Identification of Antimicrobial Peptide Genes in Black Rockfish Sebastes schlegelii and Their Responsive Mechanisms to Edwardsiella tarda Infection
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection
2.2. Libraries Construction and Sequencing
2.3. Genome Assembly and Genome Evaluation
2.4. Gene Prediction and Annotation
2.5. Genome Comparison
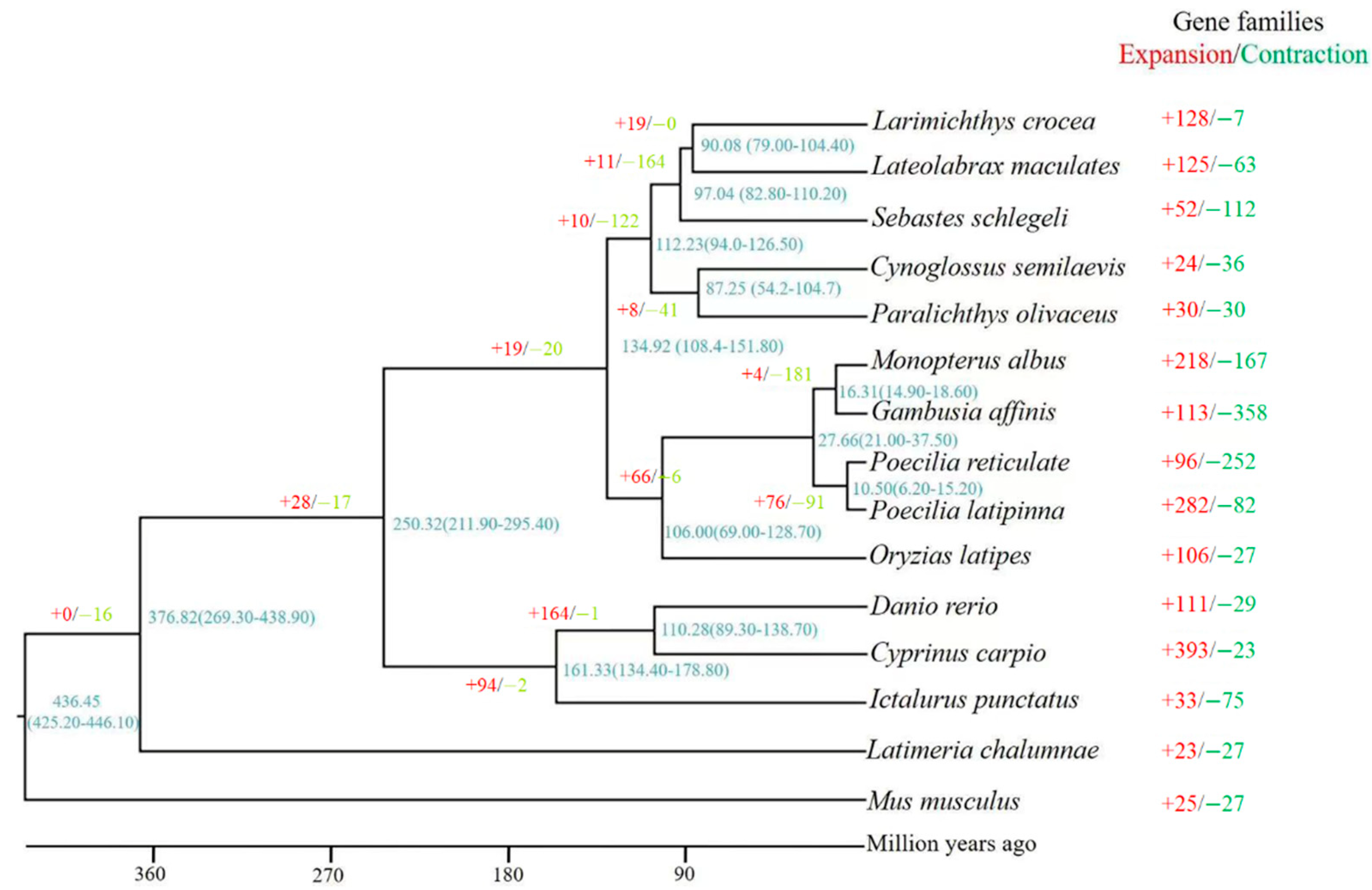
2.6. Phylogenetic Tree Reconstruction and Divergence Time Estimation
2.7. Identification and Analysis of AMPs
2.8. Healthy Tissues Extraction and Bacterial Challenge
2.9. Gene Expression Analysis of AMP Genes
2.10. Analysis of Hepcidins in Teleost
3. Results
3.1. Genome Sequencing and Assembly
3.2. Genome Prediction and Annotation
3.3. Phylogenetic Tree Construction and Divergence Time Estimation
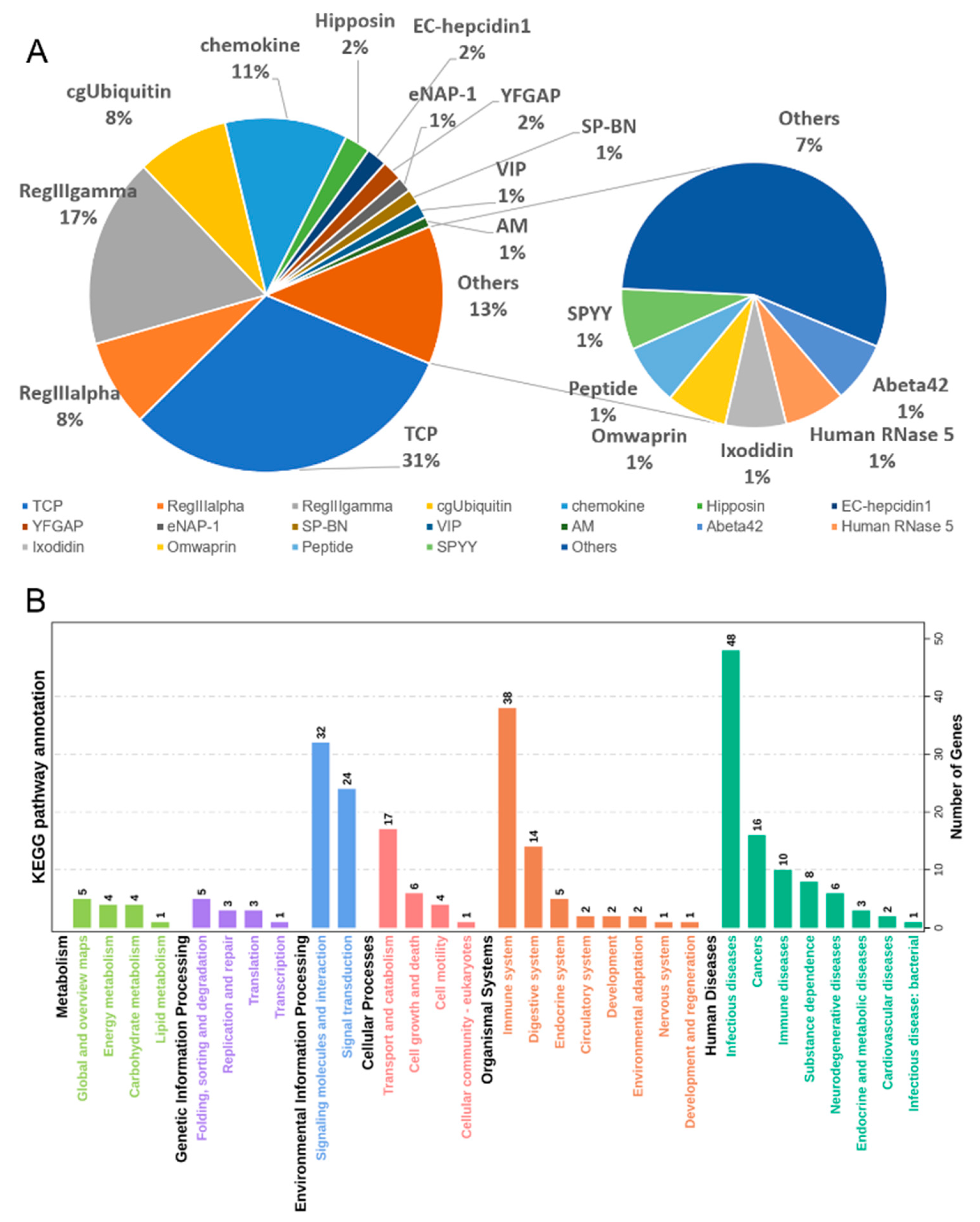
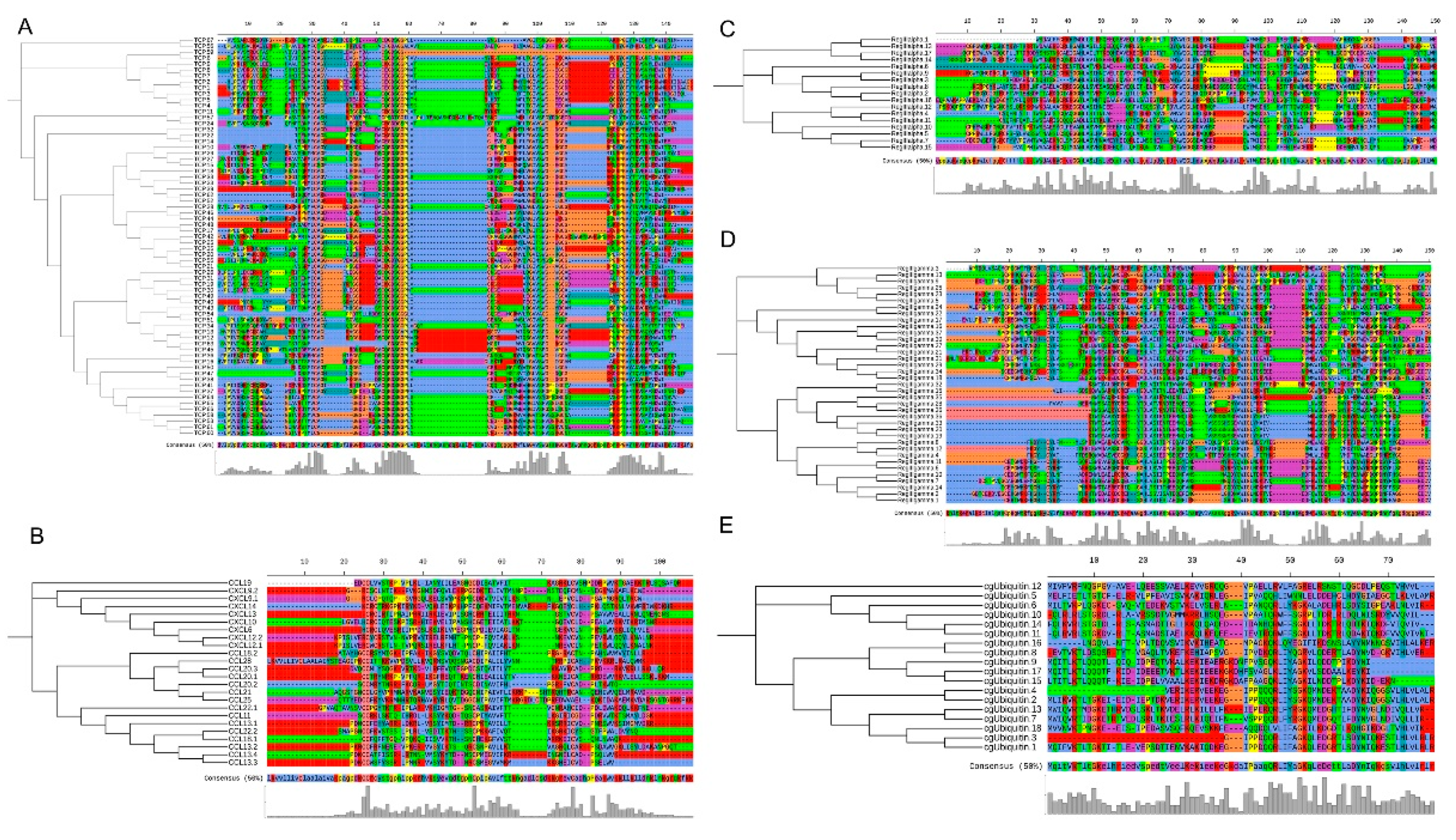
3.4. Antimicrobial Peptides (AMPs) Identification
3.5. Comparison of AMP Genes in Teleost
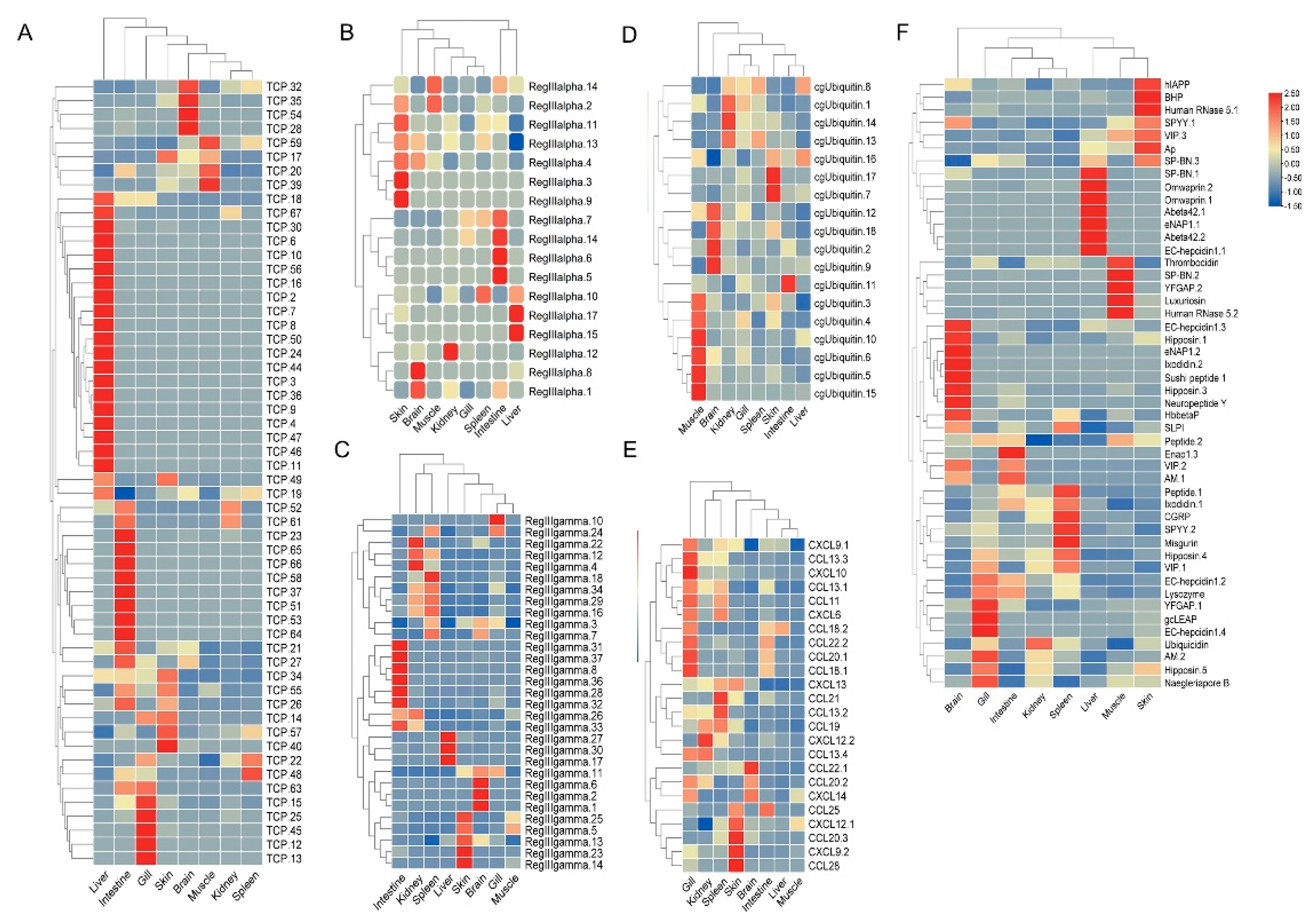
3.6. Expression Patterns of AMP Genes in Healthy Tissues
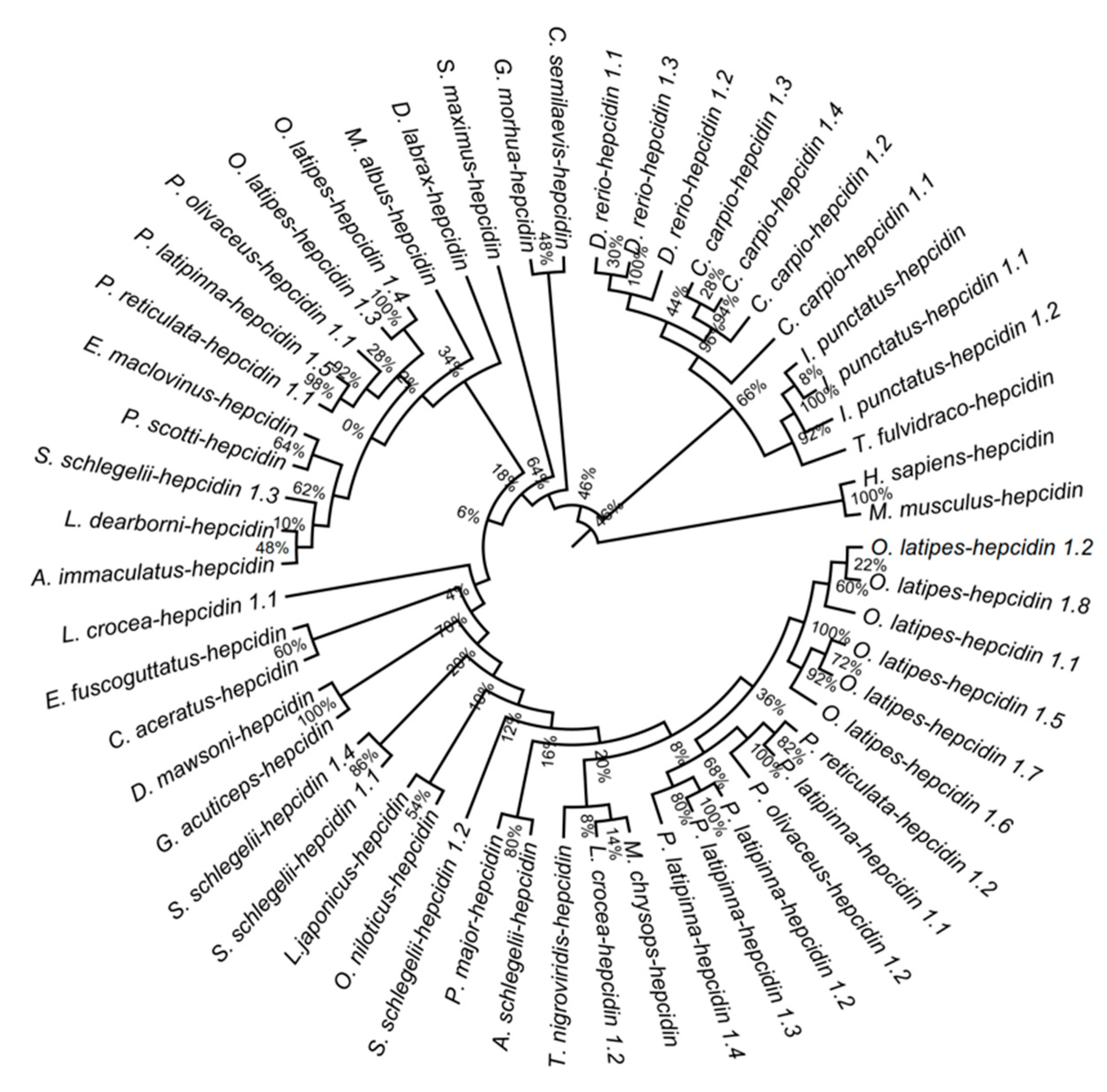
3.7. Phylogenetic Analysis of Hepcidins among Teleost
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, S.M.; Jeon, I.G.; Lee, J.Y. Effects of digestible protein and lipid levels in practical diets on growth, protein utilization and body composition of juvenile rockfish (Sebastes schlegeli). Aquaculture 2002, 211, 227–239. [Google Scholar] [CrossRef]
- Oh, S.Y.; Noh, C.H.; Kang, R.S.; Kim, C.K.; Cho, S.H.; Jo, J.Y. Compensatory growth and body composition of juvenile black rockfish Sebastes schlegeli following feed deprivation. Fish. Sci. 2008, 74, 846–852. [Google Scholar] [CrossRef]
- Cho, S.H.; Hur, S.B.; Jo, J.Y. Effect of enriched live feeds on survival and growth rates in larval Korean rockfish, Sebastes schlegeli Hilgendorf. Aquac. Res. 2001, 32, 199–208. [Google Scholar] [CrossRef]
- Mizanur, R.M.; Bai, S.C. The optimum feeding frequency in growing Korean rockfish (Sebastes schlegeli) rearing at the temperature of 15 °C and 19 °C. Asian-Australas J. Anim. Sci. 2014, 27, 1319–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehlert, G.W.; Yoklavich, M.M. Reproduction, embryonic energetics, and the maternal-fetal relationship in the viviparous genus Sebastes (Pisces: Scorpaenidae). Biol. Bull. 1984, 167, 354–370. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Lee, S.W.; Rha, S.J.; Yoon, H.S.; Park, E.S.; Han, K.H.; Kim, S.J. Dietary green tea extract improves growth performance, body composition, and stress recovery in the juvenile black rockfish, Sebastes schlegeli. Aquac. Int. 2013, 21, 525–538. [Google Scholar] [CrossRef]
- Kim, S.G.; Kang, J.C. Effect of dietary copper exposure on accumulation, growth and hematological parameters of the juvenile rockfish, Sebastes schlegeli. Mar. Environ. Res. 2004, 58, 65–82. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kang, Y.J.; Lee, S.M.; Kim, I.B. Protein requirements of the Korean rockfish Sebastes schlegeli. J. Aquac. 1993, 6, 13–27. [Google Scholar]
- Kang, S.H.; Shin, G.W.; Shin, Y.S.; Palaksha, K.J.; Kim, Y.R.; Yang, H.H.; Jung, T.S. Experimental evaluation of pathogenicity of Lactococcus garvieae in black rockfish (Sebastes schlegeli). J. Vet. Sci. 2004, 5, 387–390. [Google Scholar] [CrossRef]
- Kitamura, S.I.; Jung, S.J.; Kim, W.S.; Nishizawa, T.; Yoshimizu, M.; Oh, M.J. A new genotype of lymphocystivirus, LCDV-RF, from lymphocystis diseased rockfish. Arch. Virol. 2006, 151, 607–615. [Google Scholar] [CrossRef]
