Extracellular DNA: Insight of a Signal Molecule in Crop Protection
Abstract
:Simple Summary
Abstract
1. Introduction
2. Natural Conditions of eDNA Release and Sensing
3. Self-eDNA as a DAMP
4. eDNA as a MAMP/PAMP
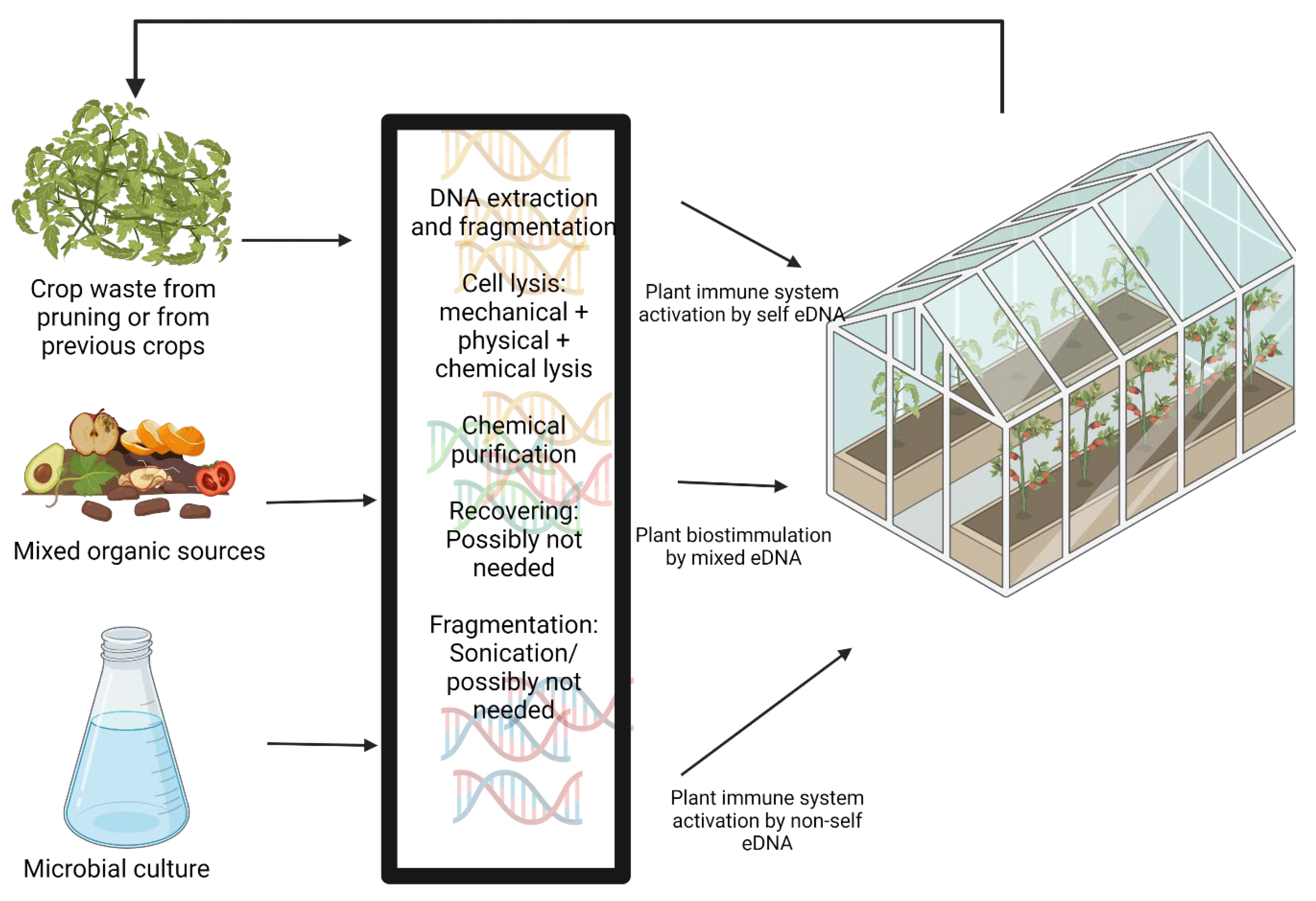
5. Technical Challenges of eDNA Application as an Agricultural Treatment
5.1. DNA Extraction
- The amplitude of the voltage or intensity of sound
- The diameter of the channel where the fluid with cells is passing
- The application time of voltage or acoustic stimulus. The time can be managed by the pump, which varies the velocity of the fluid with cells.
