Detection and Control of Fusarium oxysporum from Soft Rot in Dendrobium officinale by Loop-Mediated Isothermal Amplification Assays
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Isolation and Purification of Strain
2.3. DNA Extraction
2.4. Primers Design
2.5. Optimization of LAMP Reaction Conditions
2.6. LAMP Assays Specificity
2.7. Detection of F. axysporum by LAMP and Conventional PCR
2.8. The Feasibility Detection of LAMP Assays
2.9. Prevention and Control of F. oxysporum from Soft Rot in D. officinale by LAMP Assays
3. Results
3.1. Identification of Pathogens
3.2. Optimization of LAMP Assays
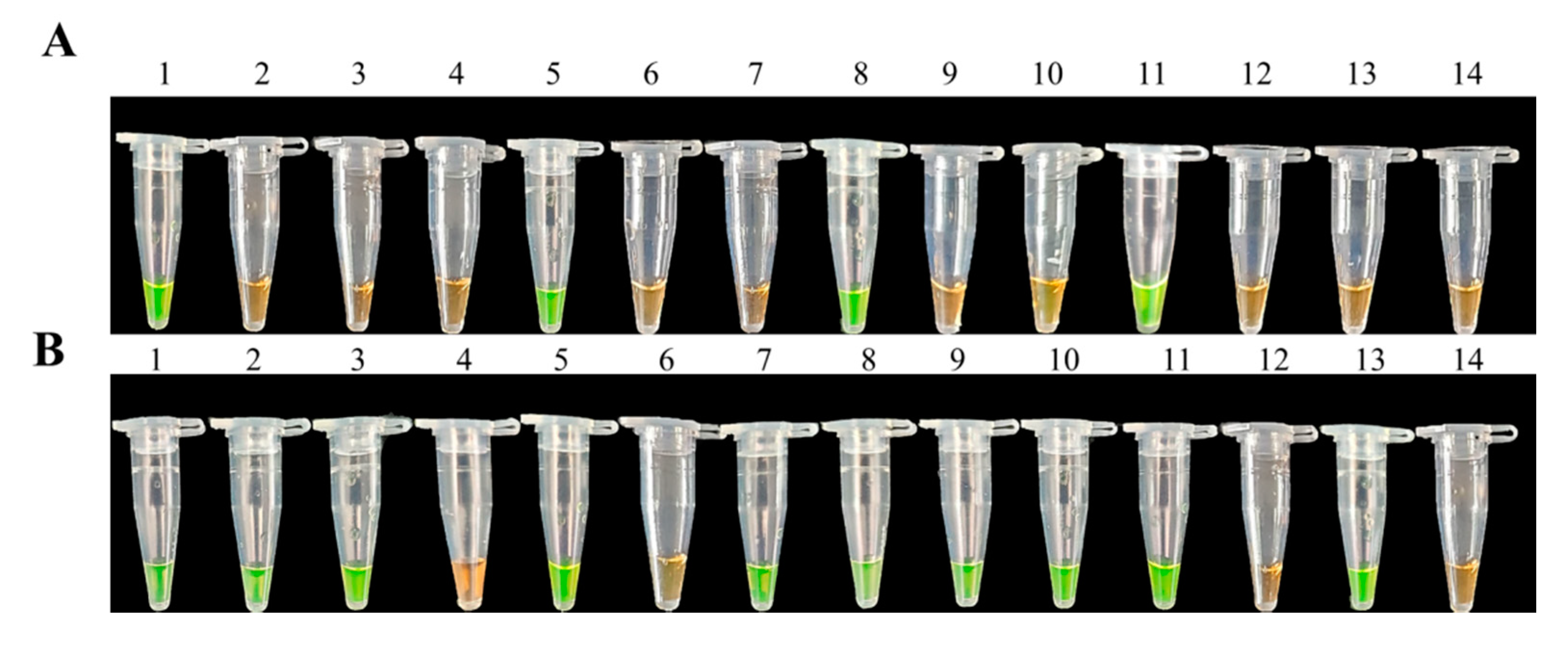
3.3. Specificity of LAMP Assays
3.4. Sensitivities of LAMP and Conventional PCR Assays
3.5. The Feasibility Detection of LAMP Assays
3.6. Prevention and Control of Soft Rot in D. officinale by LAMP Assays
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ge, D.; Li, X.; Ding, X.; Qian, L. Genetic diversity across natural populations of Dendrobium officinale, the endangered medicinal herb endemic to China, revealed by ISSR and RAPD markers. Russ. J. Genet. 2009, 45, 327–334. [Google Scholar] [CrossRef]
- Jin, Z.; Li, D.; Liu, T.; Yu, F.; Liu, Z. Cultural endophytic fungi associated with Dendrobium officinale: Identification, diversity estimation and their antimicrobial potential. Curr. Sci. 2017, 112, 1690–1697. [Google Scholar] [CrossRef]
- Luo, Q.L.; Tang, Z.H.; Zhang, X.F.; Zhong, Y.H.; Yao, S.Z.; Wang, L.S.; Lin, C.W.; Luo, X. Chemical properties and antioxidant activity of a water-soluble polysaccharide from Dendrobium officinale. Int. J. Biol. Macromol. 2016, 89, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Wu, J.J.; Li, X.F.; Lu, J.M.; Wu, W.; Sun, Y.Q.; Zhu, B.; Qin, L.P. Isolation, structural properties, bioactivities of polysaccharides from Dendrobium officinale Kimura et. Migo: A review. Int. J. Biol. Macromol. 2021, 184, 1000–1013. [Google Scholar] [CrossRef]
- Liu, X.F.; Zhu, J.; Ge, S.Y.; Xia, L.J.; Ren, F.Z. Orally Administered Dendrobium officinale and its Polysaccharides Enhance Immune Functions in BALB/c Mice. Nat. Prod. Commun. 2011, 6, 867–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.J.; Liu, Z.P.; Zhang, X.F.; Si, J.P. Effects of Various Processing Methods on the Metabolic Profile and Antioxidant Activity of Dendrobium catenatum Lindley Leaves. Metabolites 2021, 11, 351. [Google Scholar] [CrossRef]
