PPARα, δ and FOXO1 Gene Silencing Overturns Palmitate-Induced Inhibition of Pyruvate Oxidation Differentially in C2C12 Myotubes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. C2C12 Cell Culture, Differentiation into Myobutes and Small Interfering RNA (siRNA) Transfection
2.3. Gene Expression Measurements
2.4. Glucose Uptake Assay
2.5. Cell Metabolite Levels and PDC Activity Measurements
2.6. Determination of Maximal Mitochondrial ATP Production Rates in Intact C2C12 Myotubes
2.7. Western Blotting
2.8. Statistical Analysis
3. Results
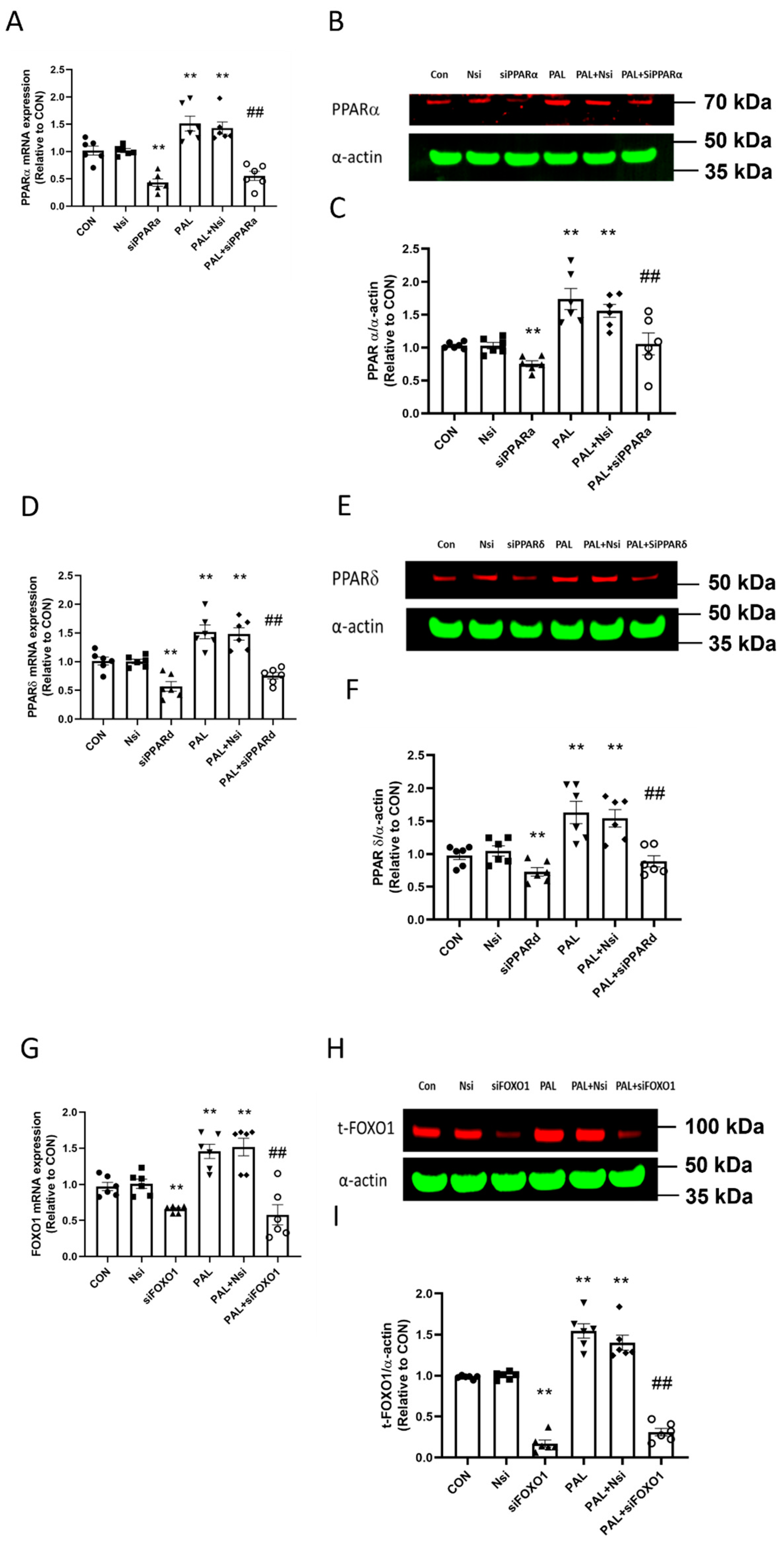
3.1. Validation of PPARα, δ and FOXO1 siRNA Gene Silencing at Transcriptional and Translational Level in C2C12 Myotubes Treated with or without Palmitate (PAL)
3.2. Metabolic Outcomes after Individual PPARα, PPARδ and FOXO1 siRNA Gene Silencing in C2C12 Myotubes Treated with or without Palmitate (PAL)
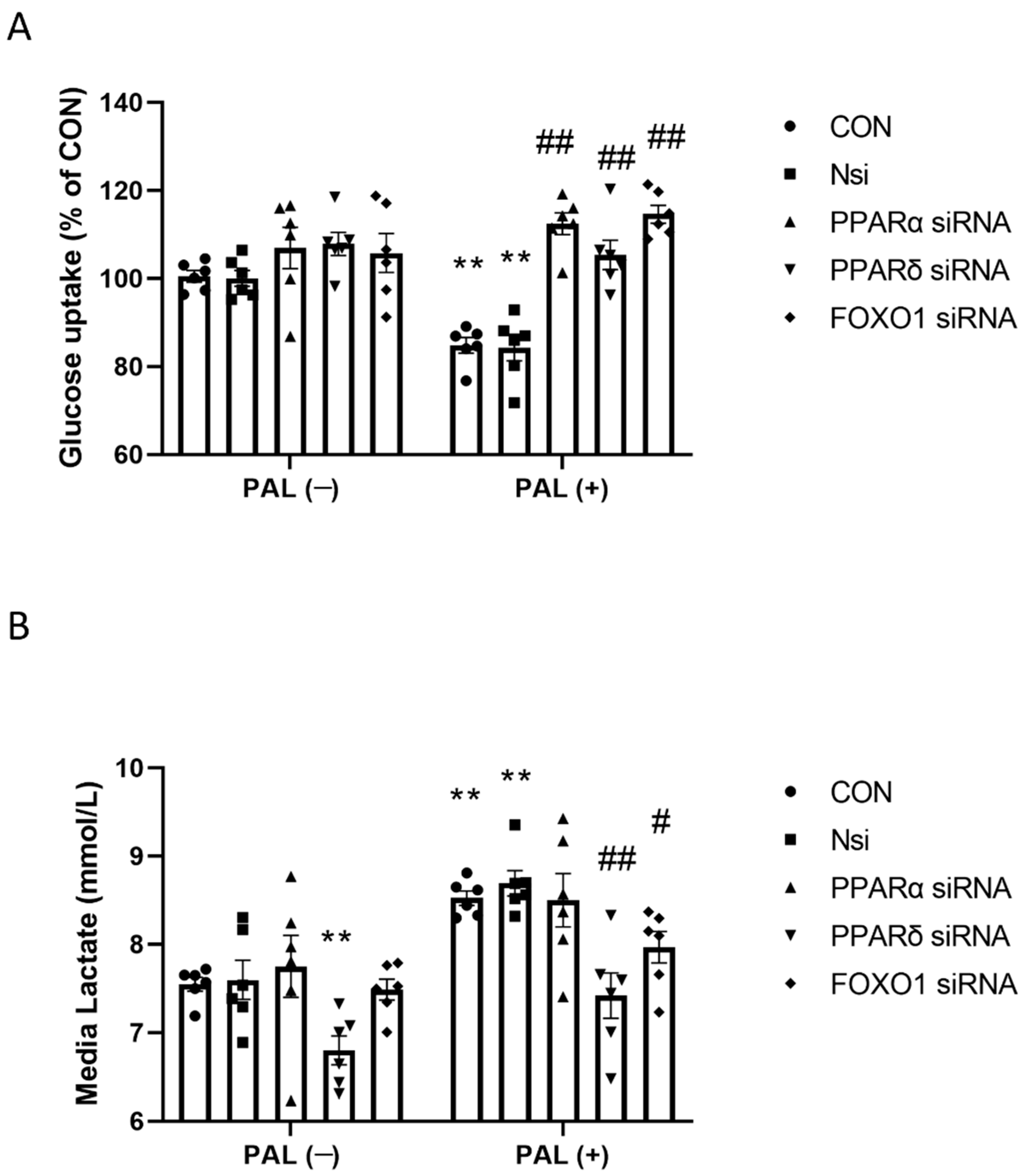
3.2.1. Glucose Uptake and Media Lactate Accumulation
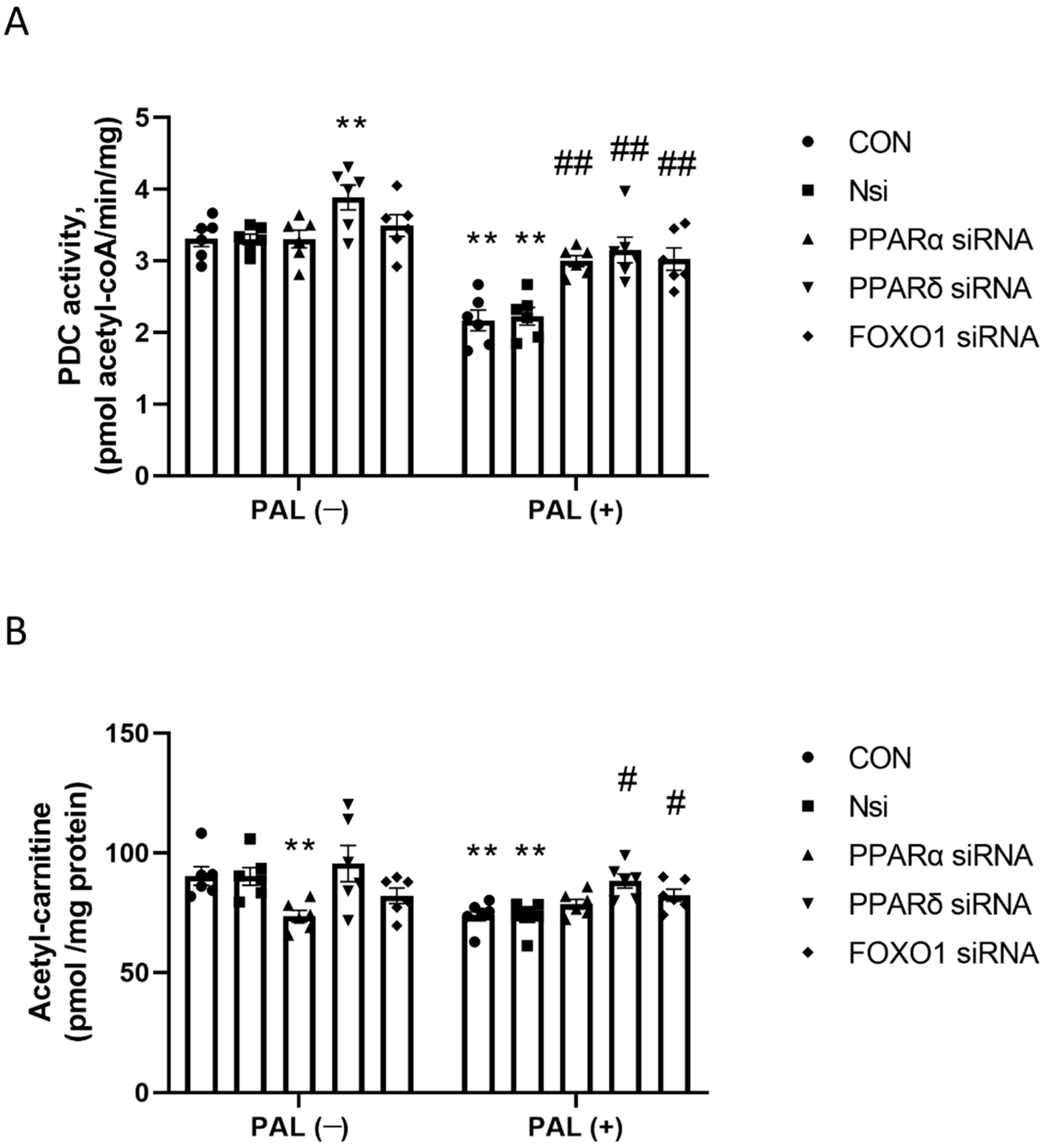
3.2.2. PDC Activity and Acetylcarnitine Concentration in C2C12 Myotubes Treated with or without Palmitate (PAL)
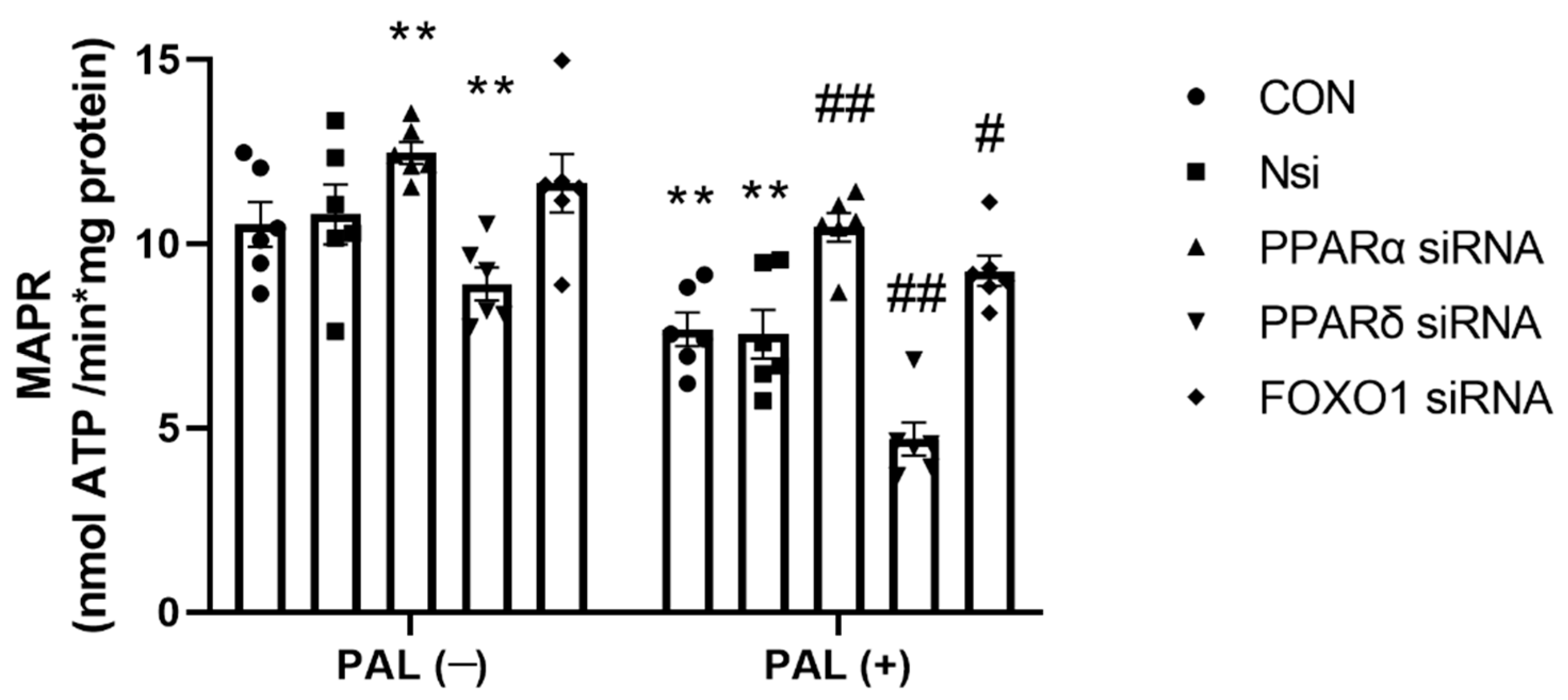
3.2.3. Maximal Rates of Mitochondrial ATP Production (MAPR)-Based on Glucose-Derived Pyruvate Oxidation in C2C12 Myotubes Treated with or without Palmitate (PAL)
3.3. Transcriptional and Translational Outcomes after Individual PPARα, PPARδ and FOXO1 siRNA Gene Silencing in C2C12 Myotubes Treated with or without Palmitate (PAL)