5.2. DNA Damaging/Fragmentation
6. Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Agrimonti, C.; Lauro, M.; Visioli, G. Smart agriculture for food quality: Facing climate change in the 21st century. Crit. Rev. Food Sci. Nutr. 2020, 61, 971–981. [Google Scholar] [CrossRef]
- FAO. Climate Change and Food Security: Risks and Responses; FAO: Rome, Italy, 2016. [Google Scholar]
- Ali, S.; Liu, Y.; Ishaq, M.; Shah, T.; Abdullah; Ilyas, A.; Din, I.U. Climate Change and Its Impact on the Yield of Major Food Crops: Evidence from Pakistan. Foods 2017, 6, 39. [Google Scholar] [CrossRef]
- Thornton, P.K.; Jones, P.G.; Alagarswamy, G.; Andresen, J.; Herrero, M. Adapting to climate change: Agricultural system and household impacts in East Africa. Agric. Syst. 2010, 103, 73–82. [Google Scholar] [CrossRef]
- Rowhani, P.; Lobell, D.B.; Linderman, M.; Ramankutty, N. Climate variability and crop production in Tanzania. Agric. For. Meteorol. 2011, 151, 449–460. [Google Scholar] [CrossRef]
- Lybbert, T.J.; Sumner, D.A. Agricultural technologies for climate change in developing countries: Policy options for innovation and technology diffusion. Food Policy 2012, 37, 114–123. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 30, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waha, K.; Muller, C.; Bondeau, A.; Dietrich, J.P.; Kurukulasuriya, P.; Heinke, J.; Lotze-Campen, H. Adaptation to climate change through the choice of cropping system and sowing date in sub-Saharan Africa. Glob. Environ. Chang. 2012, 23, 130–143. [Google Scholar] [CrossRef]
- Kabubo-Mariara, J.; Karanja, F.K. The economic impact of climate change on Kenyan crop agriculture: A Ricardian approach. Glob. Planet Chang. 2007, 57, 319–330. [Google Scholar] [CrossRef] [Green Version]
- McKune, S.; Poulsen, L.; Russo, S.; Devereux, T.; Faas, S.; McOmber, C.; Ryley, T. Reaching the end goal: Do interventions to improve climate information services lead to greater food security? Clim. Risk Manag. 2018, 22, 22–41. [Google Scholar] [CrossRef]
- Sharma, U.; Paliyal, S.S.; Sharma, S.P.; Sharma, G.D. Effects of Continuous Use of Chemical Fertilizers and Manure on Soil Fertility and Productivity of Maize–Wheat under Rainfed Conditions of the Western Himalayas. Commun. Soil Sci. Plant Anal. 2014, 45, 2647–2659. [Google Scholar] [CrossRef]
- Dhiman, D.; Sharma, R.; Sankhyan, N.K.; Sepehya, S.; Sharma, S.K.; Kumar, R. Effect of regular application of fertilizers, manure and lime on soil health and productivity of wheat in an acid Alfisol. J. Plant Nutr. 2019, 42, 2507–2521. [Google Scholar] [CrossRef]
- Lamb, A.; Green, R.; Bateman, I.; Broadmeadow, M.; Bruce, T.J.A.; Burney, J.; Carey, P.; Chadwick, D.R.; Crane, E.; Field, R.; et al. The potential for land sparing to offset greenhouse gas emissions from agriculture. Nat. Clim. Chang. 2016, 6, 488–492. [Google Scholar] [CrossRef] [Green Version]
- Mittal, S.; Hariharan, V. Mobile-based climate services impact on farmers risk management ability in India. Clim. Risk. Manag. 2018, 22, 42–51. [Google Scholar] [CrossRef]
- Rockström, J.; Steffen, W.; Noone, K.; Åsa Persson, F.; Chapin, S., III; Lambin, E.F.; Lenton, T.M.; Scheffer, M.; Folke, C.; Schellnhuber, H.J.; et al. A safe operating space for humanity. Nature 2009, 461, 472–475. [Google Scholar] [CrossRef] [PubMed]
- García-Mier, L.; Guevara-González, R.G.; Mondragón-Olguín, V.M.; Del Rocío Verduzco-Cuellar, B.; Torres-Pacheco, I. Agriculture and bioactives: Achieving both crop yield and phytochemicals. Int. J. Mol. Sci. 2013, 14, 4203–4222. [Google Scholar] [CrossRef] [PubMed]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant. 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitehead, S.R.; Poveda, K. Resource allocation trade-offs and the loss of chemical defences during apple domestication. Ann. Bot. 2019, 123, 1029–1041. [Google Scholar] [CrossRef]
- Batyrshina, Z.S.; Yaakov, B.; Shavit, R.; Singh, A.; Tzin, V. Comparative transcriptomic and metabolic analysis of wild and domesticated wheat genotypes reveals differences in chemical and physical defense responses against aphids. BMC Plant. Biol. 2020, 20, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brzozowski, L.J.; Gore, M.A.; Agrawal, A.A.; Mazourek, M. Divergence of defensive cucurbitacins in independent Cucurbita pepo domestication events leads to differences in specialist herbivore preference. Plant Cell Environ. 2020, 43, 2812–2825. [Google Scholar] [CrossRef]
- Diaz-Valenzuela, E.; Sawers, R.H.; Cibrian-Jaramillo, A. Cis- And trans-regulatory variations in the domestication of the chili pepper fruit. Mol. Biol. Evol. 2020, 37, 1593–1603. [Google Scholar] [CrossRef]
- Taha, R.S.; Alharby, H.F.; Bamagoos, A.A.; Medani, R.A.; Rady, M.M. Elevating tolerance of drought stress in Ocimum basilicum using pollen grains extract; a natural biostimulant by regulation of plant performance and antioxidant defense system. South Afr. J. Bot. 2020, 128, 42–53. [Google Scholar] [CrossRef]
