Identifying Complex lncRNA/Pseudogene–miRNA–mRNA Crosstalk in Hormone-Dependent Cancers
Abstract
:Simple Summary
Abstract
1. Introduction
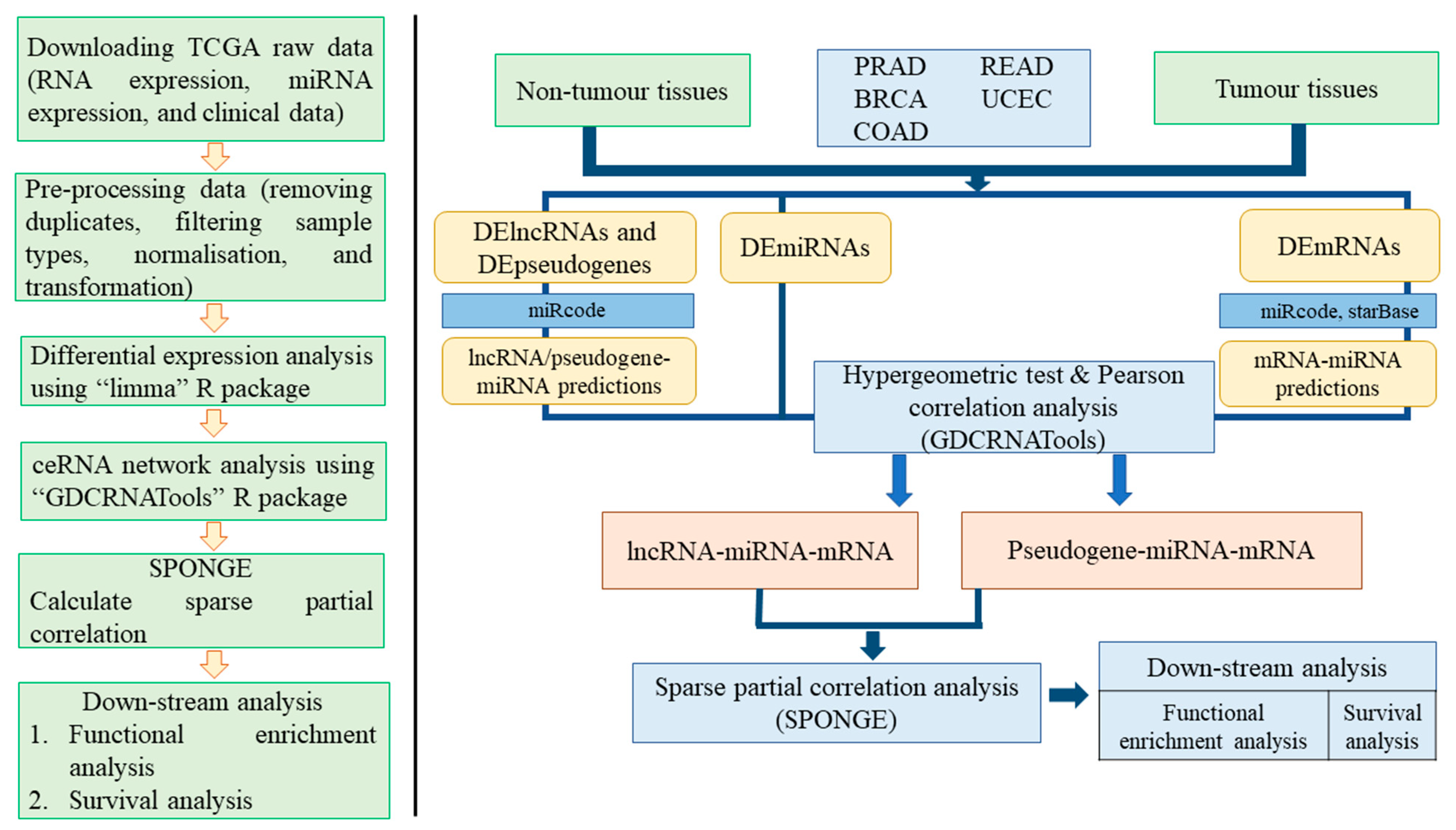
2. Materials and Methods
2.1. Ethics Statement
2.2. Patients and Samples
2.3. Differential Expression Analysis of Hormone-Dependent Cancer Data
2.4. Competing Endogenous RNA Network Analysis
2.5. Long Non-Coding RNA/Pseudogene–mRNA–microRNA Networks
2.6. Functional Enrichment Analysis
2.7. Survival Analysis
3. Results
3.1. Differential Expression Analysis Results
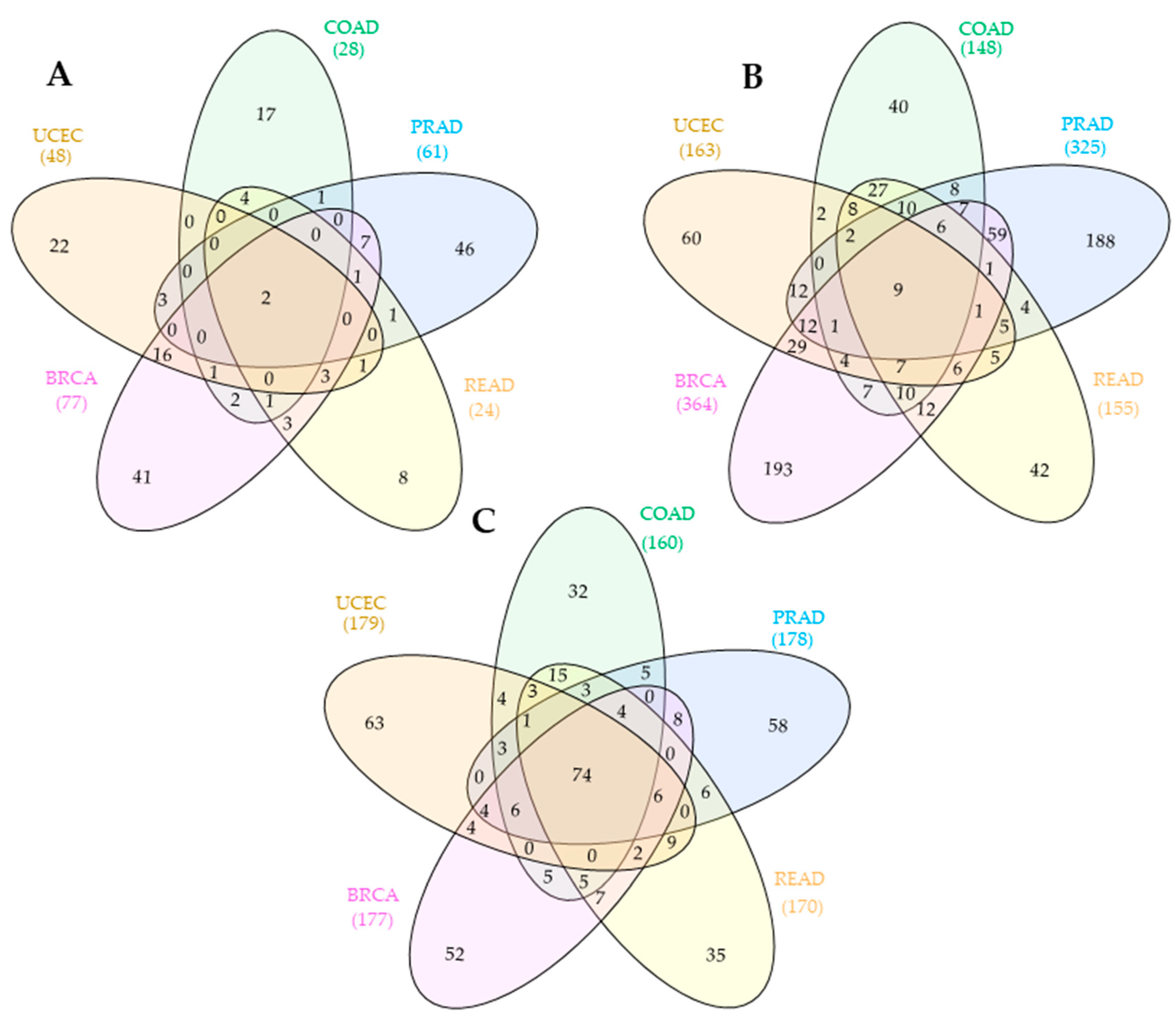
3.2. Shared Competing Endogenous RNA Networks across Hormone-Dependent Cancers
3.3. Functional Enrichment Analysis
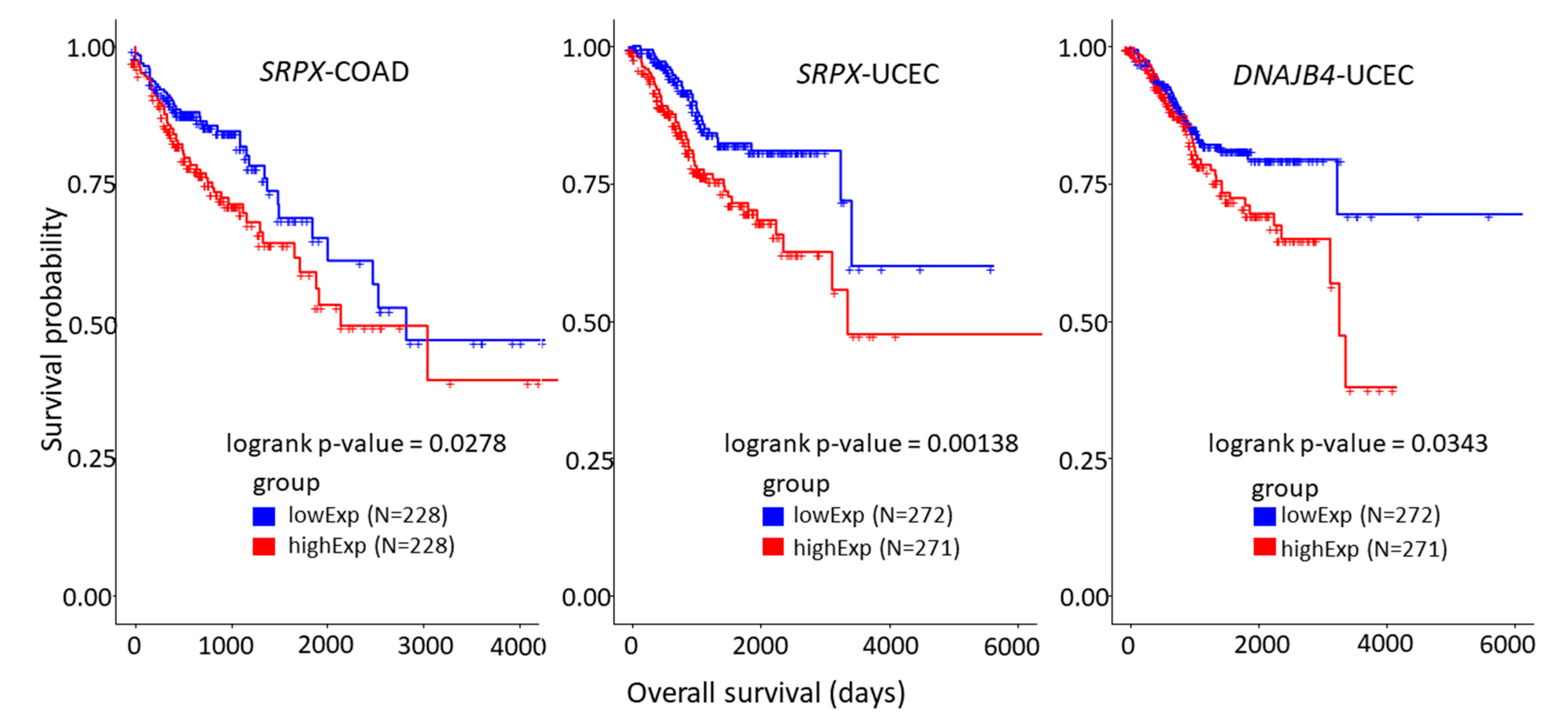
3.4. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Liu, H.; Tang, Y.; Wei, Y.; Wei, W.; Zhang, L.; Chen, J. The development and controversy of competitive endogenous RNA hypothesis in non-coding genes. Mol. Cell. Biochem. 2021, 476, 109–123. [Google Scholar] [CrossRef]
- Wei, Y.; Chang, Z.; Wu, C.; Zhu, Y.; Li, K.; Xu, Y. Identification of potential cancer-related pseudogenes in lung adenocarcinoma based on ceRNA hypothesis. Oncotarget 2017, 8, 59036–59047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Hou, J.; He, D.; Sun, M.; Zhang, P.; Yu, Y.; Chen, Y. The Emerging Function and Mechanism of ceRNAs in Cancer. Trends. Genet. 2016, 32, 211–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Yang, G.; Kang, Y.; Li, S.; Duan, R.; Shen, L.; Jiang, W.; Qian, B.; Yin, Z.; Liang, T. Construction and Analysis of a ceRNA Network Reveals Potential Prognostic Markers in Colorectal Cancer. Front. Genet. 2020, 11, 418. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, T.; Sun, X.; Wang, Y. Identification of a potential prognostic lncRNA-miRNA-mRNA signature in endometrial cancer based on the competing endogenous RNA network. J. Cell. Biochem. 