3.3.1. PDK4 mRNA and Protein Expression
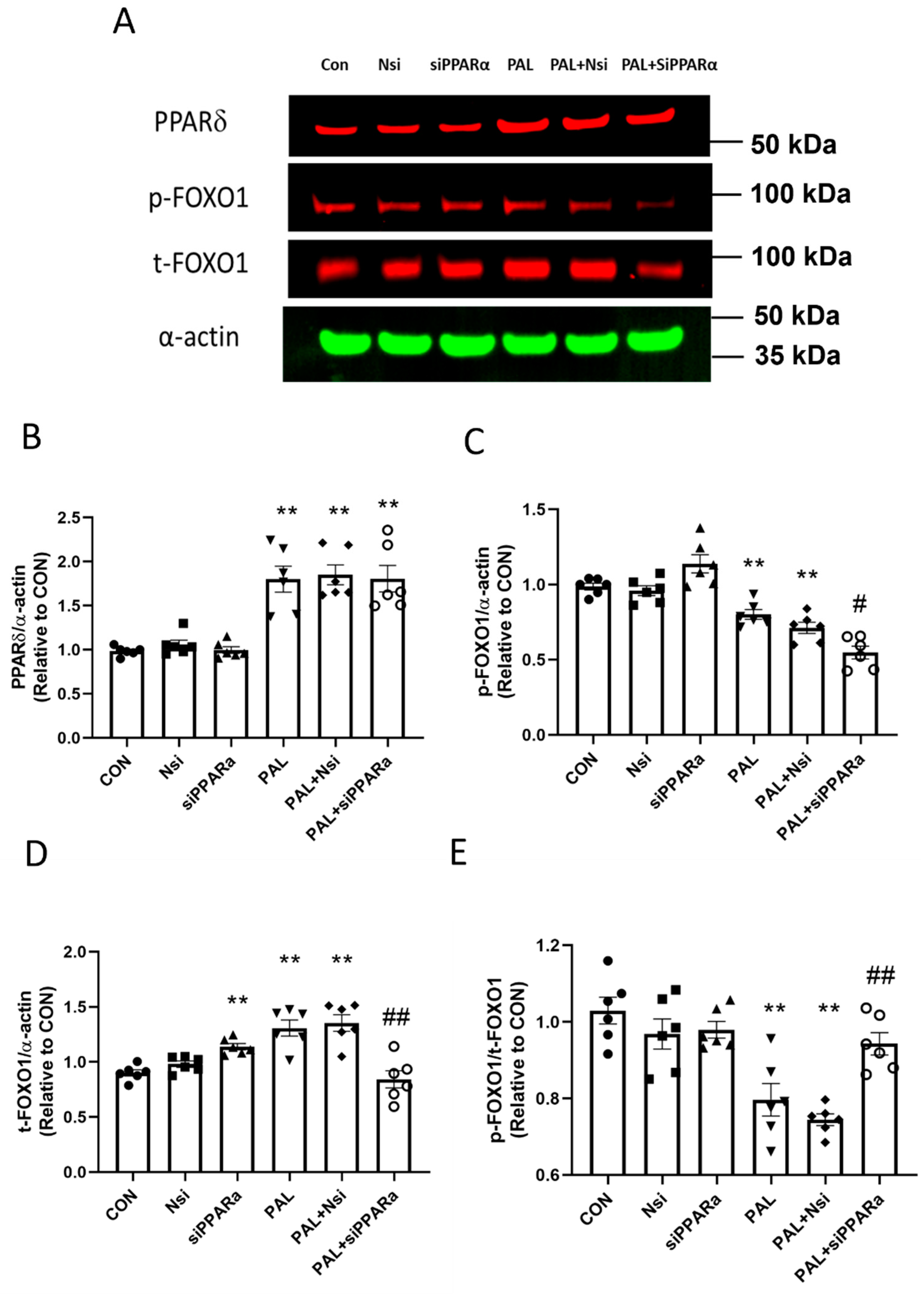
3.3.2. PPARδ and FOXO1 Translational and Post-Translational (Phosphorylation) Levels after PPARα siRNA Gene Silencing in C2C12 Myotubes Treated with or without Palmitate (PAL)
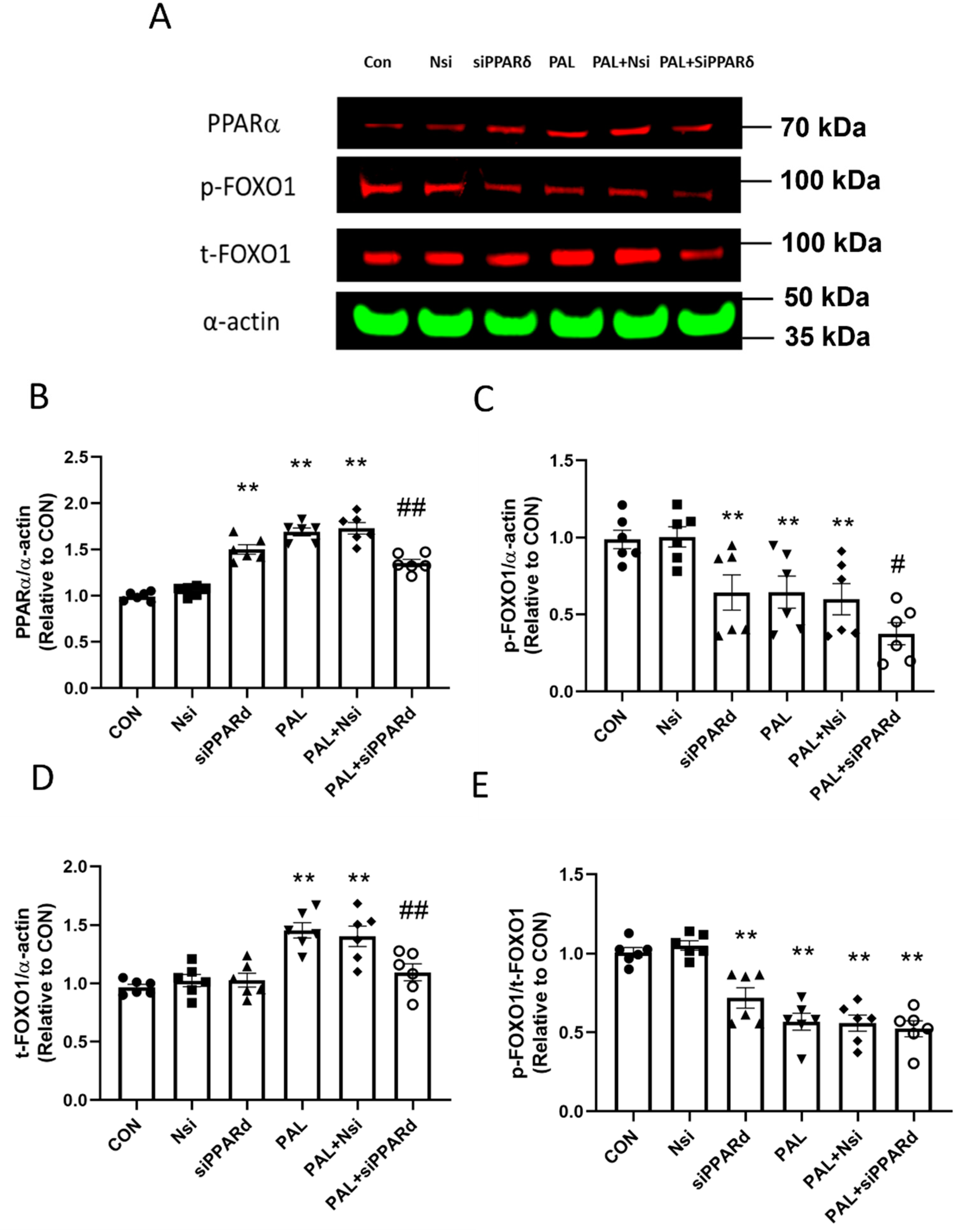
3.3.3. PPARα and FOXO1 Translational and Post-Translational (Phosphorylation) Levels after PPARδ siRNA Gene Silencing in C2C12 Myotubes Treated with or without Palmitate (PAL)
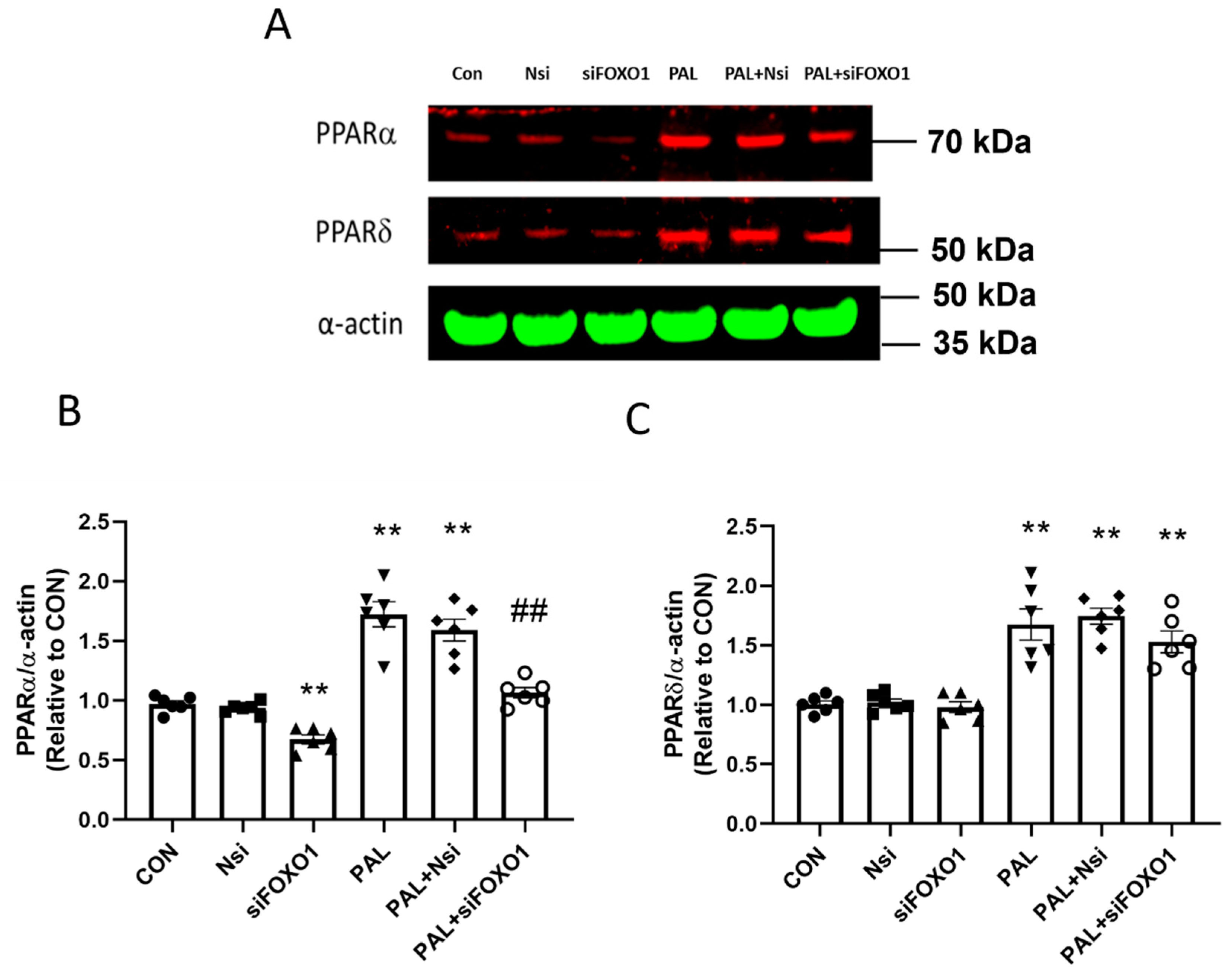
3.3.4. PPARα and δ Translational Levels after FOXO1 siRNA Gene Silencing in C2C12 Myotubes Treated with or without Palmitate (PAL)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviation
| ATCC | |
| C2C12 | Immortalized mouse myoblast cell line |
| BSA | Bovine serum albumin |
| FFA | Free fatty acid |
| FOXO1 | Forkhead Box O1 |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HMBC | Hydroxymethylbilane synthase |
| MI | Metabolic inflexibility |
| Nsi | Silencer negative control |
| PAL | Palmitate |
| PDC | Pyruvate dehydrogenase complex |
| PPARα | Peroxisome proliferator-activated receptor alpha |
| PDK4 | Pyruvate dehydrogenase isoform 4 |
| PPARδ | Peroxisome proliferator-activated receptor delta |
| PGC-1α | Peroxisome-proliferator-activated receptor γ activator-1α |
| SEM | Standard error of the mean |
| Tween-20 | Polysorbate 20 |
References
