Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Transposon Mining and Annotation
2.2. Phylogenetic Analysis
2.3. HT Detection
3. Results
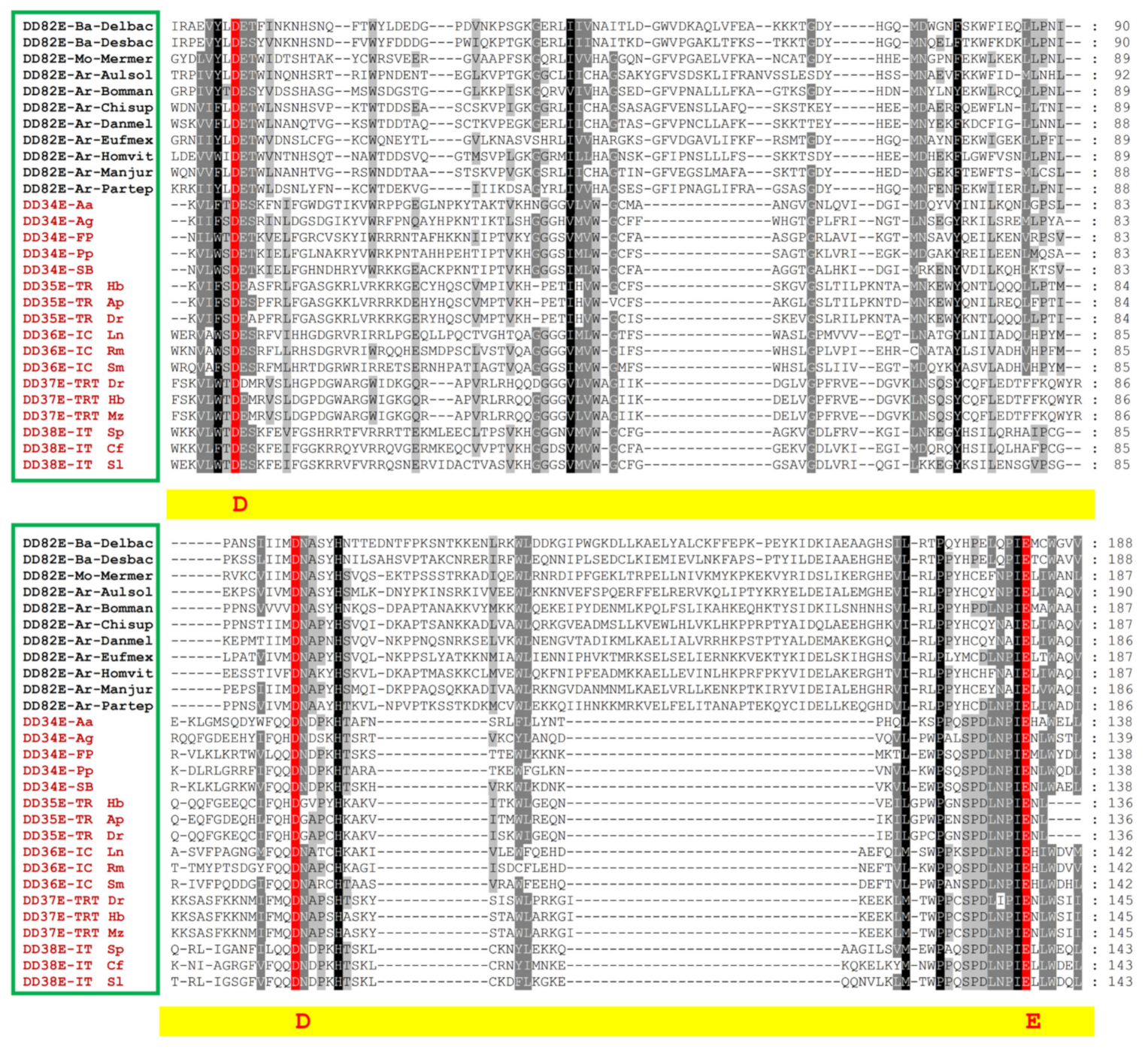
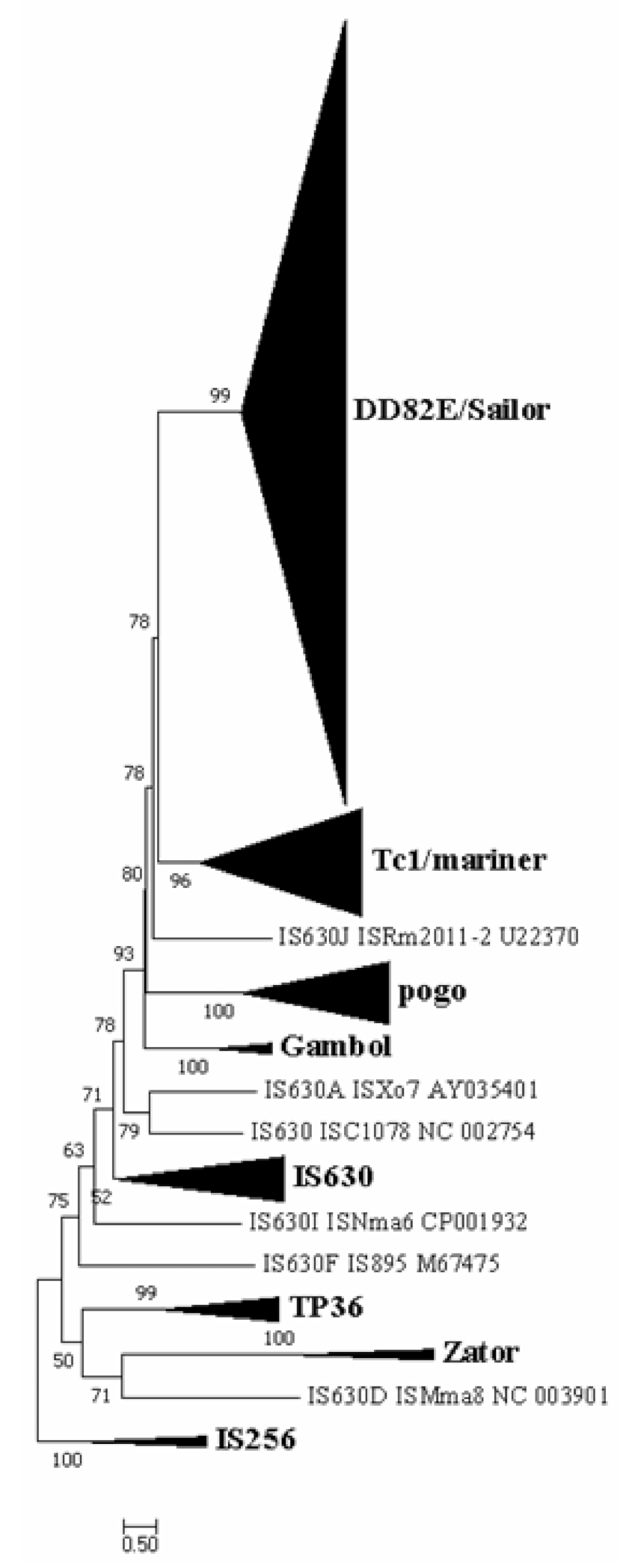
3.1. Discovery of a New Superfamily of ITm DNA Transposons, DD82E/Sailor
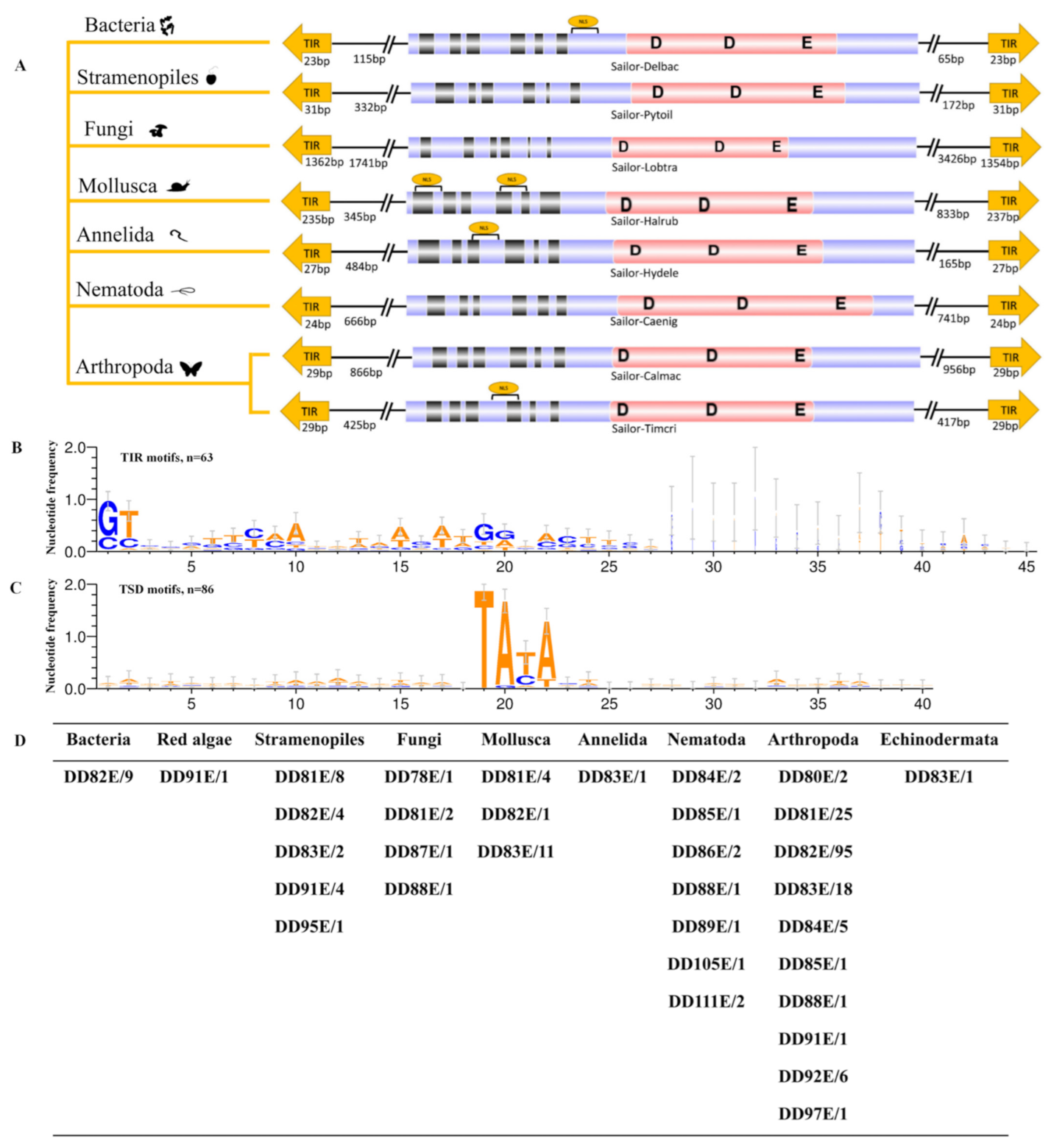
3.2. Distribution of DD82E/Sailor in Both Prokaryotes and Eukaryotes
3.3. Distinct Structural Organization of DD82E/Sailor
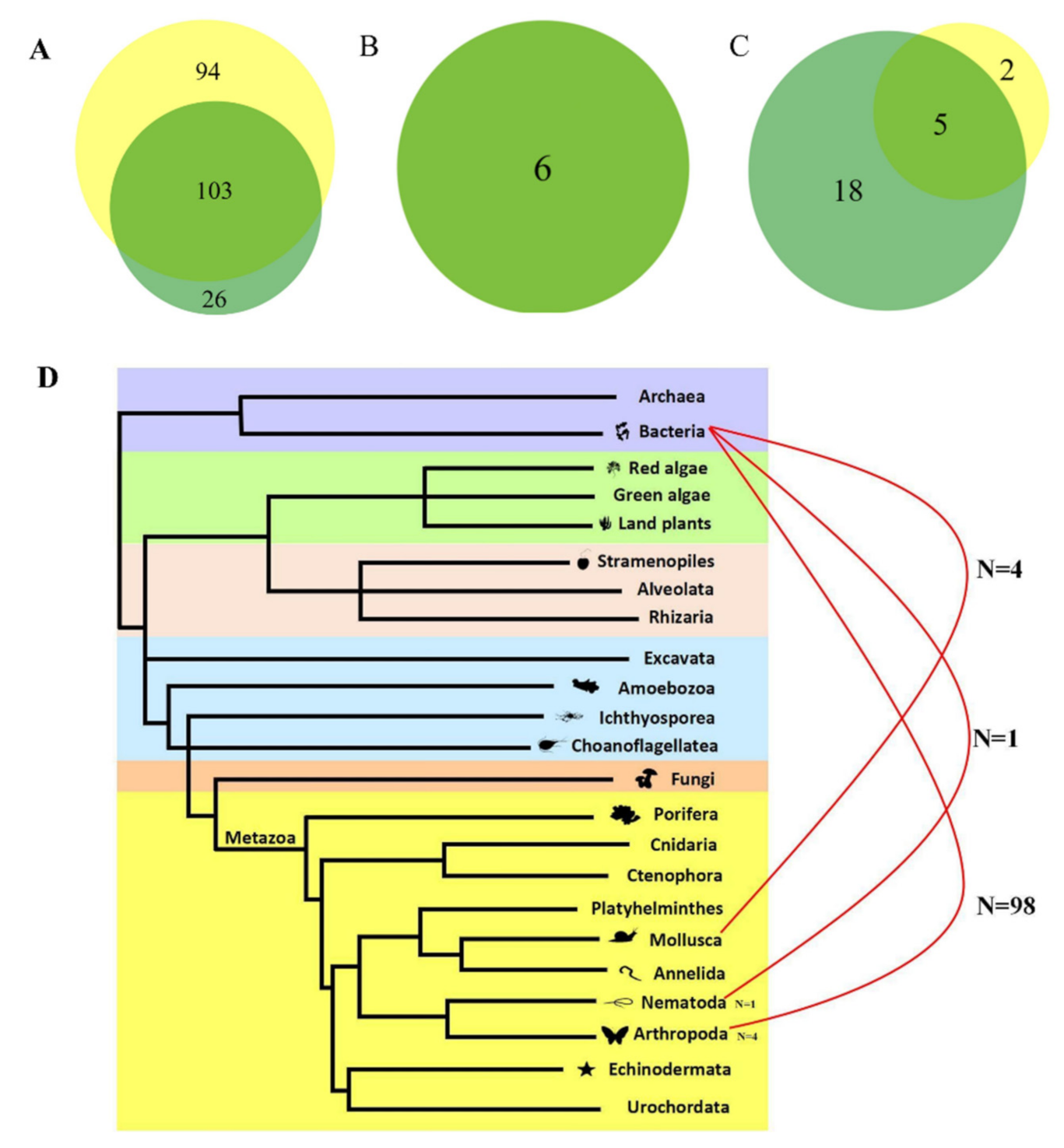
3.4. Evidence of HT Events of DD82E/Sailor between/within Prokaryotes and Eukaryotes
4. Discussion
4.1. Extensive Distribution of Sailor
4.2. Structural Organization of Sailor
4.3. HT Events of DD82E/Sailor between/within Prokaryotes and Eukaryotes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kidwell, M.G. Transposable elements and the evolution of genome size in eukaryotes. Genetica 2002, 115, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.C.; Zou, M.; Gan, X.N.; He, S.P. Genome size evolution in pufferfish: An insight from BAC clone-based Diodon holocanthus genome sequencing. BMC Genom. 2010, 11, 396. [Google Scholar] [CrossRef] [PubMed]