2019, 120, 18845–18853. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, D.; Li, R.; Li, Y.; Zhu, X. Construction of a Competitive Endogenous RNA Network in Uterine Corpus Endometrial Carcinoma. Med. Sci. Monit. 2019, 25, 7998–8010. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Wu, Y.P.; Yin, H.B.; Xue, X.Y.; Gou, X. Molecular network-based identification of competing endogenous RNAs and mRNA signatures that predict survival in prostate cancer. J. Transl. Med. 2018, 16, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuersong, T.; Li, L.; Abulaiti, Z.; Feng, S. Comprehensive analysis of the aberrantly expressed lncRNA-associated ceRNA network in breast cancer. Mol. Med. Rep. 2019, 19, 4697–4710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiannaca, A.; Paglia, L.; Rosa, M.; Rizzo, R.; Urso, A. miRTissue (ce): Extending miRTissue web service with the analysis of ceRNA-ceRNA interactions. BMC Bioinform. 2020, 21, 199. [Google Scholar] [CrossRef]
- Wang, P.; Li, X.; Gao, Y.; Guo, Q.; Wang, Y.; Fang, Y.; Ma, X.; Zhi, H.; Zhou, D.; Shen, W.; et al. LncACTdb 2.0: An updated database of experimentally supported ceRNA interactions curated from low- and high-throughput experiments. Nucleic Acids Res. 2019, 47, D121–D127. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Ghosal, S.; Sen, R.; Chakrabarti, J. lnCeDB: Database of human long noncoding RNA acting as competing endogenous RNA. PLoS ONE 2014, 9, e98965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Qiao, Z.; Jiang, J.; Zhang, J. LINC00958 promotes endometrial cancer cell proliferation and metastasis by regulating the miR-145-3p/TCF4 axis. J. Gene. Med. 2021, 23, e3345. [Google Scholar] [CrossRef]
