Insights into Emerging Begomovirus–Deltasatellite Complex Diversity: The First Deltasatellite Infecting Legumes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Sample Collection and Molecular Identification
2.2. DNA Extraction and Full-Length Begomovirus and Deltasatellite Genome Cloning from Field Samples
2.3. Sequence Analyses
2.4. Construction of Infectious Clones and Plant Agroinoculation
2.5. Begomovirus and Deltasatellite Detection
3. Results and Discussion
3.1. Known and Novel New World Begomoviruses Infect Leguminous Plants in Venezuela
3.2. Deltasatellites Belonging to a Novel Species Are Associated with CabLCV Infecting Wild and Cultivated Leguminous Plants
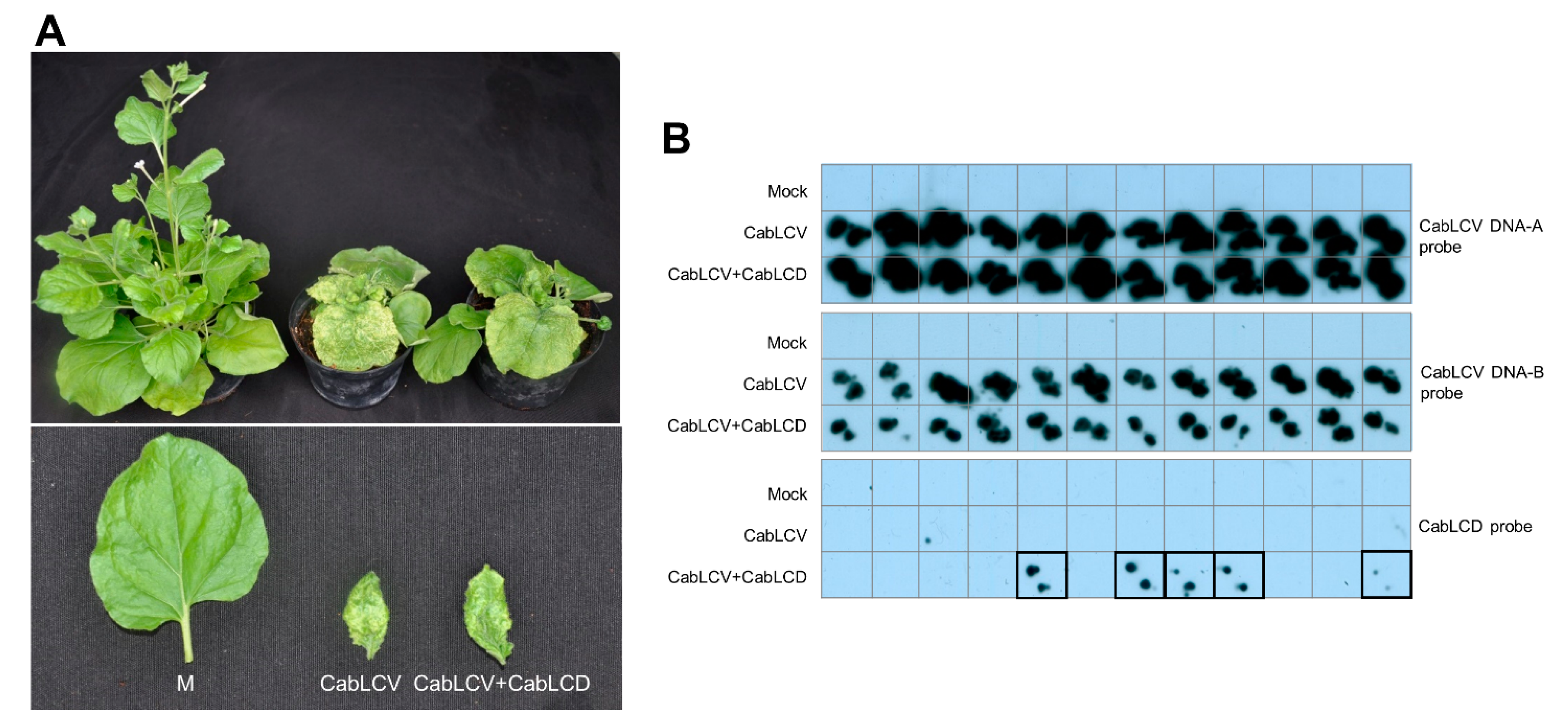
3.3. Cabbage Leaf Curl Virus Acts as a Helper Virus for CabLCD
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Chatzivassiliou, E.K. An annotated list of legume-infecting viruses in the light of metagenomics. Plants 2021, 10, 1413. [Google Scholar] [CrossRef] [PubMed]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Jadon, K.S.; Thirumalaisamy, P.P.; Kumar, R. Major seed-borne diseases in important pulses: Symptomatology, etiology and economic importance. In Seed-Borne Diseases of Agricultural Crops: Detection, Diagnosis and Management; Kumar, R., Gupta, A., Eds.; Springer: Singapore, 2020; pp. 469–542. [Google Scholar]
- Blair, M.W.; Bassett, M.J.; Abouzids, A.M.; Hiebert, E.; Polston, J.E.; McMillan, R.T., Jr.; Graves, W.; Lamberts, M. Occurrence of bean golden mosaic virus in Florida. Plant Dis. 1995, 79, 529–533. [Google Scholar] [CrossRef]
- Brown, J.K.; Ostrow, K.M.; Idris, A.M.; Stenger, D.C. Biotic, molecular, and phylogenetic characterization of bean calico mosaic virus, a distinct Begomovirus species with affiliation in the squash leaf curl virus cluster. Plant Dis. 1999, 89, 273–280. [Google Scholar] [CrossRef][Green Version]
- Carvajal-Yepes, M.; Zambrano, L.; Bueno, J.M.; Raatz, B.; Cuellar, W.J. Complete genome sequence of bean leaf crumple virus, a novel begomovirus infecting common bean in Colombia. Arch. Virol. 2017, 162, 1773–1776. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.S. Three whitefly-transmitted diseases of beans in the State of São Paulo, Brazil. FAO Plant Prot. Bull. 1965, 13, 121–130. [Google Scholar]
