Mitochondrial Translation Occurs Preferentially in the Peri-Nuclear Mitochondrial Network of Cultured Human Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
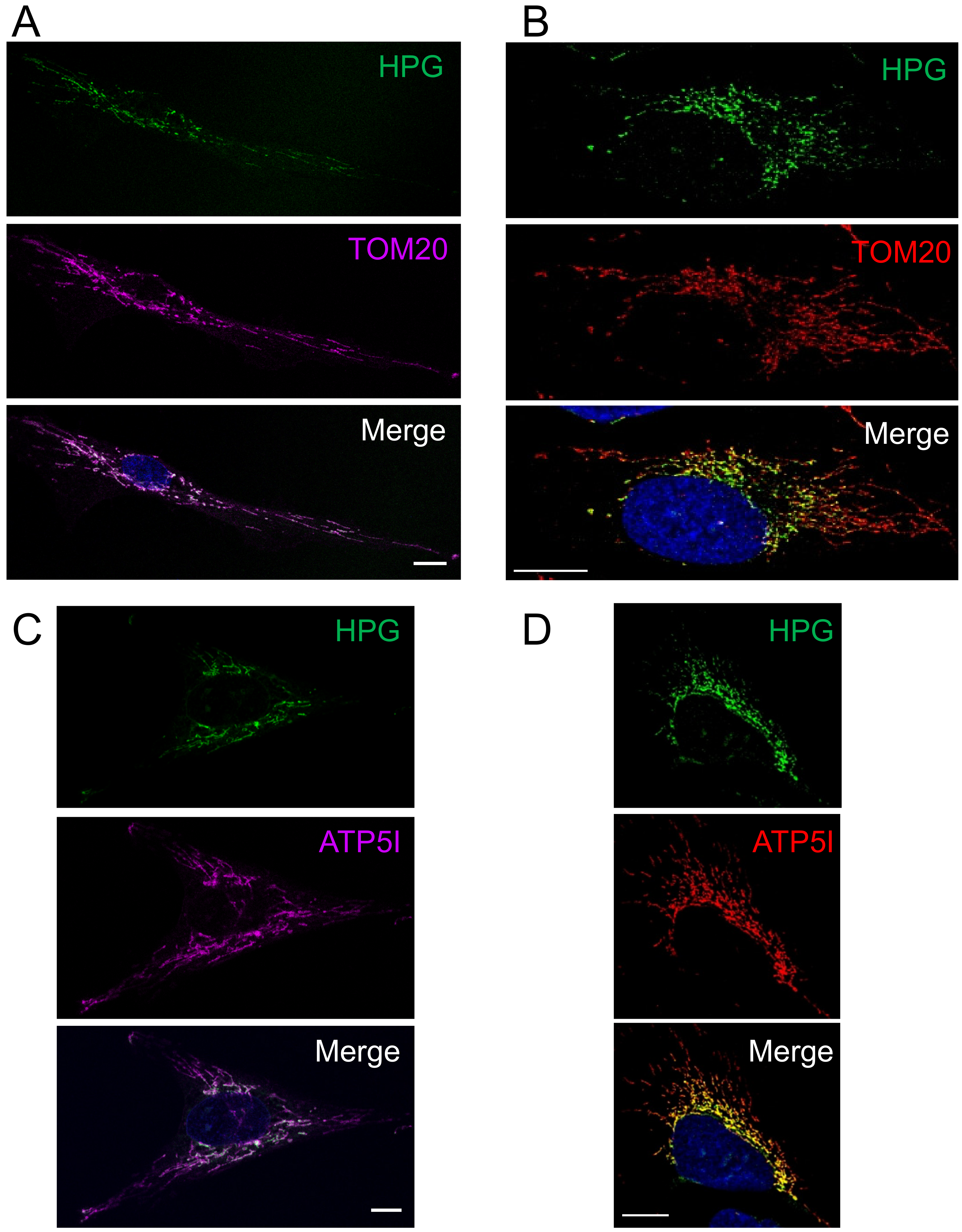
2.2. Fluorescent Non-Canonical Amino Acid Tagging (FUNCAT)
2.3. Immunofluorescent Cytochemistry
2.4. RNA FISH/Immunofluorescent Cytochemistry
2.5. Confocal Imaging
2.6. Image Analysis
3. Results
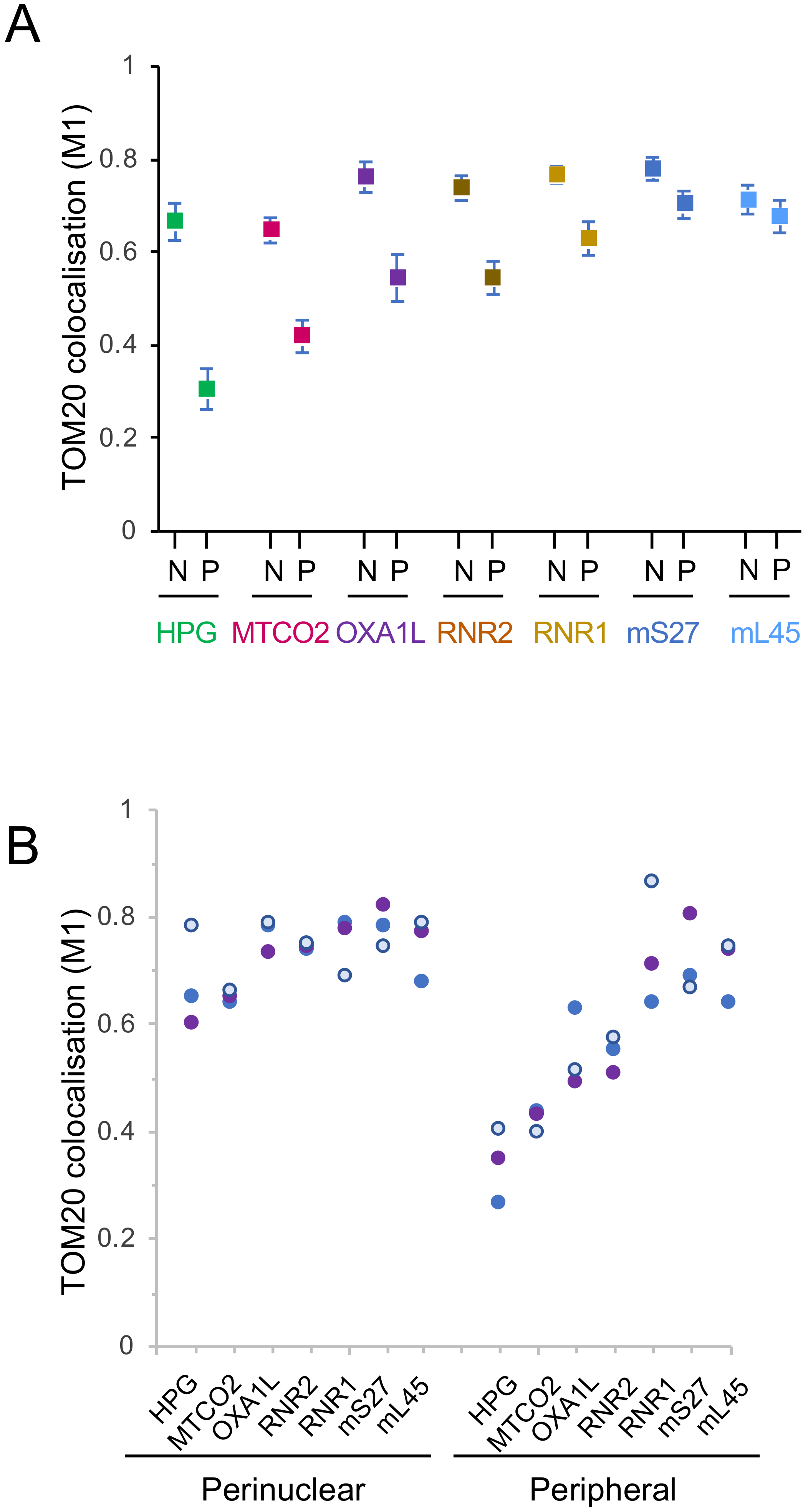
3.1. Mitochondrial Translation Occurs Preferentially in the Peri-Nuclear Network
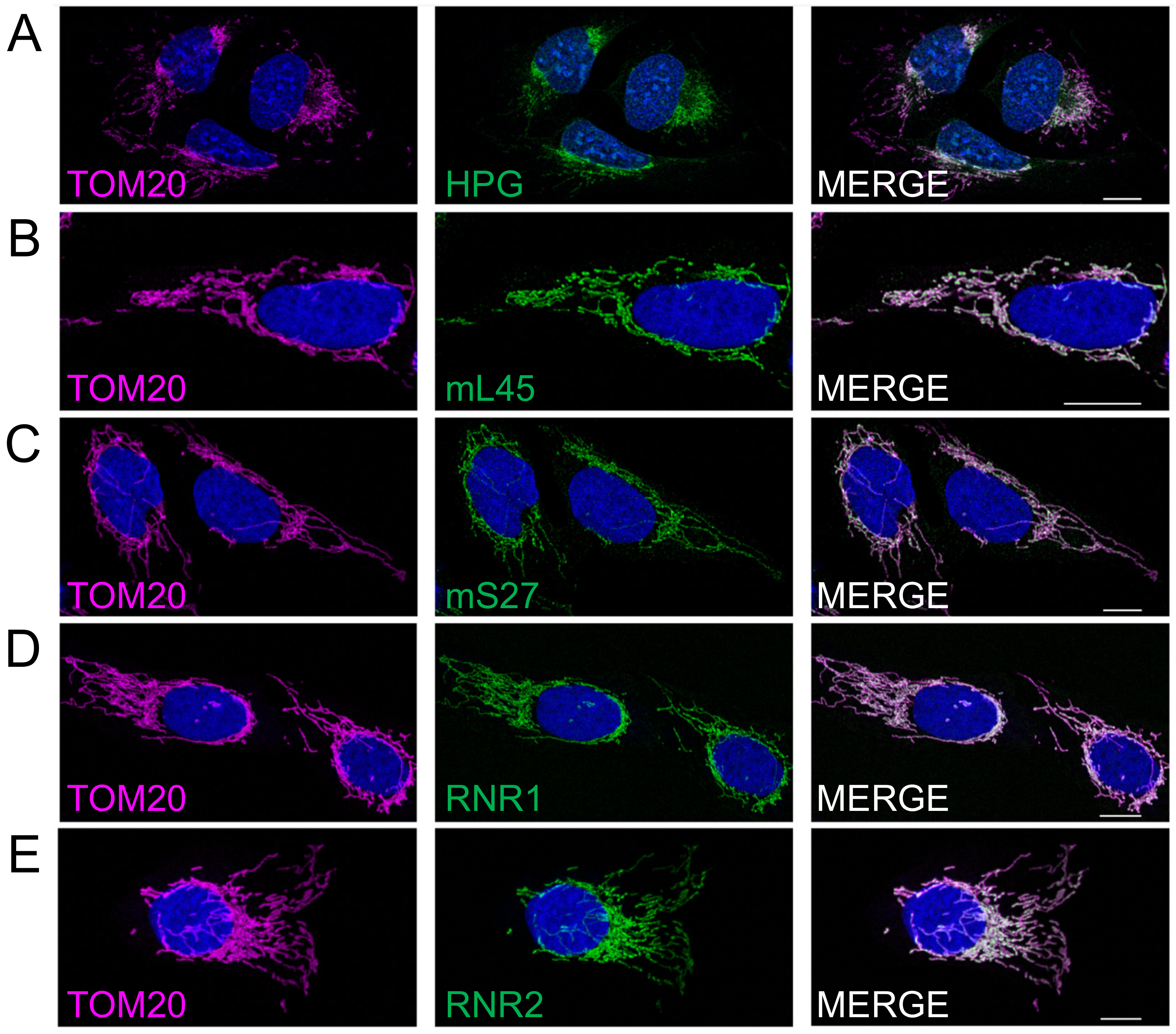
3.2. Preferential Peri-Nuclear Translation Is Not Reflective of a Similar Distribution of Mitoribosomes
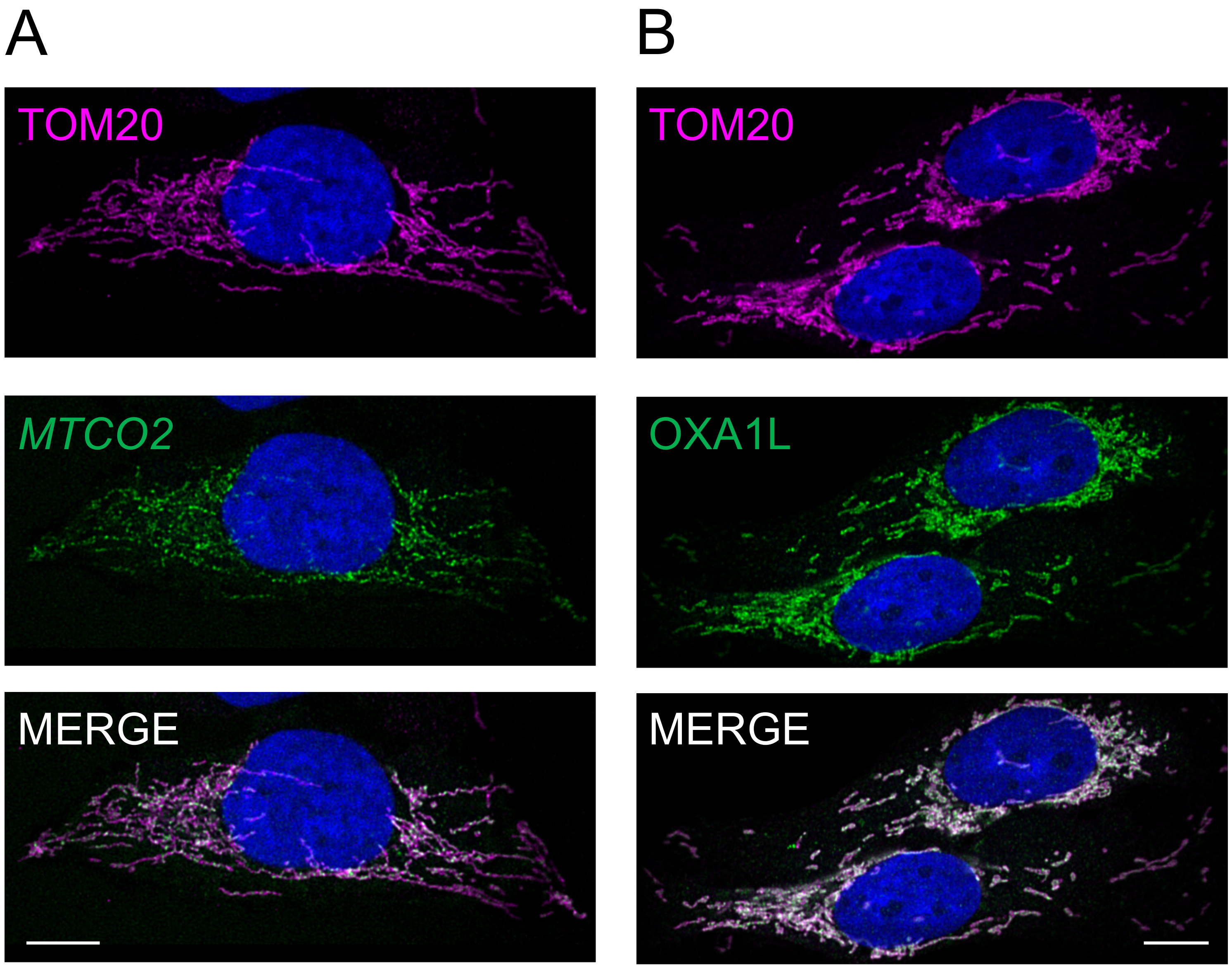
3.3. Differential Distribution of mt-Protein Synthesis Is Not Reflective of the mt-mRNA Distribution
3.4. Peripheral Depletion of Mitochondrial Translation Cannot Be Explained by the Distribution of OXAL1
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Adebayo, M.; Singh, S.; Singh, A.P.; Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 2021, 35, e21620. [Google Scholar] [CrossRef]
- Stoldt, S.; Wenzel, D.; Kehrein, K.; Riedel, D.; Ott, M.; Jakobs, S. Spatial orchestration of mitochondrial translation and OXPHOS complex assembly. Nat. Cell Biol. 2018, 20, 528–534. [Google Scholar] [CrossRef] [Green Version]
- Ikon, N.; Ryan, R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Gilkerson, R.W.; Selker, J.M.; Capaldi, R.A. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003, 546, 355–358. [Google Scholar] [CrossRef] [Green Version]