- Fang, Q.; Sang, L.; Du, S. Long noncoding RNA LINC00261 regulates endometrial carcinoma progression by modulating miRNA/FOXO1 expression. Cell. Biochem. Funct. 2018, 36, 323–330. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Qu, H.; Wang, S.; Wei, J.; Zhang, L.; Ma, R.; Lu, J.; Zhu, J.; Zhong, W.D.; Jia, Z. GDCRNATools: An R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics 2018, 34, 2515–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- List, M.; Dehghani Amirabad, A.; Kostka, D.; Schulz, M.H. Large-scale inference of competing endogenous RNA networks with sparse partial correlation. Bioinformatics 2019, 35, i596–i604. [Google Scholar] [CrossRef] [PubMed]
- Jeggari, A.; Marks, D.S.; Larsson, E. miRcode: A map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2012, 28, 2062–2063. [Google Scholar] [CrossRef] [PubMed]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paci, P.; Colombo, T.; Farina, L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 2014, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. miRCancer: A microRNA-cancer association database constructed by text mining on literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef]
- Kar, S.P.; Beesley, J.; Amin Al Olama, A.; Michailidou, K.; Tyrer, J.; Kote-Jarai, Z.; Lawrenson, K.; Lindstrom, S.; Ramus, S.J.; Thompson, D.J.; et al. Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov. 2016, 6, 1052–1067. [Google Scholar] [CrossRef] [Green Version]
- Kai-Xin, L.; Cheng, C.; Rui, L.; Zheng-Wei, S.; Wen-Wen, T.; Peng, X. Roles of lncRNA MAGI2-AS3 in human cancers. Biomed. Pharmacother. 2021, 141, 111812. [Google Scholar] [CrossRef]
- Du, S.; Hu, W.; Zhao, Y.; Zhou, H.; Wen, W.; Xu, M.; Zhao, P.; Liu, K. Long non-coding RNA MAGI2-AS3 inhibits breast cancer cell migration and invasion via sponging microRNA-374a. Cancer Biomark. 2019, 24, 269–277. [Google Scholar] [CrossRef]
- Ren, H.; Li, Z.; Tang, Z.; Li, J.; Lang, X. Long noncoding MAGI2-AS3 promotes colorectal cancer progression through regulating miR-3163/TMEM106B axis. J. Cell. Physiol. 2020, 235, 4824–4833. [Google Scholar] [CrossRef]