- Echemendía, A.L.; Ramos, P.L.; Peral, R.; Fuentes, A.; González, G.; Sanpedro, J.; Morales, F. Cuban isolate of Bean golden yellow mosaic virus is a member of the Mesoamerican BGYMV group. Plant Dis. 2001, 85, 1030. [Google Scholar] [CrossRef]
- Fernandes, F.R.; Cruz, A.R.R.; Faria, J.C.; Zerbini, F.M.; Aragão, F.J.L. Three distinct begomoviruses associated with soybean in central Brazil. Arch. Virol. 2009, 154, 1567–1570. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Márquez-Martín, B.; Hassan, I.; Chirinos, D.T.; Geraud-Pouey, F.; Moriones, E.; Navas-Castillo, J. Complete genome sequences of two novel begomoviruses infecting common bean in Venezuela. Arch. Virol. 2013, 158, 723–727. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Chirinos, D.T.; Castro, R.; Navas-Castillo, J. First report of cabbage leaf curl virus infecting common bean, cowpea, pigeon pea and Mucuna pruriens in Ecuador. Plant Dis. 2018, 102, 2667. [Google Scholar] [CrossRef]
- Garrido-Ramirez, E.R.; Sudarshana, M.R.; Gilbertson, R.L. Bean golden yellow mosaic virus from Chiapas, Mexico: Characterization, pseudorecombination with other bean-infecting geminiviruses and germplasm screening. Phytopathology 2000, 90, 1224–1232. [Google Scholar] [CrossRef]
- Gilbertson, R.L.; Faria, J.C.; Hanson, S.F.; Morales, F.J.; Ahlquist, P.G.; Maxwell, D.P.; Russell, D.R. Cloning of the complete DNA genomes of four bean-infecting geminiviruses and determining their infectivity by electric discharge particle acceleration. Phytopathology 1991, 81, 980–985. [Google Scholar] [CrossRef]
- Howarth, A.J.; Caton, J.; Bossert, M.; Goodman, R.M. Nucleotide sequence of bean golden mosaic virus and a model for gene regulation in geminiviruses. Proc. Natl. Acad. Sci. USA 1985, 82, 3572–3576. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Marrero, N.; Avalos-Calleros, J.A.; Chiquito-Almanza, E.; Acosta-Gallegos, J.A.; Ambriz-Granados, S.; Anaya-López, J.L.; Argüello-Astorga, G.R. A new begomovirus isolated from a potyvirus-infected bean plant causes asymptomatic infections in bean and N. benthamiana. Arch. Virol. 2020, 165, 1659–1665. [Google Scholar] [CrossRef]