- Azad, M.O.K.; Park, B.S.; Adnan, M.; Germ, M.; Kreft, I.; Woo, S.H.; Park, C.H. Silicon biostimulant enhances the growth characteristics and fortifies the bioactive compounds in common and Tartary buckwheat plant. J. Crop. Sci. Biotechnol. 2021, 24, 51–59. [Google Scholar] [CrossRef]
- Melo, P.; Abreu, C.; Bahcevandziev, K.; Araujo, G.; Pereira, L. Biostimulant effect of marine macroalgae bioextract on pepper grown in greenhouse. Appl. Sci. 2020, 10, 4052. [Google Scholar] [CrossRef]
- Puglisi, I.; La Bella, E.; Rovetto, E.I.; Lo Piero, A.R.; Baglieri, A. Biostimulant effect and biochemical response in lettuce seedlings treated with a scenedesmus quadricauda extract. Plants 2020, 9, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radkowski, A.; Radkowska, I.; Bocianowski, J.; Sladkovska, T.; Wolski, K. The effect of foliar application of an amino acid-based biostimulant on lawn functional value. Agronomy 2020, 10, 1656. [Google Scholar] [CrossRef]
- Vargas-Hernandez, M.; Macias-Bobadilla, I.; Guevara-Gonzalez, R.G.; Romero-Gomez, S.d.J.; Rico-Garcia, E.; Ocampo-Velazquez, R.V.; Alvarez-Arquieta, L.d.L.; Torres-Pacheco, I. Plant hormesis management with biostimulants of biotic origin in agriculture. Front. Plant. Sci. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Macías, J.P.; García, Y.C.; Núñez, M.; Díaz, K.; Olea, A.F.; Espinoza, L. Plant growth-defense trade-offs: Molecular processes leading to physiological changes. Int. J. Mol. Sci. 2021, 22, 693. [Google Scholar] [CrossRef]
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments features, relevance and applications. Appl. Microbiol. Biotechnol. 2018, 102, 6343–6356. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, P.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Morrisey, E.; McHugh, T.; Preteska, L.; Hayer, M.; Dijkstra, P.; Hungate, B.A.; Schwartz, E. Dynamics of extracellular DNA decomposition and bacterial community composition in soil. Soil Biol. Biochem. 2015, 86, 42–49. [Google Scholar] [CrossRef]
- Torti, A.; Lever, M.; Jorgensen, B. Origin, dynamics and implications of extracellular DNA pools in marine sediments. Mar. Genom. 2015, 24, 185–196. [Google Scholar] [CrossRef]
- Lorenz, M.G.; Gerjets, D.; Wackernagel, W. Release of transforming plasmid and chromosomal DNA from two cultured soil bacteria. Arch. Microbiol. 1991, 156, 319–326. [Google Scholar] [CrossRef]
- Redmile-Gordon, M.; Bookes, P.; Evershed, R.; Goulding, K.; Hirsch, P. Measuring the soil-microbial interface: Extraction of extracellular polymeric substances (EPS) from soil biofilms. Soil Biol. Biochem. 2014, 72, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef]
- Ibáñez de Aldeoca, A.; Zafra, O.; González-Pastor, J. Mechanisms and regulation of extracelular DNA release and its biological roles in microbial communities. Front. Microbiol. 2017, 8, 1390. [Google Scholar] [CrossRef] [Green Version]
- Compton, M.M. A biochemical hallmark of apoptosis: Internucleosomal degradation of the genome. Cancer Metastasis Rev. 1992, 11, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Didenko, V.V.; Ngo, H.; Baskin, D.S. Early necrotic DNA degradation: Presence of blunt-ended DNA breaks, 3′ and 5′ overhangs in apoptosis, but only 5′ overhangs in early necrosis. Am. J. Pathol. 2003, 162, 1571–1578. [Google Scholar] [CrossRef]
- Duran-Flores, D.; Heil, M. Damaged-self recognition in common bean (Phaseolus vulgaris) shows taxonomic specificity and triggers signaling via reactive oxygen species (ROS). Front. Plant. Sci. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlee, M.; Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016, 16, 566–580. [Google Scholar] [CrossRef]
- Wei, Y.; Baobao, S.; Zhang, Q.; Qiao, G.; Wang, Z.; Shao, R.; Huang, G.; Qi, Z. Molecular cloning and expression analysis of toll-like receptor genes (TLR7, TLR8 and TLR9) of golden pompano (Trachinotus ovatus). Fish. Shellfish. Immunol. 2017, 63, 270–276. [Google Scholar] [CrossRef]
- Dalpke, A.; Frank, J.; Peter, M.; Heeg, K. Activation of toll-like receptor 9 by DNA from different bacterial species. Infect. Immun. 2006, 940–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gay, N.; Symmons, M.; Gangloff, M.; Bryant, C. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol 2014, 14, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.; Ryu, C.M. Plant Perceptions of Extracellular DNA and RNA. Mol. Plant. 2016, 9, 956–958. [Google Scholar] [CrossRef] [PubMed]
- Mazzoleni, S.; Carteni, F.; Bonanomi, G.; Senatore, M.; Termolino, P.; Giannino, F.; Incerti, G.; Rietkerk, M.; Lanzotti, V.; Chiusano, M.L. Inhibitory effects of extracellular self-DNA: A general biological process? New Phytol. 2015, 206, 127–132. [Google Scholar] [CrossRef]