- Li, G.; Li, R.Y.; Gao, W.W. Occurrence situation and control strategy of Dendrobium diseases in large-scale farming system. China J. Chin. Mater. Med. 2013, 38, 485. [Google Scholar]
- Guo, M.; Li, B.; Wang, R.; Liu, P.; Chen, Q. Occurrence of dieback disease caused by Fusarium equiseti on Dendrobium officinale in China. Crop. Prot. 2020, 137, 105209. [Google Scholar] [CrossRef]
- Vellupillai, S.; Jin, G. Histological and protein changes during early stages of seed germination in the orchid, Dendrobium crumenatum. J. Hortic. Sci. 1997, 72, 941–948. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, J.Y.; Wong, J.H.; Ye, X.J.; Wing Sze, S.C.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef]
- Lin, X.; Zhou, Y.; Zhong, J.; Zhu, J.; Chen, X.; Lin, Y.; Huang, M.; Yang, L.; Zhang, Z. Imitated Ecological Cultivation Technology of Dendrobium offcinale: A Case Study of Longshitou Village, Longmen County. Plant. Dis. Pests 2021, 12, 24–26. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, H.; Liang, D.; Peng, H.; Huang, C.; Zhou, Z.; Liao, Y.; Lin, D. Wild cultivation technology of Dendrobium officinale in natural secondary forest in northern Guangdong Province. South. China For. Sci. 2018, 46, 48–50. [Google Scholar] [CrossRef]
- Tang, L.M.; Chen, Y.F.; Dai, Q.; Li, D.Y.; Jiang, L.; Deng, Y.H. The Standardized Technology Demonstration of Cultivating Dendrobium officinale Kimura et Migo under Forest. Anhui Agric. Sci. Bull. 2015, 21, 45–53. [Google Scholar] [CrossRef]
- Guan, Y.J.; Li, L.R.; Liu, S.D. Effects of Meteorological Conditions on the Growth of Dendrobium candidum under Forest. Sci. Technol. Eng. 2018, 18, 200–203. [Google Scholar]
- Oren, L.; Ezrati, S.; Cohen, D.; Sharon, A. Early events in the Fusarium verticillioides-maize interaction characterized by using a green fluorescent protein-expressing transgenic isolate. Appl. Environ. Microbiol. 2003, 69, 1695–1701. [Google Scholar] [CrossRef] [Green Version]
- Olivain, C.; Humbert, C.; Nahalkova, J.; Fatehi, J.; L’Haridon, F.; Alabouvette, C. Colonization of tomato root by pathogenic and nonpathogenic Fusarium oxysporum strains inoculated together and separately into the soil. Appl. Environ. Microbiol. 2006, 72, 1523–1531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagopodi Anastasia, L.; Ram Arthur, F.J.; Lamers, G.E.M.; Punt, P.J.; Van den Hondel, C.A.M.J.J. Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant.-Microbe Interact. 2002, 15, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Eynck, C.; Koopmann, B.; Grunewaldt-Stoecker, G.; Karlovsky, P.; Tiedemann, A.V. Differential interactions of Verticillium longisporum and V. dahliae with Brassica napus detected with molecular and histological techniques. Eur. J. Plant. Pathol. 2007, 118, 259–274. [Google Scholar] [CrossRef]
- Zhou, L.; Hu, Q.; Johansson, A.; Dixelius, C. Verticillium longisporum and V. dahliae: Infection and disease in Brassica napus. Plant. Pathol. 2006, 55, 137–144. [Google Scholar] [CrossRef]
- Herman, R.; Zvirin, Z.; Kovalski, I.; Freeman, S.; Denisov, Y.; Zuri, G.; Katzir, N.; Perl-Treves, R.; Pitrat, M. Characterization of Fusarium race 1.2 resistance in melon and mapping of a major QTL for this trait near a fruit netting locus. In Proceedings of the IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceaee, Avignon, France, 21–24 May 2008; pp. 149–156. [Google Scholar] [CrossRef]
- Song, H.H.; Lee, H.S.; Jeong, J.H.; Park, H.S.; Chan, L. Diversity in Beauvericin and Enniatins H, I, and MK1688 by Fusarium oxysporum isolated from potato. Int. J. Food Microbiol. 2008, 122, 296–301. [Google Scholar] [CrossRef]
- Son, S.W.; Kim, H.Y.; Choi, G.J.; Lim, H.K.; Jang, K.S.; Lee, S.O.; Lee, S.; Sung, N.D.; Kim, C.J. Bikaverin and fusaric acid from Fusarium oxysporum show antioomycete activity against Phytophthora infestans. J. Appl. Microbiol. 2008, 104, 692–698. [Google Scholar] [CrossRef]
- Waskiewicz, A.; Golinski, P.; Karolewski, Z.; Irzykowska, L.; Weber, Z. Formation of fumonisins and other secondary metabolites by Fusarium oxysporum and F. proliferatum: A comparative study. Food Addit. Contam. Part A 2010, 27, 608–615. [Google Scholar] [CrossRef] [Green Version]
- Zaccardelli, M.; Spasiano, A.; Bazzi, C.; Merighi, M. Identificationand in planta detection of Pseudomonas syringae pv tomatousing PCR amplification of hrpZpst. Eur. J. Plant. Pathol. 2005, 111, 85–90. [Google Scholar] [CrossRef]
- George, J.R.; Vanneste, J.L.; Cornish, D.A.; Pushparajah, I.P.S.; Yu, J.; Templeton, M.D.; Everett, K.R. Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S–23S rDNA inter transcribed spacer region and comparison with PCR primers based on other gene regions. Plant. Pathol. 2010, 59, 453–464. [Google Scholar] [CrossRef]