- Reyna, P.G.; Bejerman, N.; Laguna, I.G.; Pardina, P.R. Biological and molecular characterization of bean bushy stunt virus, a novel bipartite begomovirus infecting common bean in northwestern Argentina. Arch. Virol. 2021, 166, 1409–1414. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Chirinos, D.T.; Geraud-Pouey, F.; Moriones, E.; Navas-Castillo, J. Complete genome sequences of two begomoviruses infecting weeds in Venezuela. Arch. Virol. 2013, 158, 277–280. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Chirinos, D.T.; Geraud-Pouey, F.; Moriones, E.; Navas-Castillo, J. Complete genome sequence of Jacquemontia yellow mosaic virus, a novel begomovirus from Venezuela related to other New World bipartite begomoviruses infecting Convolvulaceae. Arch. Virol. 2014, 159, 1857–1860. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Chirinos, D.T.; Geraud-Pouey, F.; Navas-Castillo, J. Complete genome sequence of jacquemontia yellow vein virus, a novel begomovirus infecting Jacquemontia tamnifolia in Venezuela. Arch. Virol. 2017, 162, 2463–2466. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Fiallo-Olivé, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of non-coding DNA satellites associated with sweepoviruses (genus Begomovirus, Geminiviridae)—Definition of a distinct class of begomovirus-associated satellites. Front. Microbiol. 2016, 7, 162. [Google Scholar] [CrossRef] [PubMed]
- Nava, A.; Londono, A.; Polston, J.E. Characterization and distribution of tomato yellow margin leaf curl virus, a begomovirus from Venezuela. Arch. Virol. 2013, 158, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Nava, A.R.; Patte, C.P.; Hiebert, E.; Polston, J.E. Detection and variability of begomoviruses in tomato from the Andean states of Venezuela. Plant Dis. 2006, 90, 61–66. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Romay, G.; Chirinos, D.; Geraud-Pouey, F.; Desbiez, C. Association of an atypical alphasatellite with a bipartite New World begomovirus. Arch. Virol. 2010, 155, 1843–1847. [Google Scholar] [CrossRef] [PubMed]
