Species-Specific Proteins in the Oviducts of Snail Sibling Species: Proteotranscriptomic Study of Littorina fabalis and L. obtusata
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Transcriptomics Analysis
2.3. Gel-Based Proteomics
3. Results
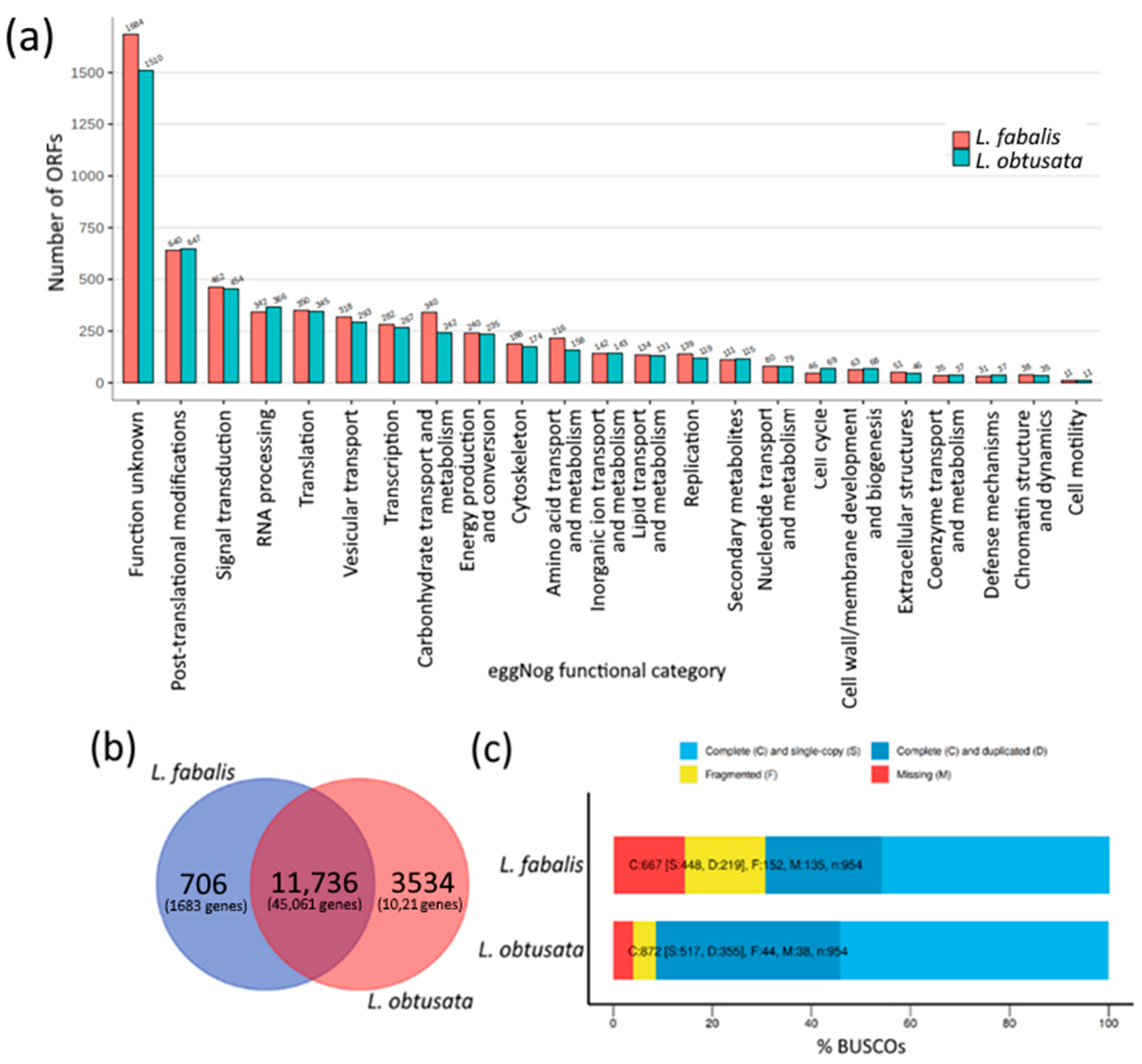
3.1. Species-Specific Orthogroups in Transcriptomes
3.2. Species-Specific Proteins in L. obtusata and L. fabalis Oviducts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reid, D.G. Systematics and Evolution of Littorina; The Ray Society: London, UK, 1996; Volume 164, 463p. [Google Scholar] [CrossRef]
- Granovitch, A.; Mikhailova, N.; Znamenskaya, O.; Petrova, Y. Species complex of mollusks of the genus Littorina (Gastropoda, Prosobranchia) from the eastern Murman coast. Zool. Zhurnal 2004, 83, 1305–1316. [Google Scholar]
- Costa, D.; Sotelo, G.; Kaliontzopoulou, A.; Carvalho, J.; Butlin, R.; Hollander, J.; Faria, R. Hybridization Patterns between Two Marine Snails, Littorina Fabalis and L. Obtusata. Ecol. Evol. 2020, 10, 1158–1179. [Google Scholar] [CrossRef]
- Maltseva, A.L.; Varfolomeeva, M.A.; Ayanka, R.V.; Gafarova, E.R.; Repkin, E.A.; Pavlova, P.A.; Shavarda, A.L.; Mikhailova, N.A.; Granovitch, A.I. Linking Ecology, Morphology, and Metabolism: Niche Differentiation in Sympatric Populations of Closely Related Species of the Genus Littorina (Neritrema). Ecol. Evol. 2021, 11, 11134–11154. [Google Scholar] [CrossRef]
- Eberhard, W.G. Evolution of Genitalia: Theories, Evidence, and New Directions. Genetica 2010, 138, 5–18. [Google Scholar] [CrossRef]
- Eberhard, W.G. Malefemale Conflict and Genitalia: Failure to Confirm Predictions in Insects and Spiders. Biol. Rev. 2004, 79, 121–186. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, W. Female Control Sexual Selection by Cryptic Female Choice; Princeton University Press: Princeton, NJ, USA, 2019; ISBN 978-0-691-20720-9. [Google Scholar]
- Lobov, A.A.; Maltseva, A.L.; Mikhailova, N.A.; Granovitch, A.I. The Molecular Mechanisms of Gametic Incompatibility in Invertebrates. Acta Nat. 2019, 11, 4–15. [Google Scholar] [CrossRef]
- Firman, R.C.; Gasparini, C.; Manier, M.K.; Pizzari, T. Postmating Female Control: 20 Years of Cryptic Female Choice. Trends Ecol. Evol. 2017, 32, 368–382. [Google Scholar] [CrossRef]
- Swanson, W.J.; Vacquier, V.D. The Rapid Evolution of Reproductive Proteins. Nat. Rev. Genet. 2002, 3, 137–144. [Google Scholar] [CrossRef]
- Zigler, K.S. The Evolution of Sea Urchin Sperm Bindin. Int. J. Dev. Biol. 2008, 52, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Geyer, L.B.; Zigler, K.S.; Tiozzo, S.; Lessios, H.A. Slow Evolution under Purifying Selection in the Gamete Recognition Protein Bindin of the Sea Urchin Diadema. Sci. Rep. 2020, 10, 9834. [Google Scholar] [CrossRef]