- Niessen, L.; Luo, J.; Denschlag, C.; Vogel, R.F. The application of loop-mediated isothermal amplification (LAMP) in food testing for bacterial pathogens and fungal contaminants. Food Microbiol. 2013, 36, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.F.; Tao, L.Y.; Yu, H.B.; Zhang, H.; Wu, Y.J.; Wu, S.G.; Zhou, J. Development of a reverse transcription loop mediated isothermal amplification assays for the detection of Mouse reovirus type 3 in laboratory mice. Sci. Rep. 2021, 11, 3508. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.A.; Huang, P.; Hou, X.Y.; Yu, D.Y. Rapid detection by a loop-mediated isothermal amplification assays based on EF-1α gene for stem rot on Cymbidium ensifolium. Eur. J. Hortic. Sci. 2021, 86, 212–218. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwamoto, T.; Sonobe, T.; Hayashi, K. Loop-Mediated Isothermal Amplification for Direct Detection of Mycobacterium tuberculosis Complex, M. avium, and M. intracellulare in Sputum Samples. J. Clin. Microbiol. 2003, 41, 2616–2622. [Google Scholar] [CrossRef] [Green Version]
- Duan, Y.B.; Yang, Y.; Wang, J.X.; Liu, C.C.; He, L.L.; Zhou, M.G. Development and application of loop-mediated isothermal amplification for detecting the highly benzimidazole-resistant isolates in Sclerotinia sclerotiorum. Sci. Rep. 2015, 5, 17278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, Y.; Notomi, T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef]
- Verma, S.; Avishek, K.; Sharma, V.; Negi, N.S.; Ramesh, V.; Salotra, P. Application of loop-mediated isothermal amplification assays for the sensitive and rapid diagnosis of visceral leishmaniasis and post-kala-azar dermal leishmaniasis. Diagn. Microbiol. Infect. Dis. 2013, 75, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.Y.; Yoshino, M.; Kojima, T.; Ikedo, M. Loop-mediated isothermal amplification for the rapid detection of Salmonella. Fems Microbiol. Lett. 2010, 253, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Nirenberg, H.I.; O′Donnell, K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia 1998, 90, 434–458. [Google Scholar] [CrossRef]
- Taylor, J.W.; Jacobson, D.J.; Fisher, M.C. The evolution of asexual fungi: Reproduction, Speciation and Classification. Immunology 1999, 121, 383–391. [Google Scholar] [CrossRef]