- Shin, K.W.; Kim, S.H.; Kim, J.H.; Hwang, S.D.; Kang, J.C. Toxic effects of ammonia exposure on growth performance, hematological parameters, and plasma components in rockfish, Sebastes schlegelii, during thermal stress. Fish. Aquat. Sci. 2016, 19, 44. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Kang, J.C. Toxic effects on bioaccumulation and hematological parameters of juvenile rockfish Sebastes schlegelii exposed to dietary lead (Pb) and ascorbic acid. Chemosphere 2017, 176, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Han, H.J.; Kim, D.Y.; Kim, W.S.; Kim, C.S.; Jung, S.J.; Oh, M.J.; Kim, D.H. Atypical Aeromonas salmonicida infection in the black rockfish, Sebastes schlegeli Hilgendorf, in Korea. J. Fish Dis. 2011, 34, 47–55. [Google Scholar] [CrossRef]
- Shin, G.; Lee, H.; Palaksha, K.J.; Kim, Y.; Lee, E.; Shin, Y.; Jung, T. Production of monoclonal antibodies against serum immunoglobulins of black rockfish (Sebastes schlegeli Higendorf). J. Vet. Sci. 2006, 7, 293–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.Y.; Jang, J.H.; Lee, J.W.; Cho, J.H. Molecular cloning and characterization of peptidoglycan recognition proteins from the rockfish, Sebastes schlegeli. Fish. Shellfish Immunol. 2010, 28, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Roopasingam, K.; Wan, Q.; Nilojan, J.; Lee, J. Identification and characterization of a calcium-dependent lily-type lectin from black rockfish (Sebastes schlegelii): Molecular antennas are involved in host defense via pathogen recognition. Dev. Comp. Immunol. 2018, 81, 54–62. [Google Scholar]
- Kwon, H.; Yang, H.; Lee, S.; Nilojan, J.; Bathige, S.D.N.K.; Wan, Q.; Lee, J. Characterization of a Kazal-type serine protease inhibitor from black rockfish Sebastes schlegelii and its possible role in hepatic immune response. Fish Shellfish Immunol. 2018, 74, 485–490. [Google Scholar] [CrossRef]
- Du, X.; Wang, G.H.; Su, Y.L.; Zhang, M.; Hu, Y.H. Black rockfish C-type lectin, SsCTL4: A pattern recognition receptor that promotes bactericidal activity and virus escape from host immune defense. Fish Shellfish Immunol. 2018, 79, 340–350. [Google Scholar]
- Li, K.; Wei, X.M.; Zhang, L.B.; Chi, H.; Yang, J.L. Raptor/mTORC1 acts as a modulatory center to regulate anti-bacterial immune response in rockfish. Front. Immunol. 2019, 10, 2953. [Google Scholar] [CrossRef] [Green Version]
- Shanaka, K.A.S.N.; Tharuka, M.D.N.; Priyathilaka, T.T.; Lee, J. Molecular characterization and expression analysis of rockfish (Sebastes schlegelii) viperin, and its ability to enervate RNA virus transcription and replication in vitro. Fish Shellfish Immunol. 2019, 92, 655–666. [Google Scholar] [CrossRef]
- Wang, J.J.; Meng, Z.Q.; Wang, G.H.; Fu, Q.; Zhang, M. A CCL25 chemokine functions as a chemoattractant and an immunomodulator in black rockfish, Sebastes schlegelii. Fish. Shellfish Immunol. 2020, 100, 161–170. [Google Scholar] [CrossRef]
- Wickramasinghe, P.D.S.U.; Kwon, H.; Elvitigala, D.A.S.; Wan, Q.; Lee, J. Identification and characterization of cystatin B from black rockfish, Sebastes schlegelii, indicating its potent immunological importance. Fish Shellfish Immunol. 2020, 104, 497–505. [Google Scholar] [CrossRef]