- Vega-Muñoz, I.; Feregrino-Pérez, A.A.; Torres-Pacheco, I.; Guevara-González, R.G. Exogenous fragmented DNA acts as a damage-Associated molecular pattern (DAMP) inducing changes in CpG DNA methylation and defence-related responses in Lactuca sativa. Funct. Plant. Biol. 2018, 45, 1065–1072. [Google Scholar] [CrossRef]
- Chiusano, M.L.; Incerti, G.; Colantuono, C.; Termolino, P.; Palomba, E.; Monticolo, F.; Benvenuto, G.; Foscari, A.; Esposito, A.; Marti, L.; et al. Arabidopsis thaliana Response to Extracellular DNA: Self Versus Nonself Exposure. Plants 2021, 10, 1744. [Google Scholar] [CrossRef]
- Brinkman, P.; Van der Putten, W.H.; Bakker, E.J.; Verhoeven, K.J.F. Plant-soil feedback: Experimental approaches, statistical analyses and ecological interpretations. J. Ecol. 2010, 98, 1063–1073. [Google Scholar] [CrossRef]
- Mangan, S.A.; Schnitzer, S.A.; Herre, E.A.; MacK, K.M.L.; Valencia, M.C.; Sanchez, E.I.; Bever, J.D. Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature 2010, 466, 752–755. [Google Scholar] [CrossRef]
- Sanabria, N.; Goring, D.; Nürnberger, T.; Dubery, I. Self/nonself perception and recognition mechanisms in plants: A comparison of self-incompatibility and innate immunity. New Phytol. 2008, 178, 503–514. [Google Scholar] [CrossRef]
- Barbero, F.; Guglielmotto, M.; Capuzzo, A.; Maffei, M.E. Extracellular self-DNA (esDNA), but not heterologous plant or insect DNA (etDNA), induces plasma membrane depolarization and calcium signaling in lima bean (Phaseolus lunatus) and maize (Zea mays). Int. J. Mol. Sci. 2016, 17, 1659. [Google Scholar] [CrossRef] [Green Version]
- Duran-Flores, D.; Heil, M. Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 2018, 72, 78–88. [Google Scholar] [CrossRef]
- Rassizadeh, L.; Cervero, R.; Flors, V.; Gamir, J. Plant Science Extracellular DNA as an elicitor of broad-spectrum resistance in Arabidopsis thaliana. Plant. Sci. 2021, 312, 111036. [Google Scholar] [CrossRef]
- Pearce, G.; Strydom, D.; Hohnson, S.; Ryan, C.A. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 1991, 253, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Jeter, C.R.; Tang, W.; Henaff, E.; Butterfield, T.; Roux, S.J. Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. Plant. Cell 2004, 16, 2652–2664. [Google Scholar] [CrossRef] [Green Version]
- Huffaker, A.; Pearce, G.; Ryan, C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 2006, 103, 10098–10103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.W.; Klessig, D.F. DAMPs, MAMPs, and NAMPs in plant innate immunity. BMC Plant. Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintana-Rodriguez, E.; Duran-Flores, D.; Heil, M.; Camacho-Coronel, X. Damage-associated molecular patterns (DAMPs) as future plant vaccines that protect crops from pests. Sci. Hortic. 2018, 237, 207–220. [Google Scholar] [CrossRef]
- Ferrusquía-Jiménez, N.I.; Chandrakasan, G.; Torres-Pacheco, I.; Rico-Garcia, E.; Feregrino-Perez, A.A.; Guevara-González, R.G. Extracellular DNA: A Relevant Plant Damage-Associated Molecular Pattern (DAMP) for Crop Protection Against Pests—A Review. J. Plant. Growth Regul. 2020, 40, 451–463. [Google Scholar] [CrossRef]
- Barbero, F.; Guglielmotto, M.; Islam, M.; Maffei, M.E. Extracellular Fragmented Self-DNA Is Involved in Plant Responses to Biotic Stress. Front. Plant. Sci. 2021, 12, 1558. [Google Scholar] [CrossRef] [PubMed]
- Pontiggia, D.; Benedetti, M.; Costantini, S.; De Lorenzo, G.; Cervone, F. Dampening the DAMPs: How Plants Maintain the Homeostasis of Cell Wall Molecular Patterns and Avoid Hyper-Immunity. Front. Plant. Sci. 2020, 11, 613259. [Google Scholar] [CrossRef]
- Di, X.; Takken, F.L.W.; Tintor, N. How phytohormones shape interactions between plants and the soil-borne fungus Fusarium oxysporum. Front. Plant. Sci. 2016, 7, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flors, V.; Ton, J.; Van Doorn, R.; Jakab, G.; García-Agustín, P.; Mauch-Mani, B. Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant. J. 2008, 54, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, Y.; Barkallah, M.; Bouazizi, E.; Gdoura, R.; Triki, M.A. Differential biochemical and physiological responses of two olive cultivars differing by their susceptibility to the hemibiotrophic pathogen Verticillium dahliae. Physiol. Mol. Plant. Path 2017, 97, 30–39. [Google Scholar] [CrossRef]
- Kabbage, M.; Yarden, O.; Dickman, M.B. Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant. Sci. 2015, 233, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yakushiji, S.; Ishiga, Y.; Inagaki, Y.; Toyoda, K.; Shiraishi, T.; Ichinose, Y. Bacterial DNA activates immunity in Arabidopsis thaliana. J. Gen. Plant. Path 2009, 75, 227–234. [Google Scholar] [CrossRef]