- Kemppainen, P.; Panova, M.; Hollander, J.; Johannesson, K. Complete Lack of Mitochondrial Divergence between Two Species of NE Atlantic Marine Intertidal Gastropods. J. Evol. Biol. 2009, 22, 2000–2011. [Google Scholar] [CrossRef]
- Marques, J.P.; Sotelo, G.; Larsson, T.; Johannesson, K.; Panova, M.; Faria, R. Comparative Mitogenomic Analysis of Three Species of Periwinkles: Littorina Fabalis, L. Obtusata and L. Saxatilis. Mar. Genom. 2017, 32, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Reid, D.G.; Dyal, P.; Williams, S.T. A Global Molecular Phylogeny of 147 Periwinkle Species (Gastropoda, Littorininae): Phylogeny of Littorininae. Zool. Scr. 2012, 41, 125–136. [Google Scholar] [CrossRef]
- Sotelo, G.; Duvetorp, M.; Costa, D.; Panova, M.; Johannesson, K.; Faria, R. Phylogeographic History of Flat Periwinkles, Littorina Fabalis and L. Obtusata. BMC Evol. Biol. 2020, 20, 23. [Google Scholar] [CrossRef]
- Maltseva, A.L.; Varfolomeeva, M.A.; Lobov, A.A.; Tikanova, P.O.; Repkin, E.A.; Babkina, I.Y.; Panova, M.; Mikhailova, N.A.; Granovitch, A.I. Premating Barriers in Young Sympatric Snail Species. Sci. Rep. 2021, 11, 5720. [Google Scholar] [CrossRef]
- Johannesson, K.; Saltin, S.H.; Charrier, G.; Ring, A.-K.; Kvarnemo, C.; André, C.; Panova, M. Non-Random Paternity of Offspring in a Highly Promiscuous Marine Snail Suggests Postcopulatory Sexual Selection. Behav. Ecol. Sociobiol. 2016, 70, 1357–1366. [Google Scholar] [CrossRef]
- Makinen, T.; Panova, M.; Andre, C. High Levels of Multiple Paternity in Littorina Saxatilis: Hedging the Bets? J. Hered. 2007, 98, 705–711. [Google Scholar] [CrossRef]
- Paterson, I.G.; Partridge, V.; Buckland-Nicks, J. Multiple Paternity in Littorina Obtusata (Gastropoda, Littorinidae) Revealed by Microsatellite Analyses. Biol. Bull. 2001, 200, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Lobov, A.A. Gamete Interaction Proteins as Factors of Reproductive Isolation of Cryptic Species of the Genus Littorina Férussac, 1822; Saint-Petersburg State University: Saint-Petersburg, Russia, 2021. [Google Scholar]
- Lobov, A.; Babkina, I.; Maltseva, A.; Mikhailova, N.; Granovitch, A. Proteins of Penial Mamilliform Glands in Closely Related Littorina Species (Mollusca, Caenogastropoda): Variability and Possible Contribution to Reproductive Isolation. Biol. Commun. 2020, 65, 200–212. [Google Scholar] [CrossRef]
- Lobov, A.A.; Maltseva, A.L.; Mikhailova, N.A.; Granovitch, A.I. LOSP: A Newly Identified Sperm Protein from Littorina Obtusata. J. Molluscan Stud. 2015, 81, 512–515. [Google Scholar] [CrossRef]
- Lobov, A.A.; Maltseva, A.L.; Starunov, V.V.; Babkina, I.Y.; Ivanov, V.A.; Mikhailova, N.A.; Granovitch, A.I. LOSP: A Putative Marker of Parasperm Lineage in Male Reproductive System of the Prosobranch Mollusk Littorina Obtusata. J. Exp. Zoolog. B Mol. Dev. Evol. 2018, 330, 193–201. [Google Scholar] [CrossRef]