- de Koning, A.P.J.; Gu, W.; Castoe, T.A.; Batzer, M.A.; Pollock, D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011, 7, e1002384. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, N.N.; Kovalenko, L.V.; Zakharov, I.K. Transposable elements: Instability of genes and genomes. Russ. J. Genet. Appl. Res. 2011, 1, 489–496. [Google Scholar] [CrossRef]
- Grabundzija, I.; Messing, S.A.; Thomas, J.; Cosby, R.L.; Bilic, I.; Miskey, C.; Döring, A.G.; Kapitonov, V.; Diem, T.; Dalda, A.; et al. A Helitron transposon reconstructed from bats reveals a novel mechanism of genome shuffling in eukaryotes. Nat. Commun. 2016, 7, 10716. [Google Scholar] [CrossRef]
- Chandler, M.; Gellert, M.; Lambowitz, A.M.; Rice, P.A.; Sandmeyer, S.B. Mobile DNA III; ASM Press: Washington, DC, USA, 2015. [Google Scholar]
- Levin, H.L.; Moran, J.V. Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 2011, 12, 615–627. [Google Scholar] [CrossRef]
- Sturm, Á.; Perczel, A.; Ivics, Z.; Vellai, T. The Piwi-piRNA pathway: Road to immortality. Aging Cell 2017, 16, 906–911. [Google Scholar] [CrossRef]
- Fedoroff, N.V. Transposable Elements, Epigenetics, and Genome Evolution. Science (New York N. Y.) 2012, 338, 758–767. [Google Scholar] [CrossRef]