- Serrano-Jamaica, L.M.; Villordo-Pineda, E.; González-Chavira, M.M.; Guevara-González, R.G.; Medina-Ramos, G. Effect of Fragmented DNA From Plant Pathogens on the Protection Against Wilt and Root Rot of Capsicum annuum L. Plants. Front. Plant. Sci. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Le Mire, G.; Siah, A.; Marolleau, B.; Gaucher, M.; Maumené, C.; Brostaux, Y.; Massart, S.; Brisset, M.N.; Haissam Jijakli, M. Evaluation of l-carrageenan, CpG-ODN, glycine betaine, spirulina platensis, and ergosterol as elicitors for control of zymoseptoria tritici in wheat. Phytopathology 2019, 109, 409–417. [Google Scholar] [CrossRef] [Green Version]
- Hadwiger, L.A.; Druffel, K.; Humann, J.L.; Schroeder, B.K. Nuclease released by Verticillium dahliae is a signal for non-host resistance. Plant. Sci. 2013, 201–202, 98–107. [Google Scholar] [CrossRef]
- Ji, Y.; Li, J.; Qin, Z.; Li, A.; Gu, Z.; Liu, X.; Lin, L.; Zhou, Y. Contribution of nuclease to the pathogenesis of aeromonas hydrophila. Virulence 2015, 6, 515–522. [Google Scholar] [CrossRef] [Green Version]
- Klosterman, S.J.; Chen, J.; Choi, J.J.; Chinn, E.E.; Hadwiger, L.A. Characterization of a 20 kDa DNase elicitor from Fusarium solani f. sp. phaseoli and its expression at the onset of induced resistance in Pisum sativum. Mol. Plant. Pathol. 2001, 2, 147–158. [Google Scholar] [CrossRef]
- Park, H.J.; Wang, W.; Curlango-Rivera, G.; Xiong, Z.; Lin, Z.; Huskey, D.A.; Hawes, M.C.; Vanetten, H.D.; Turgeon, B.G. A dnase from a fungal phytopathogen is a virulence factor likely deployed as counter defense against host-secreted extracellular DNA. MBio 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, T.M.; MacIntyre, A.; Hawes, M.; Allen, C. Escaping Underground Nets: Extracellular DNases Degrade Plant Extracellular Traps and Contribute to Virulence of the Plant Pathogenic Bacterium Ralstonia solanacearum. PLoS Pathog. 2016, 12, 26. [Google Scholar] [CrossRef]
- Huang, H.J.; Cui, J.R.; Xia, X.; Chen, J.; Ye, Y.X.; Zhang, C.X.; Hong, X.Y. Salivary DNase II from Laodelphax striatellus acts as an effector that suppresses plant defence. New Phytol. 2019, 224, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; White, G.J.; Vanetten, H.D.; Xiong, Z.; Hawes, M.C. Extracellular DNA is required for root tip resistance to fungal infection. Plant. Physiol. 2009, 151, 820–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krieg, A.M.; Yi, A.K.; Matson, S.; Waldschmidt, T.J.; Bishop, G.A.; Teasdale, R.; Koretzky, G.A.; Klinman, D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 1995, 374, 546–549. [Google Scholar] [CrossRef]
- Nehete, P.N.; Williams, L.E.; Chitta, S.; Nehete, B.P.; Patel, A.G.; Ramani, M.D.; Wisniewski, T.; Scholtzova, H. Class C CpG Oligodeoxynucleotide Immunomodulatory Response in Aged Squirrel Monkey (Saimiri Boliviensis Boliviensis). Front. Aging Neurosci. 2020, 12, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Nigar, S.; Shimosato, T. Cooperation of Oligodeoxynucleotides and Synthetic Molecules as Enhanced Immune Modulators. Front. Nutr. 2019, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Van Butselaar, T.; Van den Ackerveken, G. Salicylic Acid Steers the Growth–Immunity Tradeoff. Trends Plant. Sci. 2020, 25, 566–576. [Google Scholar] [CrossRef]
- Wang, D.; Pajerowska-Mukhtar, K.; Culler, A.H.; Dong, X. Salicylic Acid Inhibits Pathogen Growth in Plants through Repression of the Auxin Signaling Pathway. Curr. Biol. 2007, 17, 1784–1790. [Google Scholar] [CrossRef] [Green Version]
- Armengot, L.; Caldarella, E.; Marquès-Bueno, M.M.; Martínez, M.C. The protein kinase CK2 mediates cross-talk between auxin-and salicylic acid-signaling pathways in the regulation of PINOID transcription. PLoS ONE 2016, 11, 15. [Google Scholar] [CrossRef] [Green Version]
- Pajerowska-Mukhtar, K.M.; Wang, W.; Tada, Y.; Oka, N.; Tucker, C.L.; Fonseca, J.P.; Dong, X. The HSF-like Transcription Factor TBF1 Is a Major Molecular Switch for Plant Growth-to-Defense Transition. Curr. Biol. 2010, 22, 103–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carella, P.; Wilson, D.C.; Cameron, R.K. Some things get better with age: Differences in salicylic acid accumulation and defense signaling in young and mature Arabidopsis. Front. Plant. Sci. 2015, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, F.; Casadesús, A.; Brockman, H.; Munné-Bosch, S. An overview of plant-based natural biostimulants for sustainable horticulture with a particular focus on moringa leaf extracts. Plant. Sci. 2020, 295, 110194. [Google Scholar] [CrossRef] [PubMed]
- Kotchoni, S.O.; Gachomo, E.W. A rapid and hazardous reagent free protocol for genomic DNA extraction suitable for genetic studies in plants. Mol. Biol. Rep. 2009, 36, 1633–1636. [Google Scholar] [CrossRef]
- Barbosa, C.; Nogueira, S.; Gadanho, M.; Chaves, S. DNA extraction: Finding the Most Suitable Method. Molecular Microbial Diagnostic Methods: Pathways to Implementation for the Food and Water Industries; Elsevier Inc.: Amsterdam, The Netherlands, 2016; pp. 135–154. [Google Scholar]
- Islam, M.S.; Aryasomayajula, A.; Selvaganapathy, P.R. A review on macroscale and microscale cell lysis methods. Micromachines 2017, 8, 83. [Google Scholar] [CrossRef]