- Romay, G.; Chirinos, D.T.; Geraud-Pouey, F.; Gillis, A. Full-length genome sequencing of the mild strain of Tomato yellow leaf curl virus in Venezuela reveals a third introduction event of this virus in New World. Australas. Plant Dis. Notes 2014, 9, 123. [Google Scholar] [CrossRef][Green Version]
- Romay, G.; Chirinos, D.T.; Geraud-Pouey, F.; Gillis, A.; Mahillon, J.; Bragard, C. Complete genome sequence of two tomato-infecting begomoviruses in Venezuela: Evidence of a putative novel species and a novel recombinant strain. Arch. Virol. 2017, 163, 555–558. [Google Scholar] [CrossRef]
- Romay, G.; Geraud-Pouey, F.; Chirinos, D.T.; Mahillon, M.; Gillis, A.; Mahillon, J.; Bragard, C. Tomato twisted leaf virus: A novel indigenous New World monopartite begomovirus infecting tomato in Venezuela. Viruses 2019, 11, 327. [Google Scholar] [CrossRef]
- Zambrano, K.; Carballo, O.; Geraud, F.; Chirinos, D.; Fernandez, C.; Marys, E. First report of Tomato yellow leaf curl virus in Venezuela. Plant Dis. 2007, 91, 768. [Google Scholar] [CrossRef]
- Zambrano, K.; Geraud-Pouey, F.; Chirinos, D.; Romay, G.; Marys, E. Tomato chlorotic leaf distortion virus, a new bipartite begomovirus infecting Solanum lycopersicum and Capsicum chinense in Venezuela. Arch. Virol. 2011, 156, 2263–2266. [Google Scholar] [CrossRef]
- Zambrano, K.; Fernández-Rodríguez, T.; Marys, E. Molecular characterization of a new begomovirus that infects Euphorbia heterophylla and Solanum lycopersicum in Venezuela. Arch. Virol. 2012, 157, 379–382. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.A.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef]
- Gnanasekaran, P.; Kumar, R.K.; Bhattacharyya, D.; Kumar, R.V.; Chakraborty, S. Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol. Plant Pathol. 2019, 20, 1019–1033. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Mansoor, S.; Bedford, I.D.; Rishi, N.; Siwatch, S.S.; Zafar, Y.; Abdel-Salam, A.M.; Mark-ham, P.G. Diversity of DNA 1: A satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology 2004, 324, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.V.; Singh, D.; Singh, A.K.; Chakraborty, S. Molecular diversity, recombination and population structure of alphasatellites associated with begomovirus disease complexes. Infect. Genet Evol. 2017, 49, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Martínez-Zubiaur, Y.; Moriones, E.; Navas-Castillo, J. A novel class of DNA satellites associated with New World begomoviruses. Virology 2012, 426, 1–6. [Google Scholar] [CrossRef]
- Yang, X.; Guo, W.; Li, F.; Sunter, G.; Zhou, X. Geminivirus-associated betasatellites: Exploiting chinks in the antiviral arsenal of plants. Trends Plant Sci. 2019, 24, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef]
- Li, F.; Yang, X.; Bisaro, D.M.; Zhou, X. The βC1 protein of geminivirus-betasatellite complexes: A target and repressor of host defenses. Mol. Plant 2018, 11, 1424–1426. [Google Scholar] [CrossRef]
- Dry, I.B.; Krake, L.R.; Rigden, J.E.; Rezaian, M.A. A novel subviral agent associated with a geminivirus: The first report of a DNA satellite. Proc. Natl. Acad. Sci. USA 1997, 94, 7088–7093. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Tovar, R.; Navas-Castillo, J. Deciphering the biology of deltasatellites from the New World: Maintenance by New World begomoviruses and whitefly transmission. New Phytol. 2016, 212, 680–692. [Google Scholar] [CrossRef]
- Ferro, C.G.; Zerbini, F.M.; Navas-Castillo, J.; Fiallo-Olivé, E. Revealing the complexity of sweepovirus-deltasatellite-plant host interactions: Expanded natural and experimental helper virus range and effect dependence on virus-host combination. Microorganisms 2021, 9, 1018. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; Fazekas, A.J.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 1–19. [Google Scholar] [CrossRef]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- SantaLucia, J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA 1998, 95, 1460–1465. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Abouzid, A.M.; Hiebert, E.; Strandberg, J.O. Cloning, identification, and partial sequencing of the genomic components of a geminivirus infecting the Brassicaceae. Phytopathology 1992, 82, 1070. [Google Scholar]