- Galgani, J.E.; Moro, C.; Ravussin, E. Metabolic flexibility and insulin resistance. Am. J. Physiol. Metab. 2008, 295, E1009–E1017. [Google Scholar] [CrossRef] [Green Version]
- Storlien, L.; Oakes, N.D.; Kelley, D.E. Metabolic flexibility. Proc. Nutr. Soc. 2004, 63, 363–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rynders, C.A.; Blanc, S.; DeJong, N.; Bessesen, D.H.; Bergouignan, A. Sedentary behaviour is a key determinant of metabolic inflexibility. J. Physiol. 2017, 596, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 2014, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.; Thoudam, T.; Choi, E.J.; Kim, M.; Harris, R.A.; Lee, I. Loss of metabolic flexibility as a result of overexpression of pyruvate dehydrogenase kinases in muscle, liver and the immune system: Therapeutic targets in metabolic diseases. J. Diabetes Investig. 2020, 12, 21–31. [Google Scholar] [CrossRef]
- DeFronzo, R.A.; Tripathy, D. Skeletal Muscle Insulin Resistance Is the Primary Defect in Type 2 Diabetes. Diabetes Care 2009, 32, S157–S163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Constantin-Teodosiu, D. Regulation of Muscle Pyruvate Dehydrogenase Complex in Insulin Resistance: Effects of Exercise and Dichloroacetate. Diabetes Metab. J. 2013, 37, 301–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randle, P.J. Regulatory interactions between lipids and carbohydrates: The glucose fatty acid cycle after 35 years. Diabetes Metab. Rev. 1998, 14, 263–283. [Google Scholar] [CrossRef]