- Xiong, Y.; Wei, Y.; Gu, Y.; Zhang, S.; Lyu, J.; Zhang, B.; Chen, C.; Zhu, J.; Wang, Y.; Liu, H.; et al. DiseaseMeth version 2.0: A major expansion and update of the human disease methylation database. Nucleic Acids Res. 2017, 45, D888–D895. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, Q.; Wang, W.; Sun, S. MIR100HG: A credible prognostic biomarker and an oncogenic lncRNA in gastric cancer. Biosci. Rep. 2019, 39, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Yuan, F.; Zhang, X.; Chen, W.; Tang, X.; Lu, L. Elevated MIR100HG promotes colorectal cancer metastasis and is associated with poor prognosis. Oncol. Lett. 2019, 18, 6483–6490. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.Y.; Zhou, Z.Y.; Zhang, K.J.; Pang, J.; Wang, S.M. Long non-coding RNA MIR100HG promotes the migration, invasion and proliferation of triple-negative breast cancer cells by targeting the miR-5590-3p/OTX1 axis. Cancer Cell Int. 2020, 20, 508. [Google Scholar] [CrossRef]
- Tang, C.; Cai, Y.; Jiang, H.; Lv, Z.; Yang, C.; Xu, H.; Li, Z.; Li, Y. LncRNA MAGI2-AS3 inhibits bladder cancer progression by targeting the miR-31-5p/TNS1 axis. Aging 2020, 12, 25547–25563. [Google Scholar] [CrossRef]
- Werner, R.J.; Schultz, B.M.; Huhn, J.M.; Jelinek, J.; Madzo, J.; Engel, N. Sex chromosomes drive gene expression and regulatory dimorphisms in mouse embryonic stem cells. Biol. Sex. Differ. 2017, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Green, M.; Gao, G.; Santra, M.K.; Bhatnagar, S. SRPX for Treatment of Cancer. U.S. Patent, 9,290,744 B2, 22 March 2016. [Google Scholar]
- Acun, T.; Doberstein, N.; Habermann, J.K.; Gemoll, T.; Thorns, C.; Oztas, E.; Ried, T. HLJ1 (DNAJB4) Gene Is a Novel Biomarker Candidate in Breast Cancer. Omics 2017, 21, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Buttacavoli, M.; Di Cara, G.; D’Amico, C.; Geraci, F.; Pucci-Minafra, I.; Feo, S.; Cancemi, P. Prognostic and Functional Significant of Heat Shock Proteins (HSPs) in Breast Cancer Unveiled by Multi-Omics Approaches. Biology 2021, 10, 247. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Ji, P.; Jin, X. Application of the microRNA-302/367 cluster in cancer therapy. Cancer Sci. 2020, 111, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Stournaras, C.; Gravanis, A.; Margioris, A.N.; Lang, F. The actin cytoskeleton in rapid steroid hormone actions. Cytoskeleton 2014, 71, 285–293. [Google Scholar] [CrossRef] [PubMed]


| Cancer | lncRNA | Pseudogene | mRNA | miRNA | ||||
|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | |
| BRCA | 61 | 106 | 17 | 28 | 1125 | 1642 | 71 | 87 |
| COAD | 137 | 72 | 44 | 31 | 1200 | 1778 | 186 | 153 |
| PRAD | 139 | 49 | 28 | 18 | 434 | 1079 | 34 | 27 |
| READ | 181 | 53 | 52 | 18 | 1169 | 1790 | 165 | 114 |
| UCEC | 116 | 137 | 43 | 43 | 1584 | 2000 | 142 | 103 |
| lncRNA/Pseudogene | mRNA | List of MicroRNAs Associated with Each lncRNA/Pseudogene–mRNA Pair |
|---|---|---|
| MBNL1-AS1 (lncRNA) | DnaJ heat shock protein family (Hsp40) member B4 (DNAJB4) | hsa-miR-15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p |
| MAGI2-AS3* (lncRNA) | DNAJB4 * | hsa-miR-148a-3p, 152-3p, 148b-3p, 15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p, 194-5p, 204-5p, 211-5p |
| Fibroblast growth factor-2 * (FGF-2 *) | hsa-miR-15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p, 129-5p, 499a-5p | |
| Myosin light-chain kinase * (MYLK *) | hsa-miR-302a-3p, 302b-3p, 302c-3p, 302d-3p, 372-3p, 373-3p, 520e, 520a-3p, 520b, 520c-3p, 520d-3p, 302e, 200b-3p, 200c-3p, 429 | |
| Junctophilin-2 (JPH2) | hsa-miR-25-3p, 32-5p, 92a-3p, 363-3p, 367-3p, 92b-3p | |
| Cofilin-2 * (CFL2 *) | hsa-miR-212-3p, 132-3p, 302a-3p, 302b-3p, 302c-3p, 302d-3p, 372-3p, 373-3p, 520e, 520a-3p, 520b, 520c-3p, 520d-3p, 302e, 137, 141-3p, 200a-3p, 142-3p, 144-3p, 148a-3p, 152-3p, 148b-3p, 153-3p, 194-5p, 200b-3p, 200c-3p, 429, 23a-3p, 23b-3p, 25-3p, 32-5p, 92a-3p, 363-3p, 367-3p, 92b-3p, 425-5p | |