- Hernández-Zepeda, C.; Brown, J.K.; Moreno-Valenzuela, O.A.; Arguello-Astorga, G.; Idris, A.M.; Carnevali, G.; Rivera-Bustamante, R.F. Characterization of Rhynchosia yellow mosaic Yucatan virus, a new recombinant begomovirus associated with two fabaceous weeds in Yucatan, Mexico. Arch. Virol. 2010, 155, 1571–1579. [Google Scholar] [CrossRef]
- Briddon, R.W.; Patil, B.L.; Bagewadi, B.; Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 2010, 10, 97. [Google Scholar] [CrossRef] [PubMed]
- Andrade, E.C.; Manhani, G.G.; Alfenas, P.F.; Calegario, R.F.; Fontes, E.; Zerbini, F.M. Tomato yellow spot virus, a tomato-infecting begomovirus from Brazil with a closer relationship to viruses from Sida sp., forms pseudorecombinants with begomoviruses from tomato but not from Sida. J. Gen. Virol. 2006, 87, 3687–3696. [Google Scholar] [CrossRef]
- Castillo-Urquiza, G.P.; Beserra, J.E.A.; Bruckner, F.P.; Lima, A.T.; Varsani, A.; Alfenas-Zerbini, P.; Zerbini, F.M. Six novel begomoviruses infecting tomato and associated weeds in Southeastern Brazil. Arch. Virol. 2008, 153, 1985–1989. [Google Scholar] [CrossRef]
- García-Arenal, F.; Zerbini, F.M. Life on the edge: Geminiviruses at the interface between crops and wild plant hosts. Annu. Rev. Virol. 2019, 6, 411–433. [Google Scholar] [CrossRef] [PubMed]
- Souza, T.A.; Silva, J.; Nagata, T.; Martins, T.P.; Nakasu, E.; Inoue-Nagata, A.K. A temporal diversity analysis of Brazilian begomoviruses in tomato reveals a decrease in species richness between 2003 and 2016. Front. Plant Sci. 2020, 11, 1201. [Google Scholar] [CrossRef]
- Cooper, I.; Jones, R.A. Wild plants and viruses: Under-investigated ecosystems. Adv. Virus Res. 2006, 67, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Jones RAC. Disease pandemics and major epidemics arising from new encounters between indigenous viruses and introduced crops. Viruses 2020, 12, 1388. [Google Scholar] [CrossRef]
- Rosario, K.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Breitbart, M. Begomovirus-associated satellite DNA diversity captured through Vector-Enabled Metagenomic (VEM) surveys using whiteflies (Aleyrodidae). Viruses 2016, 8, 36. [Google Scholar] [CrossRef]