- Svensson, K.; Dent, J.R.; Tahvilian, S.; Martins, V.F.; Sathe, A.; Ochala, J.; Patel, M.S.; Schenk, S.; Dent, J.R. Defining the contribution of skeletal muscle pyruvate dehydrogenase α1 to exercise performance and insulin action. Am. J. Physiol. Metab. 2018, 315, E1034–E1045. [Google Scholar] [CrossRef]
- Chien, H.C.; Greenhaff, P.L.; Constantin-Teodosiu, D. PPARδ and FOXO1 Mediate Palmitate-Induced Inhibition of Muscle Pyruvate Dehydrogenase Complex and CHO Oxidation, Events Reversed by Electrical Pulse Stimulation. Int. J. Mol. Sci. 2020, 21, 5942. [Google Scholar] [CrossRef]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, H.-C.; Chan, P.-C.; Tu, C.-C.; Day, Y.-J.; Hung, L.-M.; Juan, C.-C.; Tian, Y.-F.; Hsieh, P.-S. Importance of PLC-Dependent PI3K/AKT and AMPK Signaling in RANTES/CCR5 Mediated Macrophage Chemotaxis. Chin. J. Physiol. 2018, 61, 266–279. [Google Scholar] [CrossRef] [Green Version]
- Harris, R.C.; Hultman, E.; Nordesjo, L.O. Glycogen, glycolytic intermediates and high-energy phosphates determined in biopsy samples of musculus quadriceps femoris of man at rest. Methods and variance of values. Scand. J. Clin. Lab. Investig. 1974, 33, 109–120. [Google Scholar] [CrossRef]
- Constantin-Teodosiu, D.; Carlin, J.I.; Cederblad, G.; Harrist, R.C.; Hultman, E. Acetyl group accumulation and pyruvate dehydrogenase activity in human muscle during incremental exercise. Acta Physiol. Scand. 