- Cao, M.; Yan, X.; Yang, N.; Fu, Q.; Xue, T.; Zhao, S.C.; Hu, J.; Li, Q.; Song, L.; Zhang, X.Y.; et al. Genome-wide characterization of Toll-like receptors in black rockfish Sebastes schlegelii: Evolution and response mechanisms following Edwardsiella tarda infection. Int. J. Biol. Macromol. 2020, 164, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Yan, X.; Li, Q.; Fu, Q.; Yang, N.; Song, L.; Li, C. Genome-wide identification and analysis of NOD-like receptors and their potential roles in response to Edwardsiella tarda infection in black rockfish (Sebastes schlegelii). Aquaculture 2021, 30, 736803. [Google Scholar] [CrossRef]
- Im, J.; Kim, W.R.; Lee, H.E.; Kim, A.; Kim, D.H.; Choi, Y.H.; Cha, H.J.; Kim, S.; Kim H, S. Expression analysis of LTR-derived miR-1269a and target gene, KSR2 in Sebastes schlegelii. Genes Genom. 2020, 42, 55–65. [Google Scholar] [CrossRef]
- Cao, M.; Yan, X.; Su, B.F.; Yang, N.; Fu, Q.; Xue, T.; Song, L.; Li, Q.; Li, C. Integrated Analysis of circRNA-miRNA-mRNA Regulatory networks in the intestine of Sebastes schlegelii following Edwardsiella tarda Challenge. Front. Immunol. 2021, 11, 3430. [Google Scholar] [CrossRef] [PubMed]
- Savoca, S.; Abbadi, M.; Toffan, A.; Salogni, C.; Iaria, C.; Capparucci, F.; Marino, F. Betanodavirus infection associated with larval enteropathy as a cause of mortality in cultured gilthead sea bream (Sparus aurata, Linnaeus, 1758). Aquaculture 2021, 541, 736844. [Google Scholar] [CrossRef]
- Gjurčević, E.; Kužir, S.; Žmak, L.; Obrovac, M.; Gudan Kurilj, A.; Savoca, S.; Matanović, K. A case of mycobacteriosis in farmed pikeperch (Sander lucioperca) cultured in a recirculating aquaculture system. Aquac. Res. 2020, 51, 4824–4827. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef] [Green Version]
- Lima, L.C.; Fernandes, A.A.; Costa, A.A.P.; Velasco, F.O.; Leite, R.C.; Hackett, J.L. Isolation and characterizaton of Edwardsiella tarda from pacu Myleus micans. Arq. Bras. De Med. Veterinária E Zootec. 2008, 60, 275–277. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A., Jr. Innate immune recognition and control of adaptive immune responses. Semin. Immunol. 1998, 10, 351–353. [Google Scholar] [CrossRef]
- Izadpanah, A.; Gallo, R.L. Antimicrobial peptides. J. Am. Acad. Dermatol. 2005, 52, 381–390. [Google Scholar] [CrossRef]
- Peters, B.M.; Shirtliff, M.E.; Jabra-Rizk, M.A. Antimicrobial peptides: Primeval molecules or future drugs? PLoS Pathog. 2010, 6, e1001067. [Google Scholar] [CrossRef]
- Cederlund, A.; Gudmundsson, G.H.; Agerberth, B. Antimicrobial peptides important in innate immunity. FEBS J. 2011, 278, 3942–3951. [Google Scholar] [CrossRef]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.; Mercier, C.; Koussounadis, A.; Secombes, C. Discovery of multiple beta-defensin like homologues in teleost fish. Mol. Immunol. 2007, 44, 638–647. [Google Scholar] [CrossRef]
- Cuesta, A.; Meseguer, J.; Esteban, M.A. Molecular and functional characterization of the gilthead seabream beta-defensin demonstrate its chemotactic and antimicrobial activity. Mol. Immunol. 2011, 48, 1432–1438. [Google Scholar] [CrossRef]
- Jin, J.Y.; Zhou, L.; Wang, Y.; Li, Z.; Zhao, J.G.; Zhang, Q.Y.; Gui, J.F. Antibacterial and antiviral roles of a fish beta-defensin expressed both in pituitary and testis. PLoS ONE 2010, 5, e12883. [Google Scholar] [CrossRef] [PubMed]