- Buckland-Nicks, J. Prosobranch Parasperm: Sterile Germ Cells That Promote Paternity? Micron 1998, 29, 267–280. [Google Scholar] [CrossRef]
- Maltseva, A.L.; Varfolomeeva, M.A.; Lobov, A.A.; Tikanova, P.; Panova, M.; Mikhailova, N.A.; Granovitch, A.I. Proteomic Similarity of the Littorinid Snails in the Evolutionary Context. PeerJ. 2020, 8, e8546. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. The Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate–Phenol–Chloroform Extraction: Twenty-Something Years On. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De Novo Transcript Sequence Reconstruction from RNA-Seq Using the Trinity Platform for Reference Generation and Analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. BioRxiv 2021. [Google Scholar] [CrossRef]
- Ünlü, M.; Morgan, M.E.; Minden, J.S. Difference Gel Electrophoresis. A Single Gel Method for Detecting Changes in Protein Extracts. Electrophoresis 1997, 18, 2071–2077. [Google Scholar] [CrossRef] [PubMed]
- Alban, A.; David, S.O.; Bjorkesten, L.; Andersson, C.; Sloge, E.; Lewis, S.; Currie, I. A Novel Experimental Design for Comparative Two-Dimensional Gel Analysis: Two-Dimensional Difference Gel Electrophoresis Incorporating a Pooled Internal Standard. Proteomics 2003, 3, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, S.M.; Käll, L.; Riffle, M.E.; Bilmes, J.A.; Noble, W.S. Transmembrane Topology and Signal Peptide Prediction Using Dynamic Bayesian Networks. PLoS Comput. Biol. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Brunak, S. Prediction of Glycosylation across the Human Proteome and the Correlation to Protein Function. In Proceedings of the Pac Symp Biocomput, Honolulu, HI, USA, 3–7 January 2002; pp. 310–322. [Google Scholar]
- Erdős, G.; Pajkos, M.; Dosztányi, Z. IUPred3: Prediction of Protein Disorder Enhanced with Unambiguous Experimental Annotation and Visualization of Evolutionary Conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef]
- Maltseva, A.; Varfolomeeva, M.; Lobov, A.; Mikhailova, N.; Renaud, P.; Grishankov, A.; Volovik, K.; Granovitch, A. Measuring Physiological Similarity of Closely Related Littorinid Species: A Proteomic Insight. Mar. Ecol. Prog. Ser. 2016, 552, 177–193. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 Chaperones: Cellular Functions and Molecular Mechanism. Cell. Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef]
- Munro, A.W.; McLean, K.J.; Grant, J.L.; Makris, T.M. Structure and Function of the Cytochrome P450 Peroxygenase Enzymes. Biochem. Soc. Trans. 2018, 46, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Huang, M.; Zhang, H.; Song, L. The Immune Role of C-Type Lectins in Molluscs. Invertebr. Surviv. J. 2011, 8, 241–246. [Google Scholar]