1991, 143, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.J.; Heigenhauser, G.J.F.; Stellingwerff, T.; Hargreaves, M.; Spriet, L.L. Carbohydrate ingestion reduces skeletal muscle acetylcarnitine availability but has no effect on substrate phosphorylation at the onset of exercise in man. J. Physiol. 2002, 544, 949–956. [Google Scholar] [CrossRef]
- Cederblad, G.; Carlin, J.; Constantin-Teodosiu, D.; Harper, P.; Hultman, E. Radioisotopic assays of CoASH and carnitine and their acetylated forms in human skeletal muscle. Anal. Biochem. 1990, 185, 274–278. [Google Scholar] [CrossRef]
- Wibom, R.; Hagenfeldt, L.; von Dobeln, U. Measurement of ATP production and respiratory chain enzyme activities in mitochondria isolated from small muscle biopsy samples. Anal. Biochem. 2002, 311, 139–151. [Google Scholar] [CrossRef]
- Ernst, O.; Zor, T. Linearization of the Bradford Protein Assay. J. Vis. Exp. 2010, 38, e1918. [Google Scholar] [CrossRef]
- Constantin, D.; Menon, M.K.; Houchen-Wolloff, L.; Morgan, M.D.; Singh, S.J.; Greenhaff, P.; Steiner, M. Skeletal muscle molecular responses to resistance training and dietary supplementation in COPD. Thorax 2013, 68, 625–633. [Google Scholar] [CrossRef] [Green Version]
- Degenhardt, T.; Saramaki, A.; Malinen, M.; Rieck, M.; Vaisanen, S.; Huotari, A.; Herzig, K.H.; Muller, R.; Carlberg, C. Three members of the human pyruvate dehydrogenase kinase gene family are direct targets of the peroxisome proliferator-activated receptor beta/delta. J. Mol. Biol. 2007, 372, 341–355. [Google Scholar] [CrossRef]