- Ruangsri, J.; Kitani, Y.; Kiron, V.; Lokesh, J.; Brinchmann, M.F.; Karlsen, B.O.; Fernandes, J.M. A novel beta-defensin antimicrobial peptide in Atlantic cod with stimulatory effect on phagocytic activity. PLoS ONE 2013, 8, e62302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Chang, X.; Wu, H.; Xiao, J.; Gao, Y.; Zhang, Y. Role of intestinal inflammation in predisposition of Edwardsiella tarda infection in zebrafish (Danio rerio). Fish Shellfish Immunol. 2014, 41, 271–278. [Google Scholar] [CrossRef]
- Katzenback, B.A. Antimicrobial Peptides as Mediators of innate immunity in teleosts. Biology 2015, 25, 607–639. [Google Scholar] [CrossRef] [Green Version]
- Dezfuli, B.S.; Pironi, F.; Giari, L.; Noga, E.J. Immunocytochemical localization of piscidin in mast cells of infected seabass gill. Fish Shellfish Immunol. 2010, 28, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Buonocore, F.; Randelli, E.; Casani, D.; Picchietti, S.; Belardinelli, M.C.; de Pascale, D.; De Santi, C.; Scapigliati, G. A piscidin-like antimicrobial peptide from the icefish Chionodraco hamatus (Perciformes: Channichthyidae): Molecular characterization, localization and bactericidal activity. Fish Shellfish Immunol. 2012, 33, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Shim, S.H.; Hwang, S.D.; Park, M.A.; Jee, B.Y.; An, C.M.; Kim, Y.O.; Kim, J.W.; Park, C.I. Expression analysis and biological activity of moronecidin from rock bream, Oplegnathus fasciatus. Fish Shellfish Immunol. 2014, 40, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.F.; Jin, Y.; Xu, X.; Qiao, Y.; Wu, Y.; Mao, Y.; Su, Y.Q.; Wang, J. Characterization of a novel piscidin-like antimicrobial peptide from Pseudosciaena crocea and its immune response to Cryptocaryon irritans. Fish Shellfish Immunol. 2013, 35, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Lauriano, E.R.; Capillo, G.; Icardo, J.M.; Fernandes, J.M.O.; Kiron, V.; Kuciel, M.; Zaccone, G. Neuroepithelial cells (NECs) and mucous cells express a variety of neurotransmitters and neurotransmitter receptors in the gill and respiratory air-sac of the catfish Heteropneustes fossilis (Siluriformes, Heteropneustidae): A possible role in local immune defence. Zoology 2021, 148, 125958. [Google Scholar]
- Capillo, G.; Zaccone, G.; Cupello, C.; Fernandes, J.M.O.; Viswanath, K.; Kuciel, M.; Lauriano, E.R. Expression of acetylcholine, its contribution to regulation of immune function and O2 sensing and phylogenetic interpretations of the African butterfly fish Pantodon buchholzi (Osteoglossiformes, Pantodontidae). Fish Shellfish Immunol. 2021, 111, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, H.; Zhang, X.; Luo, H.; Xue, X.; Li, Z.; Yao, B. Identification, expression and bioactivity of Paramisgurnus dabryanus β-defensin that might be involved in immune defense against bacterial infection. Fish Shellfish Immunol. 2013, 35, 399–406. [Google Scholar] [CrossRef]
- Guo, M.; Wei, J.; Huang, X.; Huang, Y.; Qin, Q. Antiviral effects of beta-defensin derived from orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2012, 32, 828–838. [Google Scholar] [CrossRef]
- Kim, Y.O.; Park, E.M.; Nam, B.H.; Kong, H.J.; Kim, W.J.; Lee, S.J. Identification and molecular characterization of two hepcidin genes from black rockfish (Sebastes schlegelii). Mol. Cell. Biochem. 2008, 315, 131. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; Tang, J. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Turner, S.W. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563. [Google Scholar] [CrossRef]