| Phospholipid scramblase 4 * (PLSCR4 *) | hsa-miR-302a-3p, 302b-3p, 302c-3p, 302d-3p, 372-3p, 373-3p, 520e, 520a-3p, 520b, 520c-3p, 520d-3p, 302e, 145-5p, 15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p | |
| Endothelin receptor type B (EDNRB) | hsa-miR-302a-3p, 302b-3p, 302c-3p, 302d-3p, 372-3p, 373-3p, 520e, 520a-3p, 520b, 520c-3p, 520d-3p, 302e | |
| Tensin 1 * (TNS1 *) | hsa-miR-302a-3p, 302b-3p, 302c-3p, 302d-3p, 372-3p, 373-3p, 520e, 520a-3p, 520b, 520c-3p, 520d-3p, 302e, 181a-5p, 181b-5p, 181c-5p, 181d-5p, 4262, 31-5p | |
| MIR100HG* (lncRNA) | FERM-domain-containing kindlin-2 * (FERMT2 *) | hsa-miR-130a-3p, 301a-3p, 130b-3p, 454-3p, 301b-3p, 4295, 3666, 135a-5p, 135b-5p, 138-5p, 15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p, 29a-3p, 29b-3p, 29c-3p, 103a-3p, 107 |
| DIX-domain-containing 1 * (DIXDC1 *) | hsa-miR-96-5p, 1271-5p, 143-3p, 145-5p, 155-5p, 15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p, 200b-3p, 200c-3p, 429, 29a-3p, 29b-3p, 29c-3p | |
| R-spondin 3 (RSPO3) | hsa-miR-15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p, 103a-3p, 107 | |
| DNAJB4 | hsa-miR-148a-3p, 152-3p, 148b-3p, 15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p, 204-5p, 211-5p, 103a-3p, 107 | |
| FGF2 | hsa-miR-15a-5p, 16-5p, 15b-5p, 195-5p, 424-5p, 497-5p, 103a-3p, 107, 129-5p | |
| Sushi repeat-containing protein X-linked * (SRPX *) | hsa-miR-130a-3p, 301a-3p, 130b-3p, 454-3p, 301b-3p, 19a-3p, 19b-3p | |
| JPH2 | hsa-miR-25-3p, 32-5p, 92a-3p, 363-3p, 367-3p, 92b-3p | |
| MEIS3P1 (pseudogene) | TNS1 | hsa-miR-138-5p, 138-1-5p, 145-5p, 204-5p, 204-3p, 211-5p, 219a-5p, 508-5p, 508-3p, 4782-3p, 23a-5p, 23b-5p, 34a-5p, 34b-5p, 449a, 449c-5p |
| KN motif and ankyrin repeat domains 2 (KANK2) | hsa-miR-138-5p, 138-1-5p, 145-5p, 204-5p, 204-3p, 211-5p, 219a-5p, 508-5p, 508-3p, 4782-3p, 34a-5p, 34b-5p, 449a, 449c-5p | |
