infect. Dis. 2007, 13, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Ballhausen, B.; Kahl, B.; Köck, R. The clinical impact of livestock-associated methicillin-resistant Staphylococcus aureus of the clonal complex 398 for humans. Vet. Microbiol. 2017, 200, 33–38. [Google Scholar] [CrossRef]
- Huber, H.; Koller, S.; Giezendanner, N.; Stephan, R.; Zweifel, C. Prevalence and characteristics of meticillin-resistant Staphy-lococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Eurosurveillance 2010, 15, 19542. [Google Scholar] [CrossRef] [PubMed]
- Budd, K.E.; Mitchell, J.; Keane, O.M. Lineage associated expression of virulence traits in bovine-adapted Staphylococcus aureus. Vet. Microbiol. 2016, 189, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chen, W.; He, J.; Jin, S.; Liu, Y.; Yi, Y.; Gao, Z.; Yang, J.; Yang, J.; Cui, J.; et al. Interplay of m 6 A and H3K27 trimethylation restrains inflammation during bacterial infection. Sci. Adv. 2020, 6, eaba0647. [Google Scholar] [CrossRef]
- Wang, D.; Liu, L.; Augustino, S.M.A.; Duan, T.; Hall, T.J.; MacHugh, D.E.; Dou, J.; Zhang, Y.; Wang, Y.; Yu, Y. Identification of novel molecular markers of mastitis caused by Staphylococcus aureus using gene expression profiling in two consecutive generations of Chinese Holstein dairy cattle. J. Anim. Sci. Biotechnol. 2020, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Hou, Y.; An, J.; Li, B.; Song, M.; Wang, X.; Sørensen, P.; Dong, Y.; Liu, C.; Wang, Y.; et al. Ge-nome-wide transcriptional and post-transcriptional regulation of innate immune and defense responses of bovine mammary gland to Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2016, 6, 193. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Bai, Y.; Li, Z.; Wang, F.; Fan, X.; Zhou, X. Bacterial extracellular vesicle-coated multi-antigenic nanovaccines protect against drug-resistant Staphylococcus aureus infection by modulating antigen processing and presentation pathways. Theranostics 2020, 10, 7131–7149. [Google Scholar] [CrossRef]
- Trivedi, U.; Madsen, J.S.; Everett, J.; Fell, C.; Russel, J.; Haaber, J.; Crosby, H.A.; Horswill, A.R.; Burmølle, M.; Rumbaugh, K.P.; et al. Staphylococcus aureus coagulases are exploitable yet stable public goods in clinically relevant conditions. Proc. Natl. Acad. Sci. USA 2018, 115, E11771–E11779. [Google Scholar] [CrossRef]
- Gutierrez, O.; Berciano, M.T.; Lafarga, M.; Fernandez-Luna, J.L. A novel pathway of TEF regulation mediated by mi-croRNA-125b contributes to the control of actin distribution and cell shape in fibroblasts. PLoS ONE 2011, 6, e17169. [Google Scholar] [CrossRef]
- Saleh, A.A.; Gohar, S.F.; Hemida, A.S.; Elgharbawy, M.; Soliman, S.E. Evaluation of ASPM and TEF gene expressions as po-tential biomarkers for bladder cancer. Biochem. Genet. 2020, 58, 490–507. [Google Scholar] [CrossRef]
- Inukai, T.; Inaba, T.; Dang, J.; Kuribara, R.; Ozawa, K.; Miyajima, A.; Wu, W.; Look, A.T.; Arinobu, Y.; Iwasaki, H.; et al. TEF, an antiapoptotic bZIP transcription factor related to the oncogenic E2A-HLF chimera, inhibits cell growth by down-regulating expression of the common beta chain of cytokine receptors. Blood 2005, 105, 4437–4444. [Google Scholar] [CrossRef][Green Version]
- Walker, R.L.; Ramaswami, G.; Hartl, C.; Mancuso, N.; Gandal, M.J.; de la Torre-Ubieta, L.; Pasaniuc, B.; Stein, J.L.; Geschwind, D.H. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell 2019, 179, 750–771. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ledue, O.; Jun, M.; Goulart, C.; Malley, R.; Lu, Y.J. Protection against Staphylococcus aureus colonization and In-FECTION by B- and T-cell-mediated mechanisms. mBio 2018, 9, e01949-18. [Google Scholar] [CrossRef]