- Mostovoy, Y.; Levy-Sakin, M.; Lam, J.; Lam, E.T.; Hastie, A.R.; Marks, P.; Cao, H. A hybrid approach for de novo human genome sequence assembly and phasing. Nat. Methods 2016, 13, 587. [Google Scholar] [CrossRef] [Green Version]
- Parra, G.; Bradnam, K.; Korf, I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007, 23, 1061–1067. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Yamasaki, M.; Araki, K.; Maruyoshi, K.; Matsumoto, M.; Nakayasu, C.; Moritomo, T.; Yamamoto, A. Comparative analysis of adaptive immune response after vaccine trials using live attenuated and formalin-killed cells of Edwardsiella tarda in ginbuna crucian carp (Carassius auratus langsdorfii). Fish Shellfish Immunol. 2015, 45, 437–442. [Google Scholar] [CrossRef]
- Krause, A.; Neitz, S.; Mägert, H.J.; Schulz, A.; Forssmann, W.G.; Schulz-Knappe, P.; Adermann, K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000, 480, 147–150. [Google Scholar] [CrossRef] [Green Version]
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinsky, M.; Svardal, H.; Tyers, A.M.; Miska, E.A.; Genner, M.J.; Turner, G.F.; Durbin, R. Whole-genome sequences of Malawi cichlids reveal multiple radiations interconnected by gene flow. Nat. Ecol. Evol. 2018, 2, 1940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vialle, R.A.; de Souza, J.E.S.; Lopes, K.D.P.; Teixeira, D.G.; Alves Sobrinho, P.D.A.; Ribeiro-dos-Santos, A.M.; Hamoy, I.G. Whole genome sequencing of the Pirarucu (Arapaima gigas) supports independent emergence of major teleost clades. Genome Biol. Evol. 2018, 10, 2366–2379. [Google Scholar] [CrossRef] [Green Version]
- Lorin, T.; Brunet, F.G.; Laudet, V.; Volff, J.N. Teleost fish-specific preferential retention of pigmentation gene-containing families after whole genome duplications in vertebrates. G3 Genes Genomes Genet. 2018, 8, 1795–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Omori, Y.; Koren, S.; Shirokiya, T.; Kuroda, T.; Miyamoto, A.; Wolfsberg, T.G. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci. Adv. 2019, 5, eaav0547. [Google Scholar] [CrossRef] [Green Version]
- Ganz, T. The role of antimicrobial peptides in innate immunity. Integr. Comp. Biol. 2003, 43, 300–304. [Google Scholar] [CrossRef] [Green Version]
- Oren, Z.; Shai, Y. A class of highly potent antibacterial peptides derived from pardaxin a pore-forming peptide isolated from Moses sole fish Pardachirus marmoratus. Eur. J. Biochem. 1996, 237, 303–310. [Google Scholar] [CrossRef]
- Bian, C.; Li, J.; Lin, X.; Chen, X.; Yi, Y.; You, X.; Shi, Q. Whole genome sequencing of the blue tilapia (Oreochromis aureus) provides a valuable genetic resource for biomedical research on tilapias. Mar. Drugs 2019, 17, 386. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Chen, X.; Zhang, X.; Li, J.; Yi, Y.; Bian, C.; You, X. Whole genome sequencing of the giant grouper (Epinephelus lanceolatus) and high-throughput screening of putative antimicrobial peptide genes. Mar. Drugs 2019, 17, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalle, M.; Papareddy, P.; Kasetty, G.; Mörgelin, M.; van der Plas, M.J.; Rydengård, V.; Schmidtchen, A. Host defense peptides of thrombin modulate inflammation and coagulation in endotoxin-mediated shock and Pseudomonas aeruginosa sepsis. PLoS ONE 2012, 7, e51313. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Yu, Y.; Huang, H.; Feng, K.; Pan, M.; Yuan, S.; Chen, S. A short-form C-type lectin from amphioxus acts as a direct microbial killing protein via interaction with peptidoglycan and glucan. J. Immunol. 