- Sinzelle, L.; Izsvák, Z.; Ivics, Z. Molecular domestication of transposable elements: From detrimental parasites to useful host genes. Cell. Mol. Life Sci. 2009, 66, 1073–1093. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten things you should know about transposable elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Piacentini, L.; Fanti, L.; Specchia, V.; Bozzetti, M.P.; Berloco, M.; Palumbo, G.; Pimpinelli, S. Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma 2014, 123, 345–354. [Google Scholar] [CrossRef]
- Auvinet, J.; Graça, P.; Belkadi, L.; Petit, L.; Bonnivard, E.; Dettaï, A.; Detrich, W.H.; Costaz, C.O.; Higuet, D. Mobilization of retrotransposons as a cause of chromosomal diversification and rapid speciation: The case for the Antarctic teleost genus Trematomus. BMC Genom. 2018, 19, 339. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Van, A.H.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Kojima, K.K. Structural and sequence diversity of eukaryotic transposable elements. Genes Genet. Syst. 2018, 94, 233–252. [Google Scholar] [CrossRef]
- Yuan, Y.W.; Wessler, S.R. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci. USA 2011, 108, 7884–7889. [Google Scholar] [CrossRef] [PubMed]
- Ivics, Z.; Hackett, P.B.; Plasterk, R.H.; Izsvák, Z. Molecular Reconstruction of Sleeping Beauty, a Tc1-like Transposon from Fish, and Its Transposition in Human Cells. Cell 1997, 91, 501–510. [Google Scholar] [CrossRef]
- Plasterk, R.H.A.; Izsvák, Z.; Ivics, Z. Resident aliens: The Tc1/mariner superfamily of transposable elements. Trends Genet. 1999, 15, 326. [Google Scholar] [CrossRef]
- Zoltán, I.; Zsuzsanna, I. Sleeping Beauty Transposition. Microbiol. Spectr. 2015, 3, 2. [Google Scholar]
- Zong, W.C.; Gao, B.; Diaby, M.; Shen, D.; Wang, S.S.; Wang, Y.L.; Sang, Y.T.; Chen, C.; Wang, X.Y.; Song, C.Y. Traveler, a New DD35E Family of Tc1/Mariner Transposons, Invaded Vertebrates Very Recently. Genome Biol. Evol. 2020, 12, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.T.; Gao, B.; Diaby, M.; Zong, W.C.; Chen, C.; Shen, D.; Wang, S.S.; Wang, Y.L.; Ivics, Z.; Song, C.Y. Incomer, a DD36E family of Tc1/mariner transposons newly discovered in animals. Mob. DNA 