- Stephan, T.; Bruser, C.; Deckers, M.; Steyer, A.M.; Balzarotti, F.; Barbot, M.; Behr, T.S.; Heim, G.; Hubner, W.; Ilgen, P.; et al. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020, 39, e104105. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.M.; Segawa, M.; Kondadi, A.K.; Anand, R.; Bailey, S.T.; Reichert, A.S.; van der Bliek, A.M.; Shackelford, D.B.; Liesa, M.; Shirihai, O.S. Individual cristae within the same mitochondrion display different membrane potentials and are functionally independent. EMBO J. 2019, 38, e101056. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.F.; Rebelo-Guiomar, P.; D’Souza, A.R.; Powell, C.A.; Van Haute, L.; Minczuk, M. Regulation of Mammalian Mitochondrial Gene Expression: Recent Advances. Trends Biochem. Sci. 2017, 42, 625–639. [Google Scholar] [CrossRef] [Green Version]
- Kotrys, A.V.; Szczesny, R.J. Mitochondrial Gene Expression and Beyond-Novel Aspects of Cellular Physiology. Cells 2019, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Garrido, N.; Griparic, L.; Jokitalo, E.; Wartiovaara, J.; van der Bliek, A.M.; Spelbrink, J.N. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 2003, 14, 1583–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feric, M.; Demarest, T.G.; Tian, J.; Croteau, D.L.; Bohr, V.A.; Misteli, T. Self-assembly of multi-component mitochondrial nucleoids via phase separation. EMBO J. 2021, 40, e107165. [Google Scholar] [CrossRef]