2007, 179, 8425–8434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weis, W.I.; Taylor, M.E.; Drickamer, K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998, 163, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Patrzykat, A.; Zhang, L.; Mendoza, V.; Iwama, G.K.; Hancock, R.E. Synergy of histone-derived peptides of coho salmon with lysozyme and flounder pleurocidin. Antimicrob. Agents Chemother 2001, 45, 1337–1342. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, J.M.; Molle, G.; Kemp, G.D.; Smith, V.J. Isolation and characterisation of oncorhyncin II, a histone H1-derived antimicrobial peptide from skin secretions of rainbow trout, Oncorhynchus mykiss. Dev. Comp. Immunol. 2004, 28, 127–138. [Google Scholar] [CrossRef]
- Luders, T.; Birkemo, G.A.; Nissen-Meyer, J.; Andersen, O.; Nes, I.F. Proline conformation-dependent antimicrobial activity of a proline-rich histone h1 N-terminal Peptide fragment isolated from the skin mucus of Atlantic salmon. Antimicrob. Agents Chemother 2005, 49, 2399–2406. [Google Scholar] [CrossRef] [Green Version]
- Shamova, O.V.; Orlov, D.S.; Balandin, S.V.; Shramova, E.I.; Tsvetkova, E.V.; Panteleev, P.V.; Leonova, Y.F.; Tagaev, A.A.; Kokryakov, V.N.; Ovchinnikova, T.V. Acipensins-novel antimicrobial peptides from Leukocytes of the Russian Sturgeon Acipenser gueldenstaedtii. Acta Nat. 2014, 6, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Birkemo, G.A.; Luders, T.; Andersen, O.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Et Biophys. Acta 2003, 1646, 207–215. [Google Scholar] [CrossRef]
- Esche, C.; Stellato, C.; Beck, L.A. Chemokines: Key players in innate and adaptive immunity. J. Investig. Dermatol. 2005, 125, 615–628. [Google Scholar] [CrossRef] [Green Version]
- Salinas, I. The mucosal immune system of teleost fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef] [Green Version]
- Emilio, J.; Magrone, T. Antimicrobial peptides as mediators of innate immunity. Curr. Pharm. Des. 2018, 24, 1041–1042. [Google Scholar] [CrossRef]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710. [Google Scholar] [CrossRef]
- Otte, J.M.; Kiehne, K.; Herzig, K.H. Antimicrobial peptides in innate immunity of the human intestine. J. Gastroenterol. 2003, 38, 717–726. [Google Scholar] [CrossRef]
- Muniz, L.R.; Knosp, C.; Yeretssian, G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front. Immunol. 2012, 3, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, K. Interleukin-8 and human cancer biology. Cytokine Growth Factor Rev. 2001, 12, 375–391. [Google Scholar] [CrossRef]
- Waugh, D.J.; Wilson, C. The interleukin-8 pathway in cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef] [Green Version]
- Sanmamed, M.F.; Carranza-Rua, O.; Alfaro, C.; Onate, C.; Martín-Algarra, S.; Perez, G.; Muñoz-Calleja, C. Serum interleukin-8 reflects tumor burden and treatment response across malignancies of multiple tissue origins. Clin. Cancer Res. 2014, 20, 5697–5707. [Google Scholar] [CrossRef] [Green Version]
- Kasahara, T.; Mukaida, N.; Yamashita, K.; Yagisawa, H.; Akahoshi, T.; Matsushima, K. IL-1 and TNF-alpha induction of IL-8 and monocyte chemotactic and activating factor (MCAF) mRNA expression in a human astrocytoma cell line. Immunology 1991, 74, 60. [Google Scholar]
- Lee, E.Y.; Park, H.H.; Kim, Y.T.; Chung, J.K.; Choi, T.J. Cloning and sequence analysis of the interleukin-8 gene from flounder (Paralichthys olivaceous). Gene 2001, 274, 237–243. [Google Scholar] [CrossRef]