- Harrison, S.T.L. Bacterial cell disruption: A key unit operation in the recovery of intracellular products. Biotechnol. Adv. 1991, 9, 217–240. [Google Scholar] [CrossRef]
- Karakousis, A.; Tan, L.; Ellis, D.; Alexiou, H.; Wormald, P.J. An assessment of the efficiency of fungal DNA extraction methods for maximizing the detection of medically important fungi using PCR. J. Microbiol. Methods 2006, 65, 38–48. [Google Scholar] [CrossRef]
- Harju, S.; Fedosyuk, H.; Peterson, K.R. Rapid isolation of yeast genomic DNA: Bust n’ Grab. BMC Biotechnol. 2004, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, P.; Venâncio, A.; Lima, N. Toxic reagents and expensive equipment: Are they really necessary for the extraction of good quality fungal DNA? Lett. Appl. Microbiol. 2018, 66, 32–37. [Google Scholar] [CrossRef] [Green Version]
- Chapman, J.; Ismail, A.E.; Dinu, C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts 2018, 8, 238. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Jang, S.H.; Oh, S.O.; Kim, J.A.; Hur, J.S. An easy, rapid, and cost-effective method for DNA extraction from various lichen taxa and specimens suitable for analysis of fungal and algal strains. Mycobiology 2014, 42, 311–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mamidala, P.; Nanna, R.S. A Simple and Rapid Method for Dna Extraction From Leaves of Tomato, Tobacco and Rape Seed. J. Phytol. 2009, 1, 388–390. [Google Scholar]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant. Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Brandfass, C.; Karlovsky, P. Upscaled CTAB-Based DNA Extraction and Real-Time PCR Assays for Fusarium culmorum and F. graminearum DNA in Plant Material with Reduced Sampling Error. Int. J. Mol. Sci. 2008, 9, 2306–2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Saha, A.; Neelavar Ananthram, A. Comparison of DNA extraction methods for non-marine molluscs: Is modified CTAB DNA extraction method more efficient than DNA extraction kits? 3 Biotech. 2020, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Turaki, A.A.; Ahmad, B.; Magaji, U.F.; Abdulrazak, U.K.; Yusuf, B.A.; Hamza, A.B. Optimised cetyltrimethylammonium bromide (CTAB) DNA extraction method of plant leaf with high polysaccharide and polyphenolic compounds for downstream reliable molecular analyses. Afr. J. Biotechnol. 2017, 16, 1354–1365. [Google Scholar] [CrossRef] [Green Version]
- Kaczerewska, O.; Martins, R.; Figueiredo, J.; Loureiro, S.; Tedim, J. Environmental behaviour and ecotoxicity of cationic surfactants towards marine organisms. J. Hazard. Mater. 2020, 392, 122299. [Google Scholar] [CrossRef]
- Isomaa, B.; Reuter, J.; Djupsund, B.M. The subacute and chronic toxicity of cetyltrimethylammonium bromide (CTAB), a cationic surfactant, in the rat. Arch. Toxicol. 1976, 35, 91–96. [Google Scholar] [CrossRef]
- Beattie, P.J. Encyclopedia of Toxicology; Elsevier: Amstedam, The Netherlands, 2005; Volume 2, pp. 32–33. [Google Scholar]
- Piccinno, F.; Hischier, R.; Seeger, S.; Som, C. From Laboratory to Industrial Scale: A Scale-up Framework for Chemical Processes in Life Cycle Assessment Studies. J. Clean Prod. 2016, 135, 1085–1097. [Google Scholar] [CrossRef]
- Pane, C.; Celano, G.; Piccolo, A.; Villecco, D.; Spaccini, R.; Palese, A.M.; Zaccardelli, M. Effects of on-farm composted tomato residues on soil biological activity and yields in a tomato cropping system. Chem. Biol. Technol. Agric. 2015, 2, 4. [Google Scholar] [CrossRef] [Green Version]
- de Koning, A.N.M. Growth of a Tomato Crop. Acta Hortic. 1993, 328, 141–146. [Google Scholar] [CrossRef]
- Heuvelink, E. Tomatoes; CAB International: Wallingford, UK, 2005. [Google Scholar] [CrossRef]
- Mann, T.L.; Krull, U.J. The application of ultrasound as a rapid method to provide DNA fragments suitable for detection by DNA biosensors. Biosens. Bioelectron. 2004, 20, 945–955. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Hernández, M.C.; Parola-Contreras, I.; Montoya-Gómez, L.M.; Torres-Pacheco, I.; Schwarz, D.; Guevara-González, R.G. Eustressors: Chemical and physical stress factors used to enhance vegetables production. Sci. Hortic. 2019, 250, 223–229. [Google Scholar] [CrossRef]
- Tabrika, I.; Azim, K.; Mayad, E.H.; Zaafrani, M. Composting of tomato plant residues: Improvement of composting process and compost quality by integration of sheep manure. Org. Agric. 2020, 10, 229–242. [Google Scholar] [CrossRef]
- Salleh, M.A.M.; Mahmoud, D.K.; Karim, W.A.W.A.; Idris, A. Cationic and anionic dye adsorption by agricultural solid wastes: A comprehensive review. Desalination 2011, 280, 1–13. [Google Scholar] [CrossRef]
- FAOSTAT. Database on Agriculture. Food and Agriculture Organization of the United Nations; Food and Agriculture Organization of the United Nations: Rome, Italy, 2021; Available online: http://www.fao.org/faostat/en/#home (accessed on 31 August 2021).