- Satoh, M.; Kuroiwa, T. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp. Cell Res. 1991, 196, 137–140. [Google Scholar] [CrossRef]
- Jourdain, A.A.; Koppen, M.; Wydro, M.; Rodley, C.D.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.; Martinou, J.C. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013, 17, 399–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonicka, H.; Sasarman, F.; Nishimura, T.; Paupe, V.; Shoubridge, E.A. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013, 17, 386–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iborra, F.J.; Kimura, H.; Cook, P.R. The functional organization of mitochondrial genomes in human cells. BMC Biol. 2004, 2, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hensen, F.; Potter, A.; van Esveld, S.L.; Tarres-Sole, A.; Chakraborty, A.; Sola, M.; Spelbrink, J.N. Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res. 2019, 47, 3680–3698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Greber, B.J.; Bieri, P.; Leibundgut, M.; Leitner, A.; Aebersold, R.; Boehringer, D.; Ban, N. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science 2015, 348, 303–308. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.; Amunts, A.; Bai, X.C.; Sugimoto, Y.; Edwards, P.C.; Murshudov, G.; Scheres, S.H.; Ramakrishnan, V. Structure of the large ribosomal subunit from human mitochondria. Science 2014, 346, 718–722. [Google Scholar] [CrossRef] [Green Version]