- Ng, T.F.; Duffy, S.; Polston, J.E.; Bixby, E.; Vallad, G.E.; Breitbart, M. Exploring the diversity of plant DNA viruses and their satellites using vector-enabled metagenomics on whiteflies. PLoS ONE 2011, 6, e19050. [Google Scholar] [CrossRef]
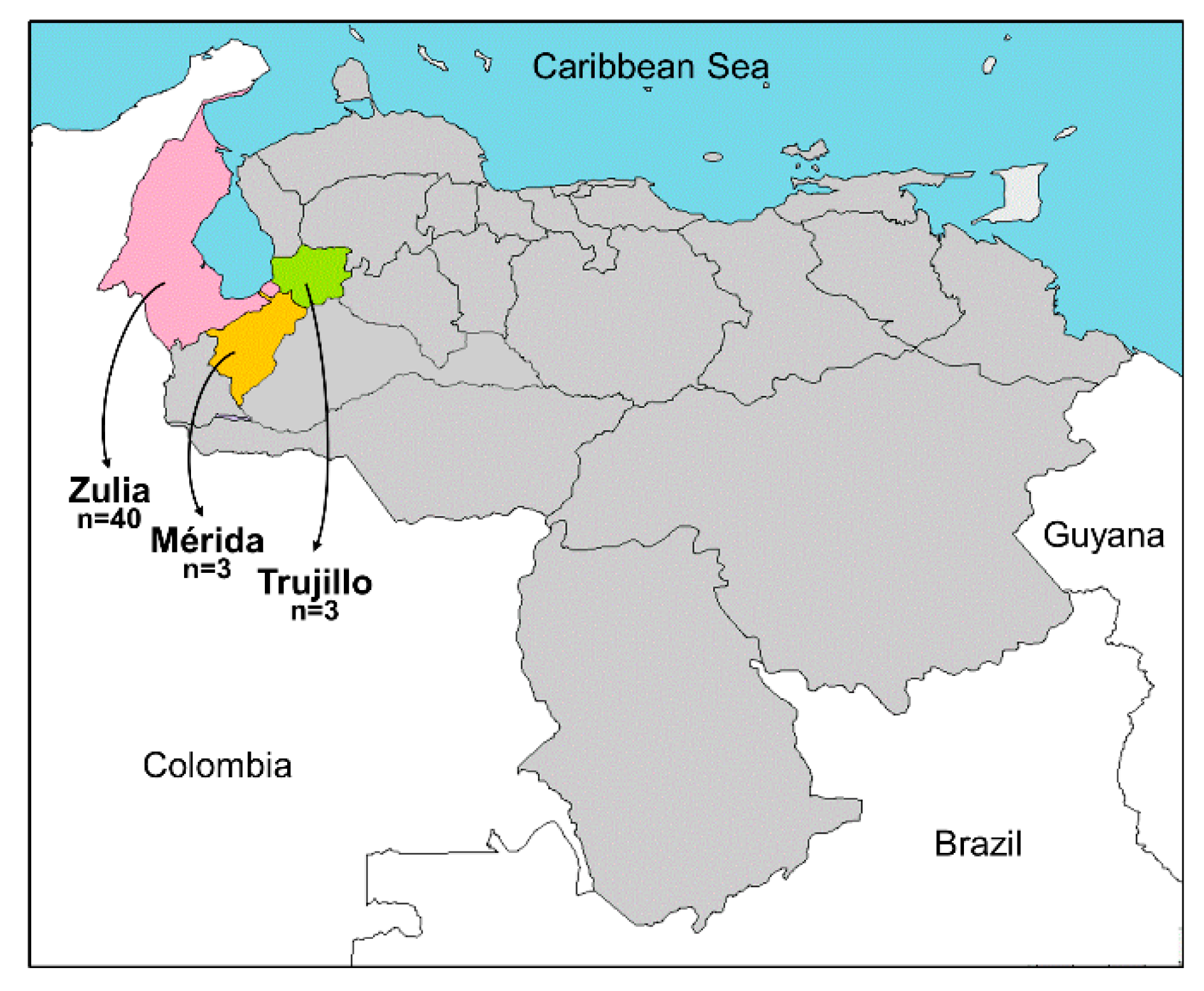


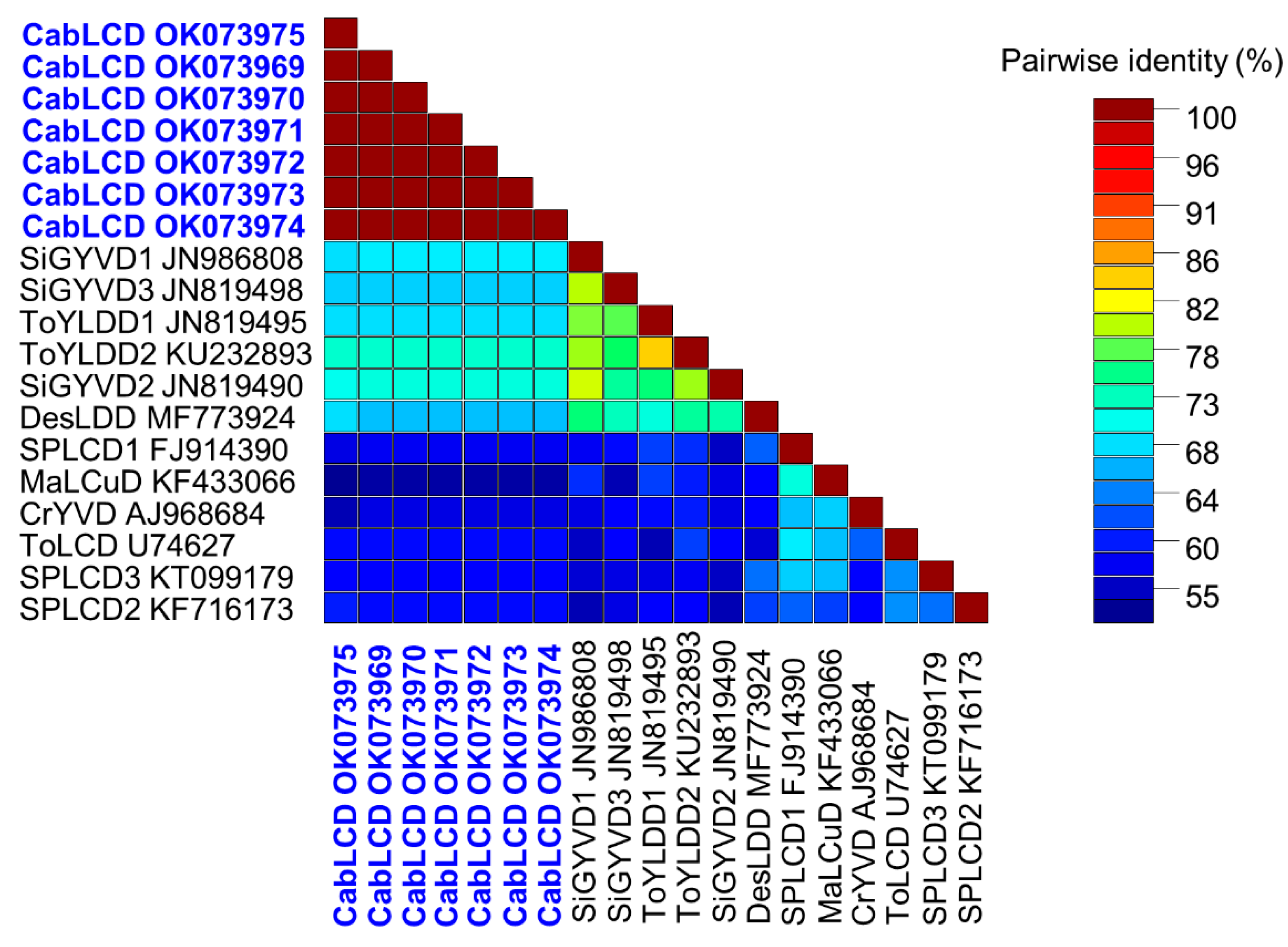
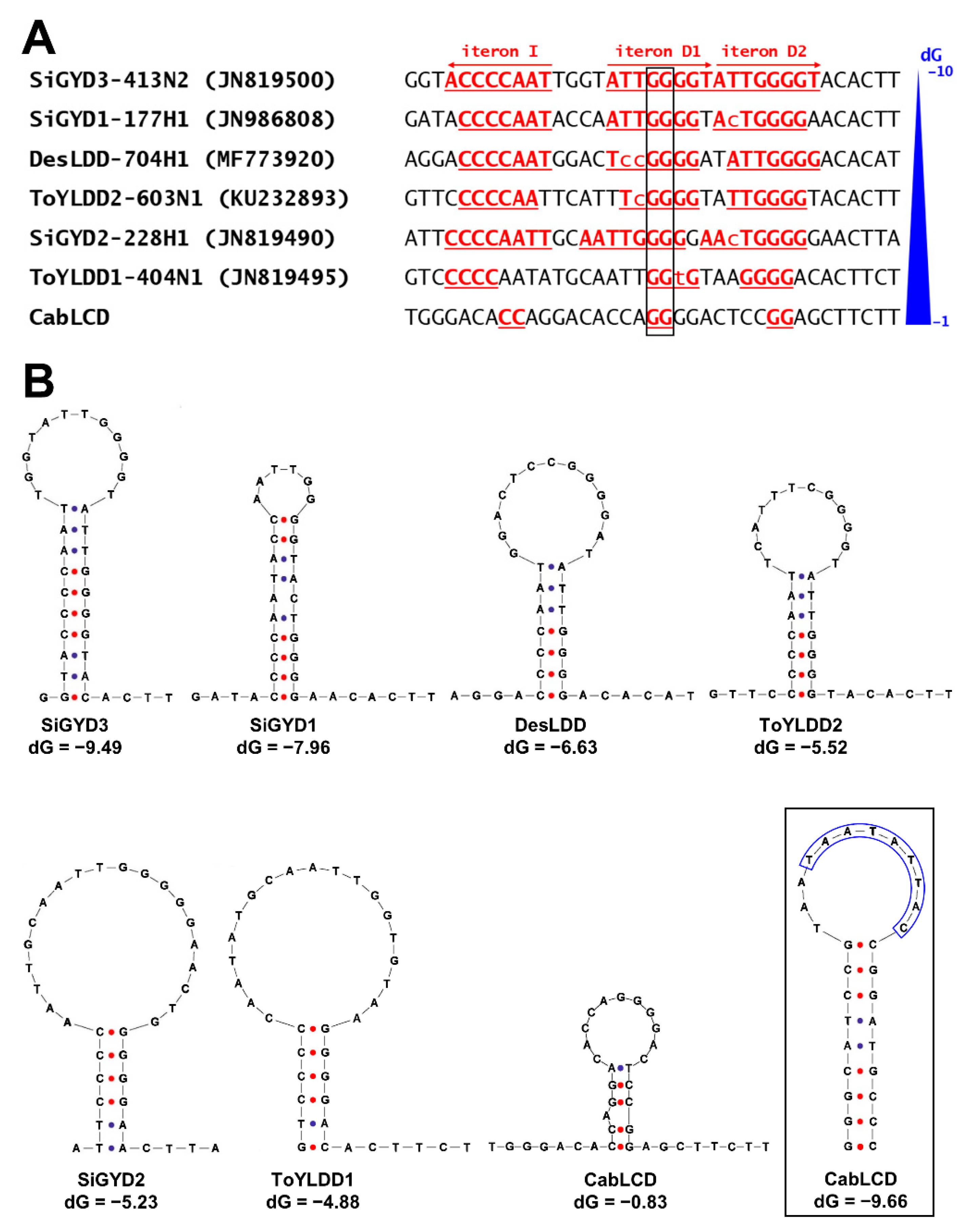
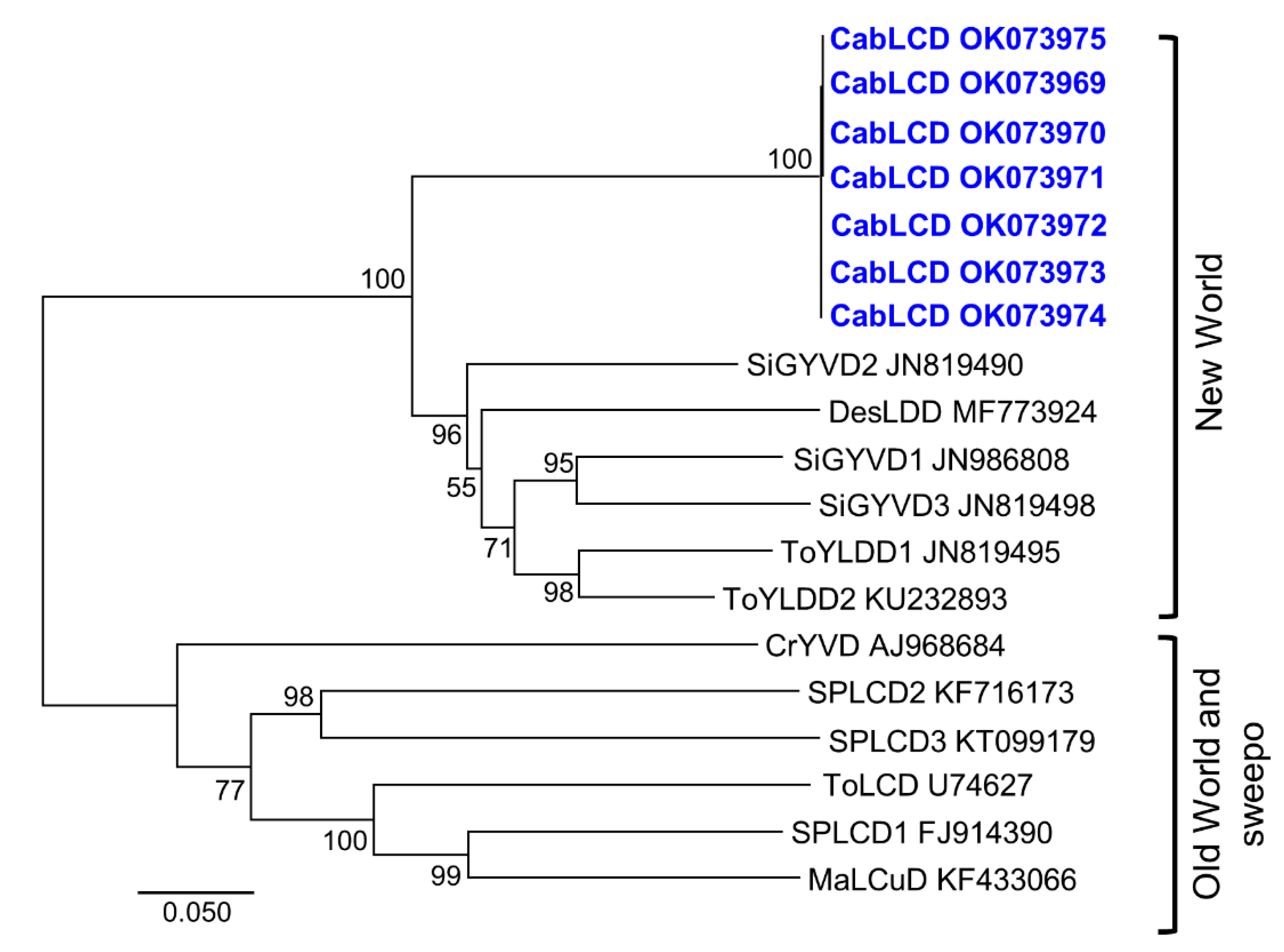
| Plant Species | State | Municipality | Sample Code | Begomovirus | Deltasatellite |
|---|---|---|---|---|---|
| Black gram | Mérida | Tulio Febres-Cordero | V12 | CabLCV * | CabLCD |
| Mérida | Tulio Febres-Cordero | V13 | CabLCV | CabLCD | |
| Common bean | Trujillo | Escuque | V14 | - | - |
| Trujillo | Escuque | V15 | - | - | |
| Trujillo | Escuque | V16 | - | - | |
| Mérida | Justo Briceño | V19 | - | - | |
| Cowpea | Zulia | Jesús Enrique Lossada | V1 | CabLCV * | CabLCD |
| Zulia | Jesús Enrique Lossada | V2 | CabLCV | CabLCD | |
| Zulia | Jesús Enrique Lossada | V24 | - | - | |
| Zulia | Jesús Enrique Lossada | V26 | - | - | |
| Zulia | Jesús Enrique Lossada | V27 | - | - | |
| Zulia | Jesús Enrique Lossada | V28 | - | - | |
| Zulia | Jesús Enrique Lossada | V29 | - | - | |
| Zulia | Jesús Enrique Lossada | V30 | - | - | |
| Zulia | Jesús Enrique Lossada | V31 | - | - | |
| Zulia | Jesús Enrique Lossada | V32 | - | - | |
| Zulia | Jesús Enrique Lossada | V33 | - | - | |
| Zulia | Jesús Enrique Lossada | V34 | - | - | |
| Zulia | Jesús Enrique Lossada | V35 | CabLCV | - | |
| Zulia | Jesús Enrique Lossada | V36 | - | - | |
| Zulia | Jesús Enrique Lossada | V37 | - | - | |
| Zulia | Jesús Enrique Lossada | V38 | - | - | |
| Zulia | Jesús Enrique Lossada | V39 | - | - | |
| Zulia | Jesús Enrique Lossada | V40 | - | - | |
| Zulia | Jesús Enrique Lossada | V41 | - | - | |
| Zulia | Jesús Enrique Lossada | V42 | - | - | |
| Zulia | Jesús Enrique Lossada | V43 | - | - | |
| Zulia | Jesús Enrique Lossada | V44 | - | - | |
| Zulia | Jesús Enrique Lossada | V45 | - | - | |