- Samadi, H.S.; Ghobadian, B.; Nosrati, M. Prediction and estimation of biomass energy from agricultural residues using air gasification technology in Iran. Renew. Energy 2018, 149, 1077–1091. [Google Scholar] [CrossRef]
- Scarlat, N.; Martinov, M.; Dallemand, J.F. Assessment of the availability of agricultural crop residues in the European Union: Potential and limitations for bioenergy use. Waste Manag. 2010, 30, 1889–1897. [Google Scholar] [CrossRef]
- CCA. Caracterización y Gestión de los Residuos Orgánicos en América del Norte; Comisión Para la Cooperación Ambiental: Montréal, QC, Canada, 2017. [Google Scholar]
- Rouphael, Y.; Colla, G. Biostimulants in Agriculture. Front. Plant. Sci. 2020, 11, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Chen, F.; Wang, X.; Rajapakse, N.C. Effect of methyl jasmonate on secondary metabolites of sweet basil (Ocimum basilicum L.). J. Agric. Food Chem. 2006, 54, 2327–2332. [Google Scholar] [CrossRef]
- Mejía-Teniente, L.; Duran-Flores, F.d.D.; Chapa-Oliver, A.M.; Torres-Pacheco, I.; Cruz-Hernández, A.; González-Chavira, M.M.; Ocampo-Velázquez, R.V.; Guevara-González, R.G. Oxidative and molecular responses in capsicum annuum L. after hydrogen peroxide, salicylic acid and chitosan foliar applications. Int. J. Mol. Sci. 2013, 14, 10178–10196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mejía-Teniente, L.; Durán-Flores, B.A.; Torres-Pacheco, I.; González-Chavira, M.M.; Rivera-Bustamante, R.F.; Feregrino-Perez, A.A.; Pérez-Ramírez, I.; Rocha-Guzmán, N.E.; Reynoso-Camacho, R.; Guevara-González, R.G. Hydrogen peroxide protects pepper (Capsicum annuum L.) against pepper golden mosaic geminivirus (PepGMV) infections. Physiol. Mol. Plant. Pathol. 2019, 106, 23–29. [Google Scholar] [CrossRef]
- Ortega-Ortiz, H.; Benavides-Mendoza, A.; Mendoza-Villarreal, R.; Ramírez-Rodríguez, H.; Romenus, K.D.A. Enzymatic Activity in Tomato Fruits as a Response to Chemical Elicitors. J. Mex. Chem. Soc. 2007, 51, 141–144. [Google Scholar]
- Zehra, A.; Meena, M.; Dubey, M.K.; Aamir, M.; Upadhyay, R.S. Synergistic effects of plant defense elicitors and Trichoderma harzianum on enhanced induction of antioxidant defense system in tomato against Fusarium wilt disease. Bot. Stud. 2017, 58, 44. [Google Scholar] [CrossRef] [PubMed]
- Agathokleous, E.; Kitao, M.; Calabrese, E.J. Hormesis: A Compelling Platform for Sophisticated Plant Science. Trends Plant. Sci. 2019, 24, 318–327. [Google Scholar] [CrossRef]
- Agathokleous, E.; Calabrese, E.J. Hormesis can enhance agricultural sustainability in a changing world. Global. Food Secur. 2019, 20, 150–155. [Google Scholar] [CrossRef]
| TLR | Activation |
|---|---|
| TLR7 | Respond to bacterial and viral single-stranded RNA (ssRNA), and are also activated by imidazoquinolines and other small synthetic immunomodulatory compounds |
| TLR8 | |
| TLR9 | Activated by DNA of viruses or bacteria with unmethylated CpG dinucleotides. |
| Reference | Plant | Source of DNA | Concentration (ppm) | Effect |
|---|---|---|---|---|
| [67] | Arabidopsis thaliana | Escherichia coli | 500 | H2O2 induction and growth inhibition, callose deposition, induced expression of FRK1 |
| [45] | Acanthus mollis | Acanthus mollis | 2 | No effect |
| 20, 200 | Reduction in root growth | |||
| Arabidopsis thaliana, Quercus ilex, Sarcophaga carnaria | 200 | No effect | ||
| [51] | Phaseolus lunatus | S. littoralis oral secretions and larvae, Zea mays | 200 | No effect |
| Zea mays | S. littoralis oral secretions and larvae, Phaseolus lunatus | 200 | No effect | |
| Zea mays | 2 | No effect | ||
| 12, 90, 120 | Increase in plasma membrane potential depolarization | |||
| 200 | Increase in plasma membrane potential depolarization and Ca2+ | |||
| Phaseolus lunatus | Phaseolus lunatus | 2, 20, 90, 120 | Increase in plasma membrane potential depolarization | |
| 200 | Increase in plasma membrane potential depolarization and Ca2+ | |||
| [52] | Phaseolus vulgaris | Phaseolus vulgaris | 2, 20 | No effect |
| 50, 100, 150, 250 | Root growth inhibition | |||
| 200 | Root growth inhibition, H2O2 increase, activation of MAPKs, induction of extrafloral nectar, lower infection rates by P. syringae. | |||
| Phaseolus lunatus | 200 | Root growth inhibition, activation of MAPKs, lower infection rates by P. syringae. | ||
| Acacia farnesiana | 200 | Lower infection rates by P. syringae. | ||
| [46] | Lactuca sativa | Lactuca sativa | 2 | Root growth inhibition, genome methylation reduction, induced expression of sod and cat |