- Zorkau, M.; Albus, C.A.; Berlinguer-Palmini, R.; Chrzanowska-Lightowlers, Z.M.A.; Lightowlers, R.N. High-resolution imaging reveals compartmentalization of mitochondrial protein synthesis in cultured human cells. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Zorkau, M.; Proctor-Kent, Y.; Berlinguer-Palmini, R.; Hamilton, A.; Chrzanowska-Lightowlers, Z.M.; Lightowlers, R.N. Visualizing Mitochondrial Ribosomal RNA and Mitochondrial Protein Synthesis in Human Cell Lines. Methods Mol. Biol. 2021, 2192, 159–181. [Google Scholar] [CrossRef]
- Paumard, P.; Vaillier, J.; Coulary, B.; Schaeffer, J.; Soubannier, V.; Mueller, D.M.; Brethes, D.; di Rago, J.P.; Velours, J. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002, 21, 221–230. [Google Scholar] [CrossRef]
- Muhleip, A.; Kock Flygaard, R.; Ovciarikova, J.; Lacombe, A.; Fernandes, P.; Sheiner, L.; Amunts, A. ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria. Nat. Commun. 2021, 12, 120. [Google Scholar] [CrossRef]
- Davies, K.M.; Strauss, M.; Daum, B.; Kief, J.H.; Osiewacz, H.D.; Rycovska, A.; Zickermann, V.; Kuhlbrandt, W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. USA 2011, 108, 14121–14126. [Google Scholar] [CrossRef] [Green Version]
- Szyrach, G.; Ott, M.; Bonnefoy, N.; Neupert, W.; Herrmann, J.M. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003, 22, 6448–6457. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Andrell, J.; Choi, A.; Richter, U.; Maiti, P.; Best, R.B.; Barrientos, A.; Battersby, B.J.; Amunts, A. Mechanism of membrane-tethered mitochondrial protein synthesis. Science 2021, 371, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Desai, N.; Yang, H.; Chandrasekaran, V.; Kazi, R.; Minczuk, M.; Ramakrishnan, V. Elongational stalling activates mitoribosome-associated quality control. Science 2020, 370, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Wurm, C.A.; Neumann, D.; Lauterbach, M.A.; Harke, B.; Egner, A.; Hell, S.W.; Jakobs, S. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc. Natl. Acad. Sci. USA 2011, 108, 13546–13551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, A.F.; Clayton, D.A. In situ localization of mitochondrial DNA replication in intact mammalian cells. J. Cell Biol. 1996, 135, 883–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnusson, J.; Orth, M.; Lestienne, P.; Taanman, J.W. Replication of mitochondrial DNA occurs throughout the mitochondria of cultured human cells. Exp. Cell Res. 2003, 289, 133–142. [Google Scholar] [CrossRef]
- Collins, T.J.; Berridge, M.J.; Lipp, P.; Bootman, M.D. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002, 21, 1616–1627. [Google Scholar] [CrossRef]
- Kuznetsov, A.V.; Margreiter, R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int. J. Mol. Sci. 2009, 10, 1911–1929. [Google Scholar] [CrossRef] [Green Version]