- Amatulli, M.; Spadaro, D.; Gullino, M.L.; Garibaldi, A. Corrigendum: Molecular identification of Fusarium spp. associated with bakanae disease of rice in Italy and assessment of their pathogenicity. Plant. Pathol. 2012, 61, 820. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, F.; White, T.J.; Lee, S.H.; Taylor, L.; Shawe-taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Li, H.S.; Ye, W.; Wang, Y.; Chen, X.H. RNA sequencing-based exploration of the effects of far-red light on lncRNAs involved in the shade-avoidance response of D. officinale. PeerJ 2021, 9, e10769. [Google Scholar] [CrossRef]
- Zhang, M.Z.; Yu, Z.M.; Zeng, D.Q.; Si, C. Transcriptome and Metabolome Reveal Salt-Stress Responses of Leaf Tissues from Dendrobium officinale. Biomolecules 2021, 11, 736. [Google Scholar] [CrossRef]
- Almasi, M.A. Development of A Colorimetric Loop-Mediated Isothermal Amplification Assays for the Visual Detection of Fusarium oxysporum f.sp. melonis. Hortic. Plant. J. 2019, 5, 41–48. [Google Scholar] [CrossRef]
- Shamsher, A.; Hasmi, S.K.; Aftab, M.A.; Khan, R.U. Biological Management of Chrysanthemum Wilt (Fusarium oxysporum f.sp. chrysanthemi. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 639–644. [Google Scholar] [CrossRef]
- Zhang, J.; Li, T.; Cai, Y.F.; Wang, Y.Z. Genetic and environmental effects on allometry of the medicinal plant Dendrobium officinale (Orchidaceae) from Yunnan, southwest China. Pak. J. Bot. 2021, 53, 1675–1682. [Google Scholar] [CrossRef]
- Bao, Y.X.; Sun, H.J.; Li, Y.F.; Duan, Z.Z.; McCord, P.H.; Cui, Y.P.; Zhang, M.Q. First Report of Fusarium oxysporum isolate gx3 Causing Sugarcane Pokkah Boeng in Guangxi of China. Plant. Dis. 2016, 100, 1785. [Google Scholar] [CrossRef]
- Pastrana, A.M.; Cline, W.; Wong, T.W.; Watson, D.C.; Mercier, J.; Ivors, K.; Broome, J.C.; Quesada-Ocampo, L.M.; Gordon, T.R. First report of Fusarium Wilt of Blackberry Caused by Fusarium oxysporum f. sp. mori in North Carolina. Plant. Dis. 2019, 104, 971. [Google Scholar] [CrossRef]
- Zhu, Y.; Lujan, P.; Wedegaertner, T.; Nichols, R.L.; Sanogo, S. First Report of Fusarium oxysporum f. sp. vasinfectum Race 4 Causing Fusarium Wilt of Cotton in New Mexico, U.S.A. Plant. Dis. 2020, 104, 588. [Google Scholar] [CrossRef]
- Bowers, J.H.; Locke, J.C. Effect of Botanical Extracts on the Population Density of Fusarium oxysporum in Soil and Control of Fusarium Wilt in the Greenhouse. Plant. Dis. 2000, 84, 300–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cal, A.D.; Pascual, S.; Larena, I.; Melgarejo, P. Biological control of Fusarium oxysporum f. sp. lycopersici. Plant. Pathol. 2010, 44, 909–917. [Google Scholar] [CrossRef]