- Zelensky, A.N.; Gready, J.E. The C-Type Lectin-like Domain Superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef] [PubMed]
- Lawniczak, M.; Barnes, A.; Linklater, J.; Boone, J.; Wigby, S.; Chapman, T. Mating and Immunity in Invertebrates. Trends Ecol. Evol. 2007, 22, 48–55. [Google Scholar] [CrossRef]
- Dodd, R.B.; Drickamer, K. Lectin-like Proteins in Model Organisms: Implications for Evolution of Carbohydrate-Binding Activity. Glycobiology 2001, 11, 71R–79R. [Google Scholar] [CrossRef] [PubMed]
- Katsu, Y.; Lubahn, D.B.; Iguchi, T. Expression of a Novel C-Type Lectin in the Mouse Vagina. Endocrinology 2003, 144, 2597–2605. [Google Scholar] [CrossRef]
- Glabe, C.G.; Grabel, L.B.; Vacquier, V.D.; Rosen, S.D. Carbohydrate Specificity of Sea Urchin Sperm Bindin: A Cell Surface Lectin Mediating Sperm-Egg Adhesion. J. Cell Biol. 1982, 94, 123–128. [Google Scholar] [CrossRef]
- Brown, A.C.; Harrison, L.M.; Kapulkin, W.; Jones, B.F.; Sinha, A.; Savage, A.; Villalon, N.; Cappello, M. Molecular Cloning and Characterization of a C-Type Lectin from Ancylostoma Ceylanicum: Evidence for a Role in Hookworm Reproductive Physiology. Mol. Biochem. Parasitol. 2007, 151, 141–147. [Google Scholar] [CrossRef]
- Wong, A.; Albright, S.N.; Giebel, J.D.; Ram, K.R.; Ji, S.; Fiumera, A.C.; Wolfner, M.F. A Role for Acp29AB, a Predicted Seminal Fluid Lectin, in Female Sperm Storage in Drosophila Melanogaster. Genetics 2008, 180, 921–931. [Google Scholar] [CrossRef][Green Version]
- Sarrias, M.R.; Gronlund, J.; Padilla, O.; Madsen, J.; Holmskov, U.; Lozano, F. The Scavenger Receptor Cysteine-Rich (SRCR) Domain: An Ancient and Highly Conserved Protein Module of the Innate Immune System. Crit. Rev. Immunol. 2004, 24, 1–38. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.C.; Lawler, J. The Thrombospondins. Cold Spring Harb. Perspect. Biol. 2011, 3, a009712. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, S.; Iwanaga, S. Role of Lectins in the Innate Immunity of Horseshoe Crab. Dev. Comp. Immunol. 1999, 23, 391–400. [Google Scholar] [CrossRef]
- Denis, M.; Thayappan, K.; Ramasamy, S.; Munusamy, A. Lectin in Innate Immunity of Crustacea. Austin Biol. 2016, 1, 1001. [Google Scholar]
- Thomas-Bulle, C.; Piednoël, M.; Donnart, T.; Filée, J.; Jollivet, D.; Bonnivard, É. Mollusc Genomes Reveal Variability in Patterns of LTR-Retrotransposons Dynamics. BMC Genom. 2018, 19, 821. [Google Scholar] [CrossRef]
- Puzakova, L.V.; Puzakov, M.V. The Tc1/Mariner DNA Transposons in the Genome of Mollusk Littorina Saxatilis. Russ. J. Genet. 2017, 53, 1358–1365. [Google Scholar] [CrossRef]