- Al-Mehdi, A.B.; Pastukh, V.M.; Swiger, B.M.; Reed, D.J.; Patel, M.R.; Bardwell, G.C.; Pastukh, V.V.; Alexeyev, M.F.; Gillespie, M.N. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci. Signal. 2012, 5, ra47. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Thai, P.N.; Lu, S.; Pu, J.; Bers, D.M. Intrafibrillar and perinuclear mitochondrial heterogeneity in adult cardiac myocytes. J. Mol. Cell Cardiol. 2019, 136, 72–84. [Google Scholar] [CrossRef]
- Wang, C.; Du, W.; Su, Q.P.; Zhu, M.; Feng, P.; Li, Y.; Zhou, Y.; Mi, N.; Zhu, Y.; Jiang, D.; et al. Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res. 2015, 25, 1108–1120. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albus, C.A.; Berlinguer-Palmini, R.; Hewison, C.; McFarlane, F.; Savu, E.-A.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.; Zorkau, M. Mitochondrial Translation Occurs Preferentially in the Peri-Nuclear Mitochondrial Network of Cultured Human Cells. Biology 2021, 10, 1050. https://doi.org/10.3390/biology10101050
Albus CA, Berlinguer-Palmini R, Hewison C, McFarlane F, Savu E-A, Lightowlers RN, Chrzanowska-Lightowlers ZM, Zorkau M. Mitochondrial Translation Occurs Preferentially in the Peri-Nuclear Mitochondrial Network of Cultured Human Cells. Biology. 2021; 10(10):1050. https://doi.org/10.3390/biology10101050
Chicago/Turabian StyleAlbus, Christin A., Rolando Berlinguer-Palmini, Caroline Hewison, Fiona McFarlane, Elisabeta-Ana Savu, Robert N. Lightowlers, Zofia M. Chrzanowska-Lightowlers, and Matthew Zorkau. 2021. "Mitochondrial Translation Occurs Preferentially in the Peri-Nuclear Mitochondrial Network of Cultured Human Cells" Biology 10, no. 10: 1050. https://doi.org/10.3390/biology10101050