- Hadian, S.; Rahnama, K.; Jamali, S.; Eskandari, A. Comparing neem extract with chemical control on Fusarium oxysporum and Meloidogyne incognita complex of tomato. Adv. Environ. Biol. 2011, 5, 2052–2057. [Google Scholar]
- Arif, M.; Chawla, S.; Zaidi, N.W.; Rayar, J.K.; Singh, U.S. Development of specific primers for genus Fusarium and F. solani using rDNA sub-unit and transcription elongation factor (TEF-1α) gene. Afr. J. Biotechnol. 2014, 11, 444–447. [Google Scholar] [CrossRef]
- Uribe-Cortés, T.B.; Silva-Rojas, H.V.; Mendoza-Onofre, L.E.; Velázquez-Cruz, C.; Rebollar-Alviter, N. Identificación de especies de Fusarium aisladas de semillas sintomáticas y asintomáticas de maíz con base en el gen TEF-1α. Rev. Fitotec. Mex. Publ. Por La Soc. Mex. De Fitogenética 2020, 43, 79. [Google Scholar] [CrossRef]
- Huang, S.; Xu, Y.; Yan, X.; Shang, Y.; Zhu, P.; Tian, W.; Xu, W. Development and application of a quantitative loop-mediated isothermal amplification method for detecting genetically modified maize MON863. J. Sci. Food Agric. 2015, 95, 253–259. [Google Scholar] [CrossRef]
- Singh, R.; Singh, D.P.; Savargaonkar, D.; Singh, O.P.; Bhatt, R.M.; Valecha, N. Evaluation of SYBR green I based visual loop-mediated isothermal amplification (LAMP) assays for genus and species-specific diagnosis of malaria in P. vivax and P. falciparum endemic regions. J. Vector Borne Dis. 2017, 54, 54–60. [Google Scholar]
- Zhang, X.; Zhang, H.; Pu, J.; Qi, Y.; Yu, Q.; Xie, Y.; Peng, J. Development of a Real-Time Fluorescence Loop-Mediated Isothermal Amplification Assays for Rapid and Quantitative Detection of Fusarium oxysporum f. sp. cubense Tropical Race 4 In Soil. PLoS ONE 2013, 349, 127–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayukawa, Y.; Hanyuda, S.; Fujita, N.; Komatsu, K.; Arie, T. Novel loop-mediated isothermal amplification (LAMP) assays with a universal QProbe can detect SNPs determining races in plant pathogenic fungi. Sci. Rep. 2017, 7, 4253. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Tarafdar, A.; Sharma, M. Rapid and sensitive diagnoses of dry root rot pathogen of chickpea (Rhizoctonia bataticola (Taub.) Butler) using loop-mediated isothermal amplification assays. Sci. Rep. 2017, 7, 42737. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhang, H.; Chen, F.P.; Zhang, X.; Xie, Y.X.; Hou, X.W.; Li, G.Y.; Pu, J.J. Rapid and quantitative detection of Fusarium oxysporum f. sp. cubense race 4 in soil by real-time fluorescence loop-mediated isothermal amplification. J. Appl. Microbiol. 2015, 117, 1740–1749. [Google Scholar] [CrossRef]