- McInerney, C.E.; Allcock, A.L.; Johnson, M.P.; Bailie, D.A.; Prodöhl, P.A. Comparative Genomic Analysis Reveals Species-Dependent Complexities That Explain Difficulties with Microsatellite Marker Development in Molluscs. Heredity 2011, 106, 78–87. [Google Scholar] [CrossRef] [PubMed]
- DeMarco, R.; Machado, A.A.; Bisson-Filho, A.W.; Verjovski-Almeida, S. Identification of 18 New Transcribed Retrotransposons in Schistosoma Mansoni. Biochem. Biophys. Res. Commun. 2005, 333, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as Regulators of Gene Expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef]
- Gorbushin, A.M.; Borisova, E.A. Lectin-like Molecules in Transcriptome of Littorina Littorea Hemocytes. Dev. Comp. Immunol. 2015, 48, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Serrato-Capuchina, A.; Matute, D.R. The role of transposable elements in speciation. Genes. 2018, 9, 254. [Google Scholar] [CrossRef] [PubMed]
- Dechaud, C.; Volff, J.N.; Schartl, M.; Naville, M. Sex and the TEs: Transposable elements in sexual development and function in animals. Mobile DNA 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Dechaud, C.; Miyake, S.; Martinez-Bengochea, A.; Schartl, M.; Volff, J.N.; Naville, M. Clustering of sex-biased genes and transposable elements in the genome of the medaka fish Oryzias latipes. Genome Biol. Evol. 2021, evab230. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lobov, A.A.; Babkina, I.Y.; Danilov, L.G.; Masharskiy, A.E.; Predeus, A.V.; Mikhailova, N.A.; Granovitch, A.I.; Maltseva, A.L. Species-Specific Proteins in the Oviducts of Snail Sibling Species: Proteotranscriptomic Study of Littorina fabalis and L. obtusata. Biology 2021, 10, 1087. https://doi.org/10.3390/biology10111087
Lobov AA, Babkina IY, Danilov LG, Masharskiy AE, Predeus AV, Mikhailova NA, Granovitch AI, Maltseva AL. Species-Specific Proteins in the Oviducts of Snail Sibling Species: Proteotranscriptomic Study of Littorina fabalis and L. obtusata. Biology. 2021; 10(11):1087. https://doi.org/10.3390/biology10111087
Chicago/Turabian StyleLobov, Arseniy A., Irina Y. Babkina, Lavrentii G. Danilov, Alexey E. Masharskiy, Alexander V. Predeus, Natalia A. Mikhailova, Andrei I. Granovitch, and Arina L. Maltseva. 2021. "Species-Specific Proteins in the Oviducts of Snail Sibling Species: Proteotranscriptomic Study of Littorina fabalis and L. obtusata" Biology 10, no. 11: 1087. https://doi.org/10.3390/biology10111087
APA StyleLobov, A. A., Babkina, I. Y., Danilov, L. G., Masharskiy, A. E., Predeus, A. V., Mikhailova, N. A., Granovitch, A. I., & Maltseva, A. L. (2021). Species-Specific Proteins in the Oviducts of Snail Sibling Species: Proteotranscriptomic Study of Littorina fabalis and L. obtusata. Biology, 10(11), 1087. https://doi.org/10.3390/biology10111087