- Lahouassa, H.; Moussay, E.; Rainard, P.; Riollet, C. Differential cytokine and chemokine responses of bovine mammary epi-thelial cells to Staphylococcus aureus and Escherichia coli. Cytokine 2007, 38, 12–21. [Google Scholar] [CrossRef]
- Zbinden, C.; Stephan, R.; Johler, S.; Borel, N.; Bünter, J.; Bruckmaier, R.M.; Wellnitz, O. The inflammatory response of primary bovine mammary epithelial cells to Staphylococcus aureus strains is linked to the bacterial phenotype. PLoS ONE 2014, 9, e87374. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Brenaut, P.; Lefèvre, L.; Rau, A.; Laloë, D.; Pisoni, G.; Moroni, P.; Bevilacqua, C.; Martin, P. Contribution of mammary epithelial cells to the immune response during early stages of a bacterial infection to Staphylococcus aureus. Vet. Res. 2014, 45, 16. [Google Scholar] [CrossRef] [PubMed]
- Marsillach, J.; Ferré, N.; Camps, J.; Riu, F.; Rull, A.; Joven, J. Moderately high folic acid supplementation exacerbates experi-mentally induced liver fibrosis in rats. Exp. Biol. Med. 2008, 233, 38–47. [Google Scholar] [CrossRef]
- Vollset, S.E.; Clarke, R.; Lewington, S.; Ebbing, M.; Halsey, J.; Lonn, E.; Armitage, J.; E Manson, J.; Hankey, G.; Spence, J.D.; et al. Effects of folic acid supplementation on overall and site-specific cancer incidence during the randomised trials: Meta-analyses of data on 50,000 individuals. Lancet 2013, 381, 1029–1036. [Google Scholar] [CrossRef]
- Yang, N.; Baumgartner, R.N.; Slattery, M.L.; Wang, C.; Giuliano, A.R.; Murtaugh, M.A.; Risendal, B.C.; Byers, T.; Baumgartner, K.B. Dietary intake of folate, B-vitamins and methionine and breast cancer risk among hispanic and non-hispanic white women. PLoS ONE 2013, 8, e54495. [Google Scholar] [CrossRef]
- Hansen, M.F.; Jensen, S.Ø.; Füchtbauer, E.M.; Martensen, P.M. High folic acid diet enhances tumour growth in PyMT-induced breast cancer. Br. J. Cancer 2017, 116, 752–761. [Google Scholar] [CrossRef]
- Bae, D.; Chon, J.-W.; Kim, D.-H.; Kim, H.; Seo, K.-H. Effect of folic acid supplementation on proliferation and apoptosis in bovine mammary epithelial (MAC-T) cells. Anim. Biotechnol. 2020, 1–9. [Google Scholar] [CrossRef]
- Burton, M.A.; Antoun, E.; Penailillo, R.S.; Burdge, G.C.; Lillycrop, K.A. Folic acid induces intake-related changes in the mammary tissue transcriptome of C57BL/6 mice. Nutrients 2020, 12, 2821. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Z.J. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 2014, 32, 461–488. [Google Scholar] [CrossRef] [PubMed]
- Kuriakose, T.; Kanneganti, T.-D. ZBP1: Innate sensor regulating cell death and inflammation. Trends Immunol. 2018, 39, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Momota, M.; Lelliott, P.; Kubo, A.; Kusakabe, T.; Kobiyama, K.; Kuroda, E.; Imai, Y.; Akira, S.; Coban, C.; Ishii, K.J. ZBP1 governs the inflammasome-independent IL-1α and neutrophil inflammation that play a dual role in anti-influenza virus im-munity. Int. Immunol. 2020, 32, 203–212. [Google Scholar] [PubMed]
- Lin, J.; Kumari, S.; Kim, C.; Van, T.M.; Wachsmuth, L.; Polykratis, A.; Pasparakis, M. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nat. Cell Biol. 2016, 540, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Panchatcharam, B.S.; Cooksley, C.M.; Ramezanpour, M.; Vediappan, R.S.; Bassiouni, A.; Wormald, P.J.; Psaltis, A.J.; Vreugde, S. Staphylococcus aureus biofilm exoproteins are cytotoxic to human nasal epithelial barrier in chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2020, 10, 871–883. [Google Scholar] [CrossRef] [PubMed]
- Malik, Z.; Roscioli, E.; Murphy, J.; Ou, J.; Bassiouni, A.; Wormald, P.J.; Vreugde, S. Staphylococcus aureus impairs the airway epithelial barrier in vitro. Int. Forum Allergy Rhinol. 2015, 5, 551–556. [Google Scholar] [CrossRef]