- Cao, Y.Y.; Wang, L.; Duan, L.; Li, J.; Ma, J.; Xie, S.; Shi, L.; Li, H. Development of a real-time fluorescence loop-mediated isothermal amplification assays for rapid and quantitative detection of Ustilago maydis. Sci. Rep. 2017, 7, 13394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Dai, T.; Zhang, H.F.; Wang, Y.C.; Zheng, X.B. Development of a Loop-Mediated Isothermal Amplification Assays to Detect Fusarium oxysporum. J. Phytopathol. 2015, 163, 63–66. [Google Scholar] [CrossRef]
- Lan, C.Z.; Ruan, H.C.; Yang, X.J.; Yao, J.A.; Jiang, J.X. Development of a loop-mediated isothermal amplification assays for sensitive and specific detection of Fusarium oxysporum f. sp. cucumerinum Owen. Phytoparasitica 2018, 46, 283–293. [Google Scholar] [CrossRef]
- Olivares, B.O.; Rey, J.C.; Lobo, D.; Navas-Cortés, J.A.; Gómez, J.A.; Landa, B.B. Fusarium Wilt of Bananas: A Review of Agro-Environmental Factors in the Venezuelan Production System Affecting Its Development. Agronomy 2021, 11, 986. [Google Scholar] [CrossRef]
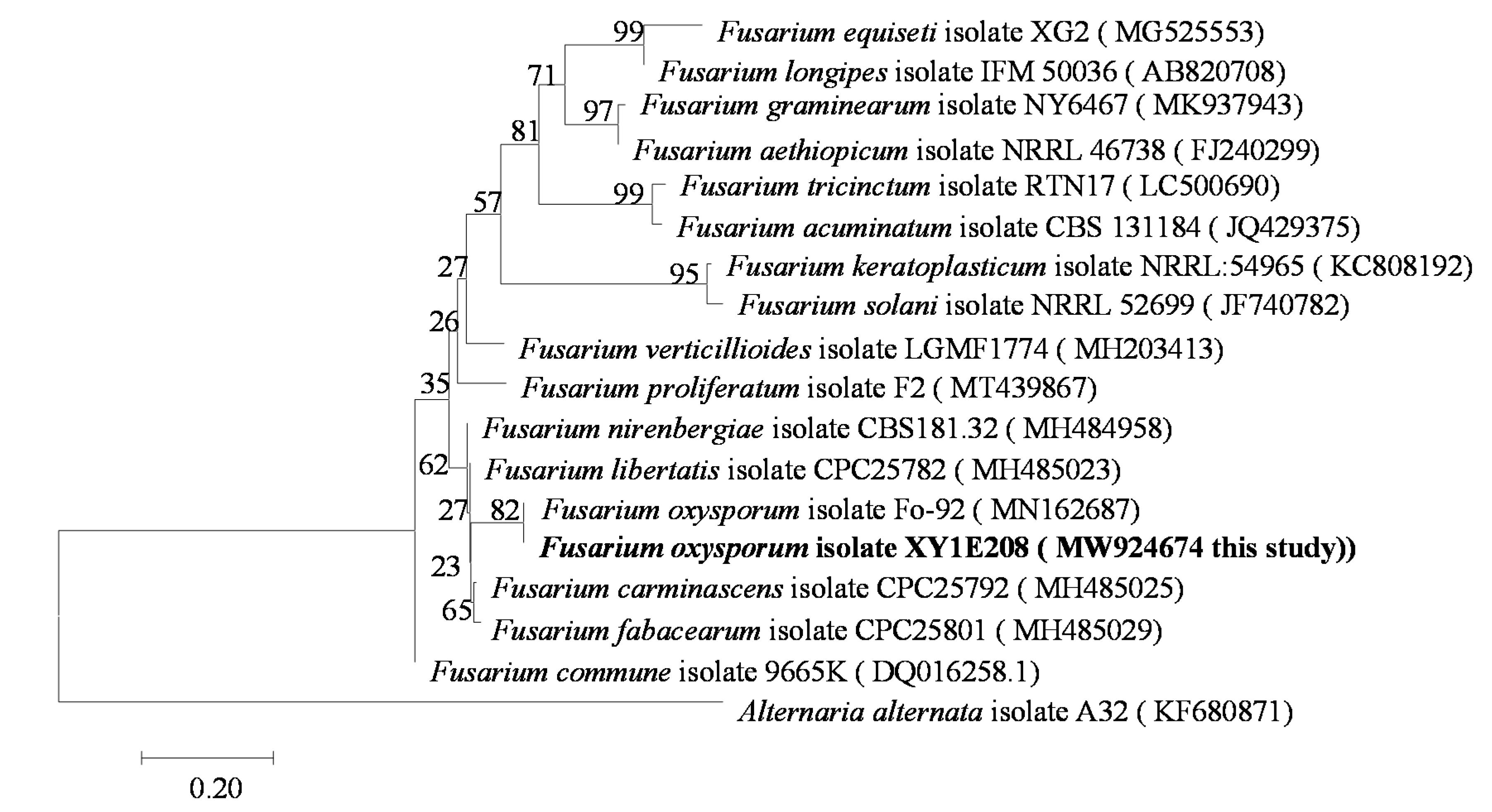

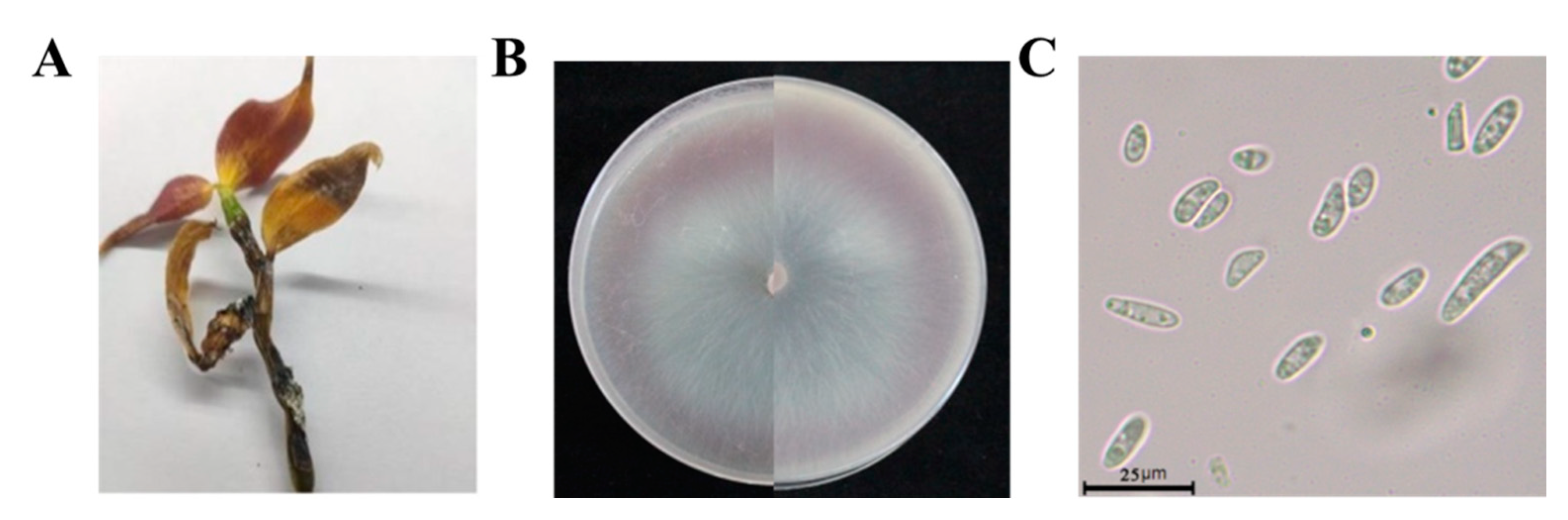
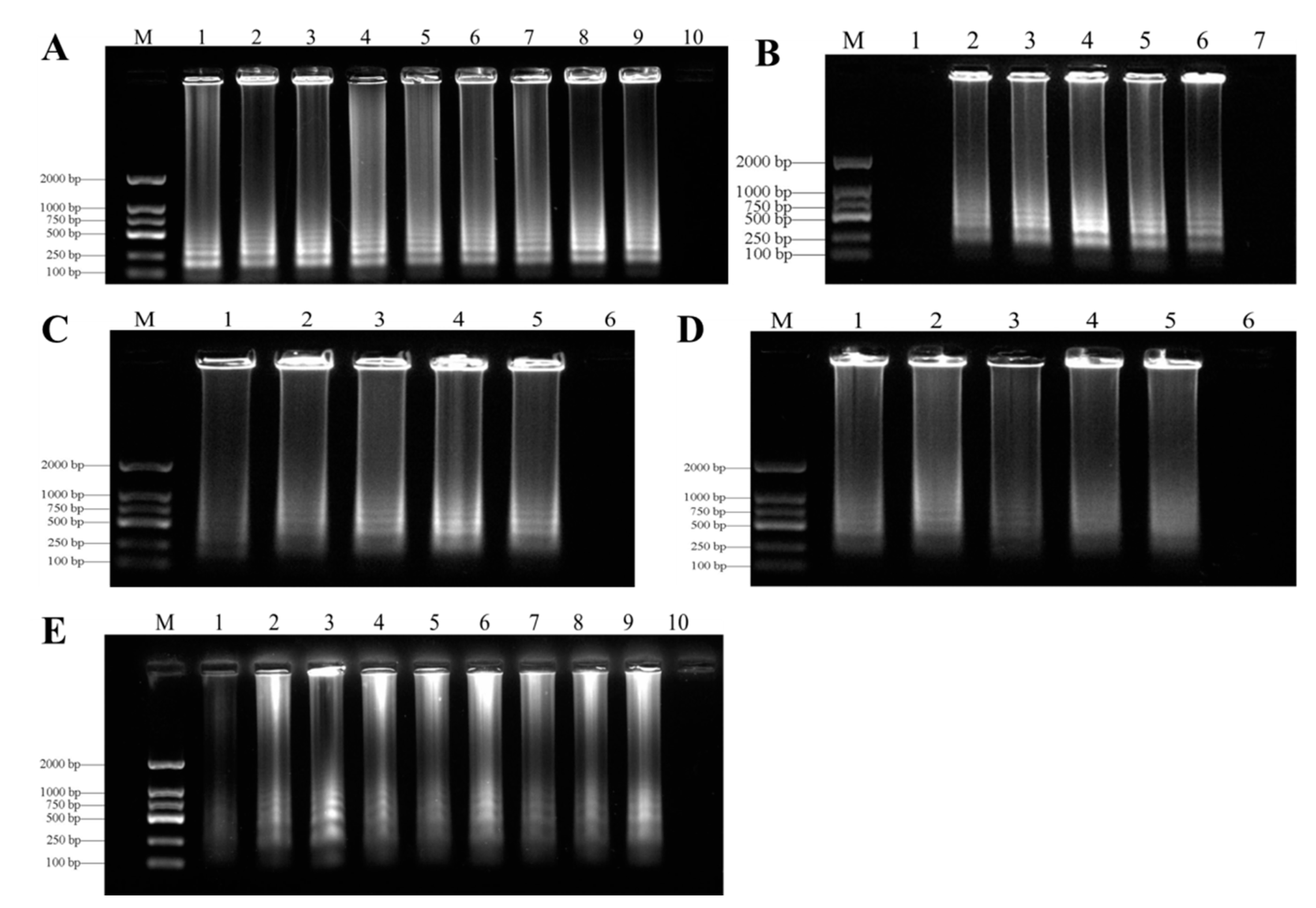

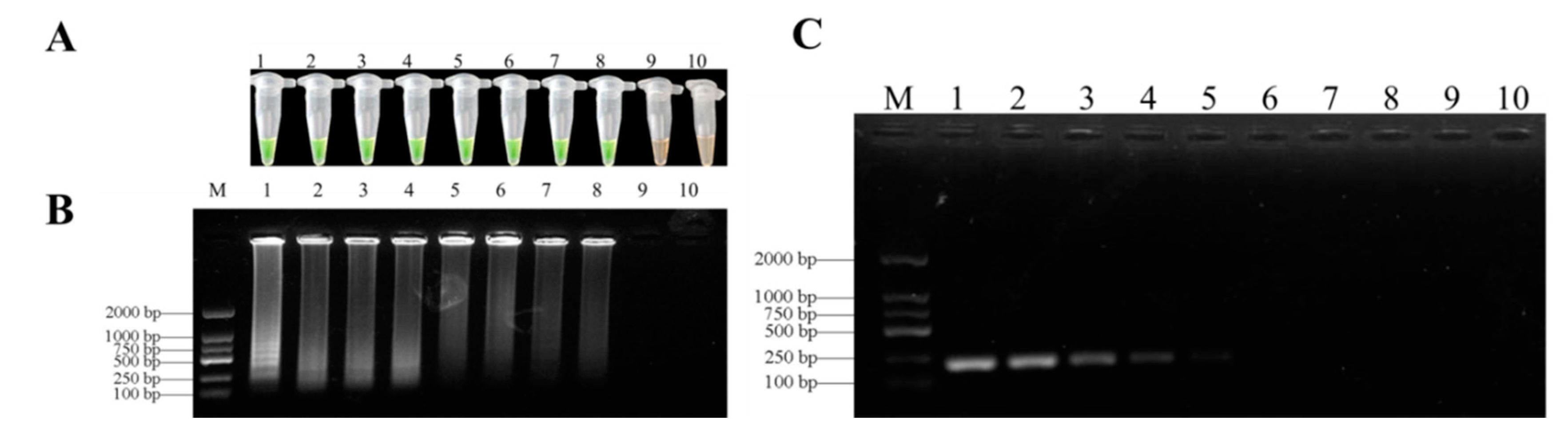

| Species | Host | Geographical Location | Number of Strains | LAMP detection | |
|---|---|---|---|---|---|
| Agarose Gel | SYBR Green I | ||||
| Fusarium oxysporum | Dendrobium officinale | Xingyi, Guizhou | 1 | + | + |
| Fusarium proliferatum | Dendrobium officinale | Huaxi, Guiyang | 1 | − | − |
| Fusarium equiseti | Dendrobium officinale | Huaxi, Guiyang | 1 | − | − |
| Fusarium solani | Dendrobium officinale | Libo, Guiyang | 1 | − | − |
| Fusarium chlamydosporum | Dendrobium officinale | Anlong, Guiyang | 1 | − | − |
| Fusarium fujikuroi | Plum | Huishui, Guiyang | 1 | − | − |
| Fusarium graminearum | Kiwi Fruit | Xifeng, Guiyang | 1 | − | − |
| Colletotrichum fructicola | Dendrobium officinale | Sansui, Guizhou | 1 | − | − |
| Epicoccum sorghinum | Dendrobium officinale | Xingyi, Guizhou | 1 | − | − |
| Neurospora sitophila | Dendrobium officinale | Jinping, Guizhou | 1 | − | − |