- Chen, L.; He, C.; Baoprasertkul, P.; Xu, P.; Li, P.; Serapion, J.; Liu, Z. Analysis of a catfish gene resembling interleukin-8: cDNA cloning, gene structure, and expression after infection with Edwardsiella ictaluri. Dev. Comp. Immunol. 2005, 29, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Nam, Y.K.; Cho, Y.S.; Lee, S.Y.; Kim, B.S.; Kim, D.S. Molecular characterization of hepcidin gene from mud loach (Misgurnus mizolepis; Cypriniformes). Fish Shellfish Immunol. 2011, 31, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Hilton, K.B.; Lambert, L.A. Molecular evolution and characterization of hepcidin gene products in vertebrates. Gene 2008, 415, 40–48. [Google Scholar] [CrossRef] [PubMed]

| Genome Assembly | Length | Number | ||
|---|---|---|---|---|
| Contig (bp) | Scaffold (bp) | Contig | Scaffold | |
| Total | 844,742,280 | 848,872,105 | 2113 | 744 |
| Max | 22,693,483 | 81,503,846 | - | - |
| Number ≥ 2000 | - | - | 2081 | 714 |
| N50 | 5,473,395 | 35,728,622 | 44 | 10 |
| N60 | 4,291,763 | 33,987,958 | 62 | 13 |
| N70 | 2,952,698 | 33,130,827 | 85 | 15 |
| N80 | 1,543,679 | 28,667,948 | 123 | 18 |
| N90 | 290,346 | 24,877,842 | 243 | 21 |
| AMPs | Ictalurus punctatus | Cyprinus carpio | Danio rerio | Oryzias latipes | Poecilia reticulata | Poecilia latipinna | Monopterus albus | Paralichthys olivaceus | Cynoglossus semilaevis | Sebastes schlegelii | Larimichthys crocea | Latimeria chalumnae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RegIIIalpha | 57 | 58 | 31 | 121 | 72 | 79 | 23 | 18 | 22 | 17 | 150 | 7 |
| RegIIIgamma | 83 | 89 | 95 | 129 | 74 | 87 | 57 | 31 | 34 | 37 | 25 | 42 |
| TCP | 134 | 84 | 126 | 113 | 109 | 109 | 93 | 59 | 108 | 67 | 127 | 102 |
| Chemokine | 71 | 150 | 72 | 200 | 40 | 39 | 30 | 17 | 22 | 24 | 42 | 33 |
| cgUbiquitin | 36 | 71 | 30 | 37 | 31 | 29 | 27 | 13 | 26 | 18 | 46 | 27 |
| Hipposin | 30 | 28 | 41 | 23 | 15 | 17 | 19 | 9 | 28 | 5 | 26 | 13 |
| Lysozyme | 27 | 9 | 5 | 3 | 7 | 8 | 4 | 3 | 2 | 1 | 5 | 2 |
| Histone H2B-1 | 20 | 1 | 17 | 0 | 0 | 0 | 4 | 0 | 3 | 0 | 0 | 0 |
| HbbetaP-1 | 11 | 18 | 8 | 5 | 4 | 4 | 2 | 3 | 3 | 1 | 6 | 1 |
| Vasoactive intestinal polypeptide | 10 | 15 | 7 | 10 | 7 | 8 | 7 | 2 | 8 | 3 | 7 | 2 |
| eNAP-1 | 7 | 12 | 10 | 39 | 5 | 6 | 5 | 3 | 3 | 3 | 4 | 2 |
| BHP | 7 | 2 | 1 | 2 | 9 | 7 | 4 | 0 | 1 | 1 | 0 | 1 |
| Misgurin | 6 | 0 | 4 | 6 | 0 | 4 | 0 | 0 | 5 | 0 | 5 | 0 |
| SK84 | 5 | 1 | 1 | 0 | 2 | 2 | 1 | 1 | 2 | 0 | 1 | 1 |
| Porcine NK-Lysin | 4 | 8 | 5 | 2 | 0 | 0 | 2 | 1 | 0 | 0 | 2 | 0 |
| Beta-amyloid peptide | 4 | 4 | 2 | 10 | 3 | 4 | 4 | 2 | 11 | 2 | 4 | 1 |
| Neuropeptide Y | 3 | 1 | 1 | 5 | 2 | 2 | 2 | 2 | 1 | 1 | 5 | 2 |
| Mouse Ang4 | 3 | 4 | 3 | 2 | 5 | 3 | 4 | 1 | 1 | 0 | 1 | 0 |
| Adrenomedullin | 3 | 7 | 3 | 10 | 2 | 2 | 2 | 2 | 3 | 2 | 6 | 2 |
| YFGAP | 2 | 3 | 2 | 2 | 3 | 4 | 2 | 3 | 2 | 4 | 1 | 1 |
| SP-BN | 2 | 9 | 5 | 4 | 5 | 6 | 4 | 3 | 4 | 3 | 6 | 3 |
| Omwaprin | 2 | 0 | 1 | 0 | 1 | 2 | 3 | 1 | 1 | 2 | 3 | 0 |
| Human TC | 2 | 1 | 1 | 5 | 2 | 3 | 0 | 1 | 0 | 0 | 0 | 4 |
| HMGN | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Hepcidin | 2 | 4 | 3 | 8 | 2 | 5 | 1 | 2 | 1 | 4 | 2 | 1 |
| gcLEAP | 2 | 3 | 2 | 3 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 |
| Calcitonin gene-related peptide | 2 | 6 | 1 | 5 | 3 | 3 | 1 | 1 | 2 | 1 | 2 | 2 |