- Asselstine, V.; Miglior, F.; Suárez-Vega, A.; Fonseca, P.; Mallard, B.; Karrow, N.; Islas-Trejo, A.; Medrano, J.; Cánovas, A. Genetic mechanisms regulating the host response during mastitis. J. Dairy Sci. 2019, 102, 9043–9059. [Google Scholar] [CrossRef]
- Murphy, J.; Ramezanpour, M.; Drilling, A.; Roscioli, E.; Psaltis, A.; Wormald, P.-J.; Vreugde, S. In vitro characteristics of an airway barrier-disrupting factor secreted by Staphylococcus aureus. Int. Forum Allergy Rhinol. 2019, 9, 187–196. [Google Scholar] [CrossRef]
- Murphy, J.; Ramezanpour, M.; Stach, N.; Dubin, G.; Psaltis, A.; Wormald, P.-J.; Vreugde, S. Staphylococcus aureus V8 protease disrupts the integrity of the airway epithelial barrier and impairs IL-6 production in vitro. Laryngoscope 2018, 128, E8–E15. [Google Scholar] [CrossRef]
- Huynh, H.T.; Robitaille, G.; Turner, J.D. Establishment of bovine mammary epithelial cells (MAC-T): An in vitro model for bovine lactation. Exp. Cell Res. 1991, 197, 191–199. [Google Scholar] [CrossRef]
- Günther, J.; Koy, M.; Berthold, A.; Schuberth, H.-J.; Seyfert, H.-M. Comparison of the pathogen species-specific immune response in udder derived cell types and their models. Vet. Res. 2016, 47, 22. [Google Scholar] [CrossRef] [PubMed]
- Ryman, V.E.; Packiriswamy, N.; Sordillo, L.M. Role of endothelial cells in bovine mammary gland health and disease. Anim. Health Res. Rev. 2015, 16, 135–149. [Google Scholar] [CrossRef] [PubMed]


| Gene Names | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| Nuc | GCGATTGATGGTGATACGGTT | AGCCAAGCCTTGACGAACTAAAGC |
| MecA | GTAGAAATGACTGAACGTCCGATAA | CCAATTCCACATTGTTTCGGTCTAA |
| TEF-AS | GGTGGCTGAGCTAGAAGGG | CAGGGTCGGGATTGAAGT |
| TEF | GCCTCCGAACAGACAAATC | AATCAAGGGTCACTGCTACG |
| RBBP8-AS | GTTAGTCAGGGAACGAGG | AGGGCTTCCACAACTGCT |
| RBBP8 | AGCCAAGGATGTGAGA | TGGACGAAGAGGGATT |
| ZBP1 | CAGGAGACACAGACCTTGAGCAG | CATCTTGTGGAGGAGCTGGTT |
| GAPDH | GGTGCTGAGTATGTGGTGGA | GGCATTGCTGACAATCTTGA |
| IFNAR1 | TCTGCGTCCTTTGCC | CACAGGGCTGCTTACA |
| IFNAR2 | CTGTGTGTGTGAGAGCCCTT | GGCCACCAAAACGCTTGATT |
| IRF3 | CGACCCAACACTTAGGCCAG | GTCGGGCTTATCCTTCCCAG |
| IRF7 | GACTTCGGCACCTTCTTCCA | TAGATGGTGTAGTGCGGGGA |
| TBK1 | GGATGAGGGACAATTCTGTGTCT | ACCAAAACCTAATCCTTCTGGG |
| IKBKE | GGCCTCTCTGCTCAACACAT | ATCTCCACGAACCAGTGCAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mi, S.; Tang, Y.; Shi, L.; Liu, X.; Si, J.; Yao, Y.; Augustino, S.M.A.; Fang, L.; Yu, Y. Protective Roles of Folic Acid in the Responses of Bovine Mammary Epithelial Cells to Different Virulent Staphylococcus aureus Strains. Biology 2021, 10, 1164. https://doi.org/10.3390/biology10111164
Mi S, Tang Y, Shi L, Liu X, Si J, Yao Y, Augustino SMA, Fang L, Yu Y. Protective Roles of Folic Acid in the Responses of Bovine Mammary Epithelial Cells to Different Virulent Staphylococcus aureus Strains. Biology. 2021; 10(11):1164. https://doi.org/10.3390/biology10111164
Chicago/Turabian StyleMi, Siyuan, Yongjie Tang, Liangyu Shi, Xueqin Liu, Jingfang Si, Yuelin Yao, Serafino M. A. Augustino, Lingzhao Fang, and Ying Yu. 2021. "Protective Roles of Folic Acid in the Responses of Bovine Mammary Epithelial Cells to Different Virulent Staphylococcus aureus Strains" Biology 10, no. 11: 1164. https://doi.org/10.3390/biology10111164
APA StyleMi, S., Tang, Y., Shi, L., Liu, X., Si, J., Yao, Y., Augustino, S. M. A., Fang, L., & Yu, Y. (2021). Protective Roles of Folic Acid in the Responses of Bovine Mammary Epithelial Cells to Different Virulent Staphylococcus aureus Strains. Biology, 10(11), 1164. https://doi.org/10.3390/biology10111164