| Lasiodiplodia pseudotheobromae | Dendrobium officinale | Jinping, Guizhou | 1 | − | − |
| Trichoderma harzianum | Soil | Jinping, Guizhou | 1 | − | − |
| Botryosphaeria dothidea | Dendrobium officinale | Jinping, Guizhou | 1 | − | − |
| Phomopsis sp. | Kiwi Fruit | Xifeng, Guizhou | 1 | − | − |
| Pythium ultimum | Dendrobium officinale | Huaxi, Guizhou | 1 | − | − |
| Magnaporthe grisea | Oryza sativa | Huaxi, Guizhou | 1 | − | − |
| Rhizoctonia solani | Oryza sativa | Huaxi, Guizhou | 1 | − | − |
| Botrytis cinerea | Kiwi Fruit | Xifeng, Guizhou | 1 | − | − |
| Primer Name | Sequence (5′–3′) | Length |
|---|---|---|
| F3 | ACTGCTTGACACGTGACG | 18 |
| B3 | CACTTTCCCTTCGATCGCG | 19 |
| FIP | ACTTACCCCGCCACTTGAGCACGCACTCATTGAGGTTGTG | 40 |
| BIP | TTGGTCTCGAGCGGGGTAGCTCCTTTGCCCATCGATTTCC | 40 |
| LF | CGTTTGCCCTCTTAACCATTCT | 22 |
| LB | GGGCACATTTCGAGTCGTAGG | 21 |
| Treatment | Concentration (g a.i./hm2) | Average Disease Index before Spraying Fungicide | Average Disease Index 14 Days after Spraying Fungicide | Control Efficacy (%) |
|---|---|---|---|---|
| CK | / | 32.08 | 87.41 | / |
| Pyraclostrobin(A) | 4.9635 | 35.83 | 44.44 | 84.25 a |
| Picoxystrobin(B) | 11.0865 | 43.47 | 55.65 | 78.07 c |
| A:B(1:3) | 4.128 | 47.78 | 57.5 | 82.39 ab |
| Osthole(C) | 16.839 | 41.59 | 53.06 | 79.29 bc |
| Physcion(D) | 192.3 | 35.65 | 54.21 | 66.47 d |
| C:D(7:1) | 18.195 | 38.33 | 51.11 | 76.74 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, C.; Li, R. Detection and Control of Fusarium oxysporum from Soft Rot in Dendrobium officinale by Loop-Mediated Isothermal Amplification Assays. Biology 2021, 10, 1136. https://doi.org/10.3390/biology10111136
Xiao C, Li R. Detection and Control of Fusarium oxysporum from Soft Rot in Dendrobium officinale by Loop-Mediated Isothermal Amplification Assays. Biology. 2021; 10(11):1136. https://doi.org/10.3390/biology10111136
Chicago/Turabian StyleXiao, Caiyun, and Rongyu Li. 2021. "Detection and Control of Fusarium oxysporum from Soft Rot in Dendrobium officinale by Loop-Mediated Isothermal Amplification Assays" Biology 10, no. 11: 1136. https://doi.org/10.3390/biology10111136
APA StyleXiao, C., & Li, R. (2021). Detection and Control of Fusarium oxysporum from Soft Rot in Dendrobium officinale by Loop-Mediated Isothermal Amplification Assays. Biology, 10(11), 1136. https://doi.org/10.3390/biology10111136