| 20 | Root growth inhibition, genome methylation reduction, induced expression of sod, cat and pal | |||
| 50, 100, 150 | Root growth inhibition, genome methylation reduction, induced expression of pal | |||
| 200 | Root growth inhibition, induced expression of sod, cat, and pal | |||
| Acaciella angustissima | 2, 20, 50, 100, 150 | No effect | ||
| 200 | Genome methylation reduction, induced expression of pal | |||
| Capsicum chinense | 2 | No effect | ||
| 20, 50 | Inhibited root growth | |||
| 100, 150 | Inhibited germination and root growth | |||
| 200 | Inhibited germination, genome methylation, induced expression of sod and cat | |||
| [68] | Capsicum annum | P. capsici, F. oxysporum and R. solani mixed | 20, 60, 100 | Resistance to pathogens and increase of total phenols and flavonoids |
| [53] | Arabidopsis thaliana | Arabidopsis thaliana | 150 | MPKs, ROS and Ca2+ signalling, SA and JA related genes expression upregulation, increase in H2O2 and callose accumulation, resistance against pathogens |
| Brassica oleracea | 150 | Upregulation of MPK3, OXI1, and CML37 gene expression | ||
| C. aurantrum, Solanum lycopersicum, S. oleraceae | 150 | No effect | ||
| Phaseolus vulgaris | 150 | Upregulation of MPK3, OXI1, and CML37 gene expression | ||
| Zea mays | 150 | Upregulation of MPK3 and OXI1 genes | ||
| [47] | Arabidopsis thaliana | Arabidopsis thaliana | 200 | Differential expression of less than 2.5% of total genes (upregulation: brassinosteroids and cytokinins, downregulation: abscisic acid and gibberellins) |
| Clupea harengus | 200 | Differential expression of more than 15% of total genes (upregulation: salicylic acid, downregulation: abscisic acid and auxins) | ||
| [60] | Solanum lycopersicum | Solanum lycopersicum | 50 | Plasma transmembrane potential depolarization, ligand-gated K+ channels, and H2O2 production activationPlasma activation |
| 100 | ||||
| 200 | Plasma transmembrane potential depolarization, ligand-gated K+ channels, and H2O2 production activation, downregulation: Myo-inositol, NO, ROS, cell wall, JA and sucrose biosynthetic and metabolic process, upregulation: oxygen transport, defence responses to gram-negative bacteria, lactate and adenine biosynthetic process, auxin influx |
| Technique | Advantages | Disadvantages |
|---|---|---|
| Mechanical | Several devices are already commercially available at an industrial scale [88]. Suitable for several kind of tissues with high efficiency [89]. | Production of small cell debris so next purification steps become harder [88]. High capital investment and energy costs [89]. |
| Physical | It has shown high efficiency [90,91]. | Some methods are expensive and so it is not widely used for macroscale application [88]. |
| Chemical | Use effective buffers that also protects DNA [90]. | Must be coupled with other techniques [90]. Some chemicals are toxic and need special waste disposition [92]. |
| Biological | High specificity. Use of enzymatical products to lyse cell wall and membrane components [90]. | Must be coupled with other techniques [90]. Depending on the needed enzymes it could be expensive in bigger scales [89,93]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carbajal-Valenzuela, I.A.; Medina-Ramos, G.; Caicedo-Lopez, L.H.; Jiménez-Hernández, A.; Ortega-Torres, A.E.; Contreras-Medina, L.M.; Torres-Pacheco, I.; Guevara-González, R.G. Extracellular DNA: Insight of a Signal Molecule in Crop Protection. Biology 2021, 10, 1022. https://doi.org/10.3390/biology10101022
Carbajal-Valenzuela IA, Medina-Ramos G, Caicedo-Lopez LH, Jiménez-Hernández A, Ortega-Torres AE, Contreras-Medina LM, Torres-Pacheco I, Guevara-González RG. Extracellular DNA: Insight of a Signal Molecule in Crop Protection. Biology. 2021; 10(10):1022. https://doi.org/10.3390/biology10101022
Chicago/Turabian StyleCarbajal-Valenzuela, Ireri Alejandra, Gabriela Medina-Ramos, Laura Helena Caicedo-Lopez, Alejandra Jiménez-Hernández, Adrian Esteban Ortega-Torres, Luis Miguel Contreras-Medina, Irineo Torres-Pacheco, and Ramón Gerardo Guevara-González. 2021. "Extracellular DNA: Insight of a Signal Molecule in Crop Protection" Biology 10, no. 10: 1022. https://doi.org/10.3390/biology10101022
APA StyleCarbajal-Valenzuela, I. A., Medina-Ramos, G., Caicedo-Lopez, L. H., Jiménez-Hernández, A., Ortega-Torres, A. E., Contreras-Medina, L. M., Torres-Pacheco, I., & Guevara-González, R. G. (2021). Extracellular DNA: Insight of a Signal Molecule in Crop Protection. Biology, 10(10), 1022. https://doi.org/10.3390/biology10101022