| SLPI | 2 | 11 | 2 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 1 | 1 |
| Skin peptide tyrosine-tyrosine | 2 | 4 | 3 | 5 | 2 | 2 | 4 | 0 | 2 | 2 | 0 | 0 |
| Ubiquicidin | 1 | 3 | 2 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Thrombocidin | 1 | 1 | 1 | 5 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
| pCM19 | 1 | 2 | 1 | 0 | 1 | 1 | 1 | 0 | 2 | 0 | 1 | 1 |
| Naegleriapore A | 1 | 2 | 1 | 6 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 |
| Human islet amyloid polypeptide | 1 | 2 | 1 | 4 | 2 | 0 | 0 | 0 | 1 | 1 | 4 | 2 |
| Ap | 0 | 2 | 0 | 2 | 16 | 2 | 3 | 0 | 2 | 1 | 2 | 2 |
| Peptide 3910 | 0 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 1 |
| Sushi peptide | 0 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 |
| Luxuriosin | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 4 |
| Buforin II | 0 | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| hPF4 | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BD | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ixodidin | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 1 |
| gcLEAP | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| hGAPDH | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 |
| Ceratoxin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Chemerin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| Psoriasin | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 2 | 0 |
| Chrombacin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| Piscidin 4 | 0 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| Misgurin | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Kaliocin-1 | 0 | 0 | 1 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 |
| LEAP-2 | 0 | 1 | 1 | 3 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Chicken LEAP-2 | 1 | 1 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| Amoebapore A | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| Fa-AMP2 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Human Rnase | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 | 0 | 0 |
| Enkelytin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Elafin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 547 | 633 | 501 | 817 | 435 | 446 | 319 | 182 | 310 | 214 | 504 | 281 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Cao, M.; Xiu, Y.; Fu, Q.; Yang, N.; Su, B.; Li, C. Identification of Antimicrobial Peptide Genes in Black Rockfish Sebastes schlegelii and Their Responsive Mechanisms to Edwardsiella tarda Infection. Biology 2021, 10, 1015. https://doi.org/10.3390/biology10101015
Zhang M, Cao M, Xiu Y, Fu Q, Yang N, Su B, Li C. Identification of Antimicrobial Peptide Genes in Black Rockfish Sebastes schlegelii and Their Responsive Mechanisms to Edwardsiella tarda Infection. Biology. 2021; 10(10):1015. https://doi.org/10.3390/biology10101015
Chicago/Turabian StyleZhang, Min, Min Cao, Yunji Xiu, Qiang Fu, Ning Yang, Baofeng Su, and Chao Li. 2021. "Identification of Antimicrobial Peptide Genes in Black Rockfish Sebastes schlegelii and Their Responsive Mechanisms to Edwardsiella tarda Infection" Biology 10, no. 10: 1015. https://doi.org/10.3390/biology10101015
APA StyleZhang, M., Cao, M., Xiu, Y., Fu, Q., Yang, N., Su, B., & Li, C. (2021). Identification of Antimicrobial Peptide Genes in Black Rockfish Sebastes schlegelii and Their Responsive Mechanisms to Edwardsiella tarda Infection. Biology, 10(10), 1015. https://doi.org/10.3390/biology10101015