- Furuyama, T.; Kitayama, K.; Yamashita, H.; Mori, N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem. J. 2003, 375, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Wua, P.; Peters, J.; Harris, R.A. Adaptive Increase in Pyruvate Dehydrogenase Kinase 4 during Starvation Is Mediated by Peroxisome Proliferator-Activated Receptor α. Biochem. Biophys. Res. Commun. 2001, 287, 391–396. [Google Scholar] [CrossRef]
- Sugden, M.C.; Holness, M.J. Mechanisms underlying regulation of the expression and activities of the mammalian pyruvate dehydrogenase kinases. Arch. Physiol. Biochem. 2006, 112, 139–149. [Google Scholar] [CrossRef]
- Hopkins, T.A.; Sugden, M.C.; Holness, M.J.; Kozak, R.; Dyck, J.R.B.; Lopaschuk, G.D. Control of cardiac pyruvate dehydrogenase activity in peroxisome proliferator-activated receptor-α transgenic mice. Am. J. Physiol. Circ. Physiol. 2003, 285, H270–H276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rena, G.; Woods, Y.L.; Prescott, A.R.; Peggie, M.; Unterman, T.G.; Williams, M.R.; Cohen, P. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 2002, 21, 2263–2271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.I.; Lee, F.N.; Choi, W.S.; Lee, S.; Youn, J.H. Insulin Regulation of Skeletal Muscle PDK4 mRNA Expression Is Impaired in Acute Insulin-Resistant States. Diabetes 2006, 55, 2311–2317. [Google Scholar] [CrossRef] [Green Version]