| TUBAP5 (pseudogene) | MYB proto-oncogene-like 2 (MYBL2) | hsa-miR-130a-3p, 301a-5p, 301b-5p, 301b-3p, 454-5p, 721, 4295, 3666, 7-5p, 7-1-3p, 148a-3p, 152-5p, 15a-5p, 16-5p, 16-1-3p, 195-5p, 322, 424-5p, 497-3p, 1907, 214-5p, 761, 3619-5p, 22-5p, 22-3p, 122-5p, 122-3p, 1352, 24-3p, 24-1-5p, 24-2-5p, 29a-3p, 103a-3p, 107, 107ab, 124-5p, 124-3p, 506-5p, 338-5p, 338-3p |
| Cancer | miRNA (High/Low Expression Levels Associated with Survival) | Hazard Ratio | p-Value | Cancer | miRNA (High/Low Expression Levels Associated with Survival) | Hazard Ratio | p-Value |
|---|---|---|---|---|---|---|---|
| BRCA | hsa-miR-16-5p (high) 1,2,3,4 | 0.672 | 0.0136 | UCEC | hsa-miR-142-3p (high) 1,2,3,4,5 | 0.5634 | 0.0078 |
| BRCA | hsa-miR-181c-5p (high) 1,2,3,4,5 | 0.6578 | 0.0114 | UCEC | hsa-miR-148a-3p (high) 1,2,3,4,5 | 0.55 | 0.0055 |
| BRCA | hsa-miR-195-5p (high) 1,2,3,4,5 | 0.6859 | 0.0212 | UCEC | hsa-miR-152-3p (low) 1,2,3,4,5 | 1.6863 | 0.0156 |
| BRCA | hsa-miR-200c-3p (high) 1,2,3,4,5 | 0.7097 | 0.04 | UCEC | hsa-miR-212-3p (low) 1,2,3,4 | 1.7536 | 0.0096 |
| BRCA | hsa-miR-204-5p (high) 1,2,3,4,5 | 0.6294 | 0.0052 | UCEC | hsa-miR-25-3p (low) 2,3,4,5 | 1.5573 | 0.0365 |
| BRCA | hsa-miR-29a-3p (high) 1,2,3,4,5 | 0.7168 | 0.0429 | UCEC | hsa-miR-301a-3p (low) 2,3,4,5 | 1.8982 | 0.0032 |
| BRCA | hsa-miR-29c-3p (high) 1,3,4,5 | 0.6313 | 0.0061 | UCEC | hsa-miR-301b-3p (low) 1,2,5 | 1.6064 | 0.0277 |
| BRCA | hsa-miR-301b-3p (low) 1,2,5 | 1.3884 | 0.0478 | UCEC | hsa-miR-302a-3p (high) 1,2,3,4,5 | 0.5663 | 0.0071 |
| BRCA | hsa-miR-31-5p (high) 1,2,3,4 | 0.5542 | 0.0003 | UCEC | hsa-miR-302b-3p (high) 1,2,3,4,5 | 0.5608 | 0.0061 |
| BRCA | hsa-miR-363-3p (high) 2,3,4,5 | 0.6961 | 0.0279 | UCEC | hsa-miR-302c-3p (high) 1,2,3,4,5 | 0.5498 | 0.0049 |
| BRCA | hsa-miR-372-3p (low) 1,2,3,4 | 1.409 | 0.0392 | UCEC | hsa-miR-302d-3p (high) 1,2,3,4,5 | 0.5531 | 0.0053 |