| Zulia | Jesús Enrique Lossada | V46 | - | - | |
| Faba bean | Zulia | Sucre | V18 | - | - |
| Mung bean | Zulia | Sucre | V17 | - | - |
| Desmodium scorpiurus | Zulia | Sucre | V6 | DesMV | - |
| Zulia | Sucre | V7 | DesYSV | - | |
| Zulia | Sucre | V8 | CabLCV * | CabLCD | |
| Zulia | Maracaibo | V47 | - | - | |
| Macroptilium bracteatum | Zulia | Jesús Enrique Lossada | V3 | MacMoV | - |
| Zulia | Jesús Enrique Lossada | V4 | BLCrV | - | |
| Zulia | Jesús Enrique Lossada | V5 | - | - | |
| Rhynchosia minima | Zulia | Sucre | V9 | CabLCV | CabLCD |
| Zulia | Sucre | V10 | CabLCV | CabLCD | |
| Zulia | Sucre | V11 | RhMoV | - | |
| Zulia | Sucre | V20 | CabLCV | - | |
| Zulia | Maracaibo | V21 | RhMoV | - | |
| Zulia | Maracaibo | V22 | CabLCV | - | |
| Zulia | Maracaibo | V23 | CabLCV | - |
| Virus/Deltasatellite | No. Infected Plants/No. Agroinoculated Plants | |||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | |||
| CabLCV | CabLCD | CabCV | CabLCD | |
| CabLCV | 12/12 | - | 12/12 | - |
| CabLCV + CabLCD | 12/12 | 5/12 | 12/12 | 3/12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiallo-Olivé, E.; Bastidas, L.; Chirinos, D.T.; Navas-Castillo, J. Insights into Emerging Begomovirus–Deltasatellite Complex Diversity: The First Deltasatellite Infecting Legumes. Biology 2021, 10, 1125. https://doi.org/10.3390/biology10111125
Fiallo-Olivé E, Bastidas L, Chirinos DT, Navas-Castillo J. Insights into Emerging Begomovirus–Deltasatellite Complex Diversity: The First Deltasatellite Infecting Legumes. Biology. 2021; 10(11):1125. https://doi.org/10.3390/biology10111125
Chicago/Turabian StyleFiallo-Olivé, Elvira, Liseth Bastidas, Dorys T. Chirinos, and Jesús Navas-Castillo. 2021. "Insights into Emerging Begomovirus–Deltasatellite Complex Diversity: The First Deltasatellite Infecting Legumes" Biology 10, no. 11: 1125. https://doi.org/10.3390/biology10111125
APA StyleFiallo-Olivé, E., Bastidas, L., Chirinos, D. T., & Navas-Castillo, J. (2021). Insights into Emerging Begomovirus–Deltasatellite Complex Diversity: The First Deltasatellite Infecting Legumes. Biology, 10(11), 1125. https://doi.org/10.3390/biology10111125