- Constantin, D.; Constantin-Teodosiu, D.; Layfield, R.; Tsintzas, K.; Bennett, A.J.; Greenhaff, P.L. PPARdelta agonism induces a change in fuel metabolism and activation of an atrophy programme, but does not impair mitochondrial function in rat skeletal muscle. J. Physiol. 2007, 583 Pt 1, 381–390. [Google Scholar] [CrossRef]
- Abbot, E.L.; McCormack, J.G.; Reynet, C.; Hassall, D.G.; Buchan, K.W.; Yeaman, S.J. Diverging regulation of pyruvate dehydrogenase kinase isoform gene expression in cultured human muscle cells. FEBS J. 2005, 272, 3004–3014. [Google Scholar] [CrossRef] [PubMed]
- Constantin-Teodosiu, D.; Baker, D.J.; Constantin, D.; Greenhaff, P. PPARδ agonism inhibits skeletal muscle PDC activity, mitochondrial ATP production and force generation during prolonged contraction. J. Physiol. 2009, 587, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Hondares, E.; Pineda-Torra, I.; Iglesias, R.; Staels, B.; Villarroya, F.; Giralt, M. PPARdelta, but not PPARalpha, activates PGC-1alpha gene transcription in muscle. Biochem. Biophys. Res. Commun. 2007, 354, 1021–1027. [Google Scholar] [CrossRef]
- Remels, A.H.; Schrauwen, P.; Broekhuizen, R.; Willems, J.; Kersten, S.; Gosker, H.R.; Schols, A.M. Peroxisome proliferator-activated receptor expression is reduced in skeletal muscle in COPD. Eur. Respir. J. 2007, 30, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jornayvaz, F.R.; Shulman, G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010, 47, 69–84. [Google Scholar] [PubMed] [Green Version]