| COAD | hsa-miR-1271-5p (low) 1,2,3,4,5 | 1.6083 | 0.0166 | UCEC | hsa-miR-302e (high) 1,2,3,4,5 | 0.4897 | 0.0008 |
| COAD | hsa-miR-130a-3p (low) 1,2,3,4,5 | 1.8346 | 0.0021 | UCEC | hsa-miR-367-3p (high) 1,2,3,4,5 | 0.5204 | 0.0021 |
| COAD | hsa-miR-145-5p (low) 1,2,3,4 | 1.5823 | 0.0214 | UCEC | hsa-miR-425-5p (low) 1,2,3,4 | 1.6045 | 0.0301 |
| COAD | hsa-miR-181b-5p (low) 1,2,5 | 1.5294 | 0.0326 | UCEC | hsa-miR-4262 (high) 1,2,3,4,5 | 0.4897 | 0.0008 |
| COAD | hsa-miR-32-5p (low) 1,2,3,4,5 | 1.5932 | 0.0213 | UCEC | hsa-miR-497-5p (high) 1,2,3,4 | 0.5285 | 0.0037 |
| COAD | hsa-miR-497-5p (low) 1,2,3,4 | 1.5895 | 0.0206 | UCEC | hsa-miR-520b (high) 1,2,3,4 | 0.5896 | 0.0129 |
| COAD | hsa-miR-96-5p (low) 1,2,3,4 | 1.4888 | 0.0474 | UCEC | hsa-miR-520c-3p (high) 1,2,3,4 | 0.5791 | 0.0099 |
| PRAD | hsa-miR-19a-3p (low) 1,2,3,4 | 6.9585 | 0.026 | UCEC | hsa-miR-520d-3p (high) 2,3,4 | 0.5043 | 0.0012 |
| PRAD | hsa-miR-29b-3p (high) 1,2,3,4 | 0.2343 | 0.0434 | UCEC | hsa-miR-520e (high) 2,3,4 | 0.626 | 0.0273 |
| READ | hsa-miR-155-5p (high) 1,2,3,4,5 | 0.4544 | 0.0426 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayarathna, D.K.; Rentería, M.E.; Sauret, E.; Batra, J.; Gandhi, N.S. Identifying Complex lncRNA/Pseudogene–miRNA–mRNA Crosstalk in Hormone-Dependent Cancers. Biology 2021, 10, 1014. https://doi.org/10.3390/biology10101014
Jayarathna DK, Rentería ME, Sauret E, Batra J, Gandhi NS. Identifying Complex lncRNA/Pseudogene–miRNA–mRNA Crosstalk in Hormone-Dependent Cancers. Biology. 2021; 10(10):1014. https://doi.org/10.3390/biology10101014
Chicago/Turabian StyleJayarathna, Dulari K., Miguel E. Rentería, Emilie Sauret, Jyotsna Batra, and Neha S. Gandhi. 2021. "Identifying Complex lncRNA/Pseudogene–miRNA–mRNA Crosstalk in Hormone-Dependent Cancers" Biology 10, no. 10: 1014. https://doi.org/10.3390/biology10101014