- Wang, P.; Liu, J.; Li, Y.; Wu, S.; Luo, J.; Yang, H.; Subbiah, R.; Chatham, J.; Zhelyabovska, O.; Yang, Q. Peroxisome proliferator-activated receptor {delta} is an essential transcriptional regulator for mitochondrial protection and biogenesis in adult heart. Circ. Res. 2010, 106, 911–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chien, H.-C.; Constantin, D.; Greenhaff, P.L.; Constantin-Teodosiu, D. PPARα, δ and FOXO1 Gene Silencing Overturns Palmitate-Induced Inhibition of Pyruvate Oxidation Differentially in C2C12 Myotubes. Biology 2021, 10, 1098. https://doi.org/10.3390/biology10111098
Chien H-C, Constantin D, Greenhaff PL, Constantin-Teodosiu D. PPARα, δ and FOXO1 Gene Silencing Overturns Palmitate-Induced Inhibition of Pyruvate Oxidation Differentially in C2C12 Myotubes. Biology. 2021; 10(11):1098. https://doi.org/10.3390/biology10111098
Chicago/Turabian StyleChien, Hung-Che, Despina Constantin, Paul L. Greenhaff, and Dumitru Constantin-Teodosiu. 2021. "PPARα, δ and FOXO1 Gene Silencing Overturns Palmitate-Induced Inhibition of Pyruvate Oxidation Differentially in C2C12 Myotubes" Biology 10, no. 11: 1098. https://doi.org/10.3390/biology10111098
APA StyleChien, H.-C., Constantin, D., Greenhaff, P. L., & Constantin-Teodosiu, D. (2021). PPARα, δ and FOXO1 Gene Silencing Overturns Palmitate-Induced Inhibition of Pyruvate Oxidation Differentially in C2C12 Myotubes. Biology, 10(11), 1098. https://doi.org/10.3390/biology10111098