2019, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zong, W.C.; Miskey, C.; Ullah, N.; Diaby, M.; Chen, C.; Wang, X.Y.; Ivics, Z.; Song, C.Y. Intruder (DD38E), a recently evolved sibling family of DD34E/Tc1 transposons in animals. Mob. DNA 2020, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Diabya, M.; Puzakov, M.; Ullaha, N.; Wang, Y.l.; Danley, P.; Chen, C.; YanWang, X.; Gao, B.; Song, C.Y. Divergent evolution profiles of DD37D and DD39D families of Tc1/mariner transposons in eukaryotes. Mol. Phylogenetics Evol. 2021, 107143. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Gao, B.; Miskey, C.; Chen, C.; Sang, Y.T.; Zong, W.C.; Wang, S.S.; Wang, Y.L.; Wang, X.Y.; Ivics, Z.; et al. Multiple Invasions of Visitor, a DD41D Family of Tc1/mariner Transposons, throughout the Evolution of Vertebrates. Genome Biol. Evol. 2020, 12, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.G.; Tu, Z.J. Expanding the diversity of the IS630-Tc1-mariner superfamily: Discovery of a unique DD37E transposon and reclassification of the DD37D and DD39D transposons. Genetics 2001, 159, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Tellier, M.; Bouuaert, C.C.; Chalmers, R. Mariner and the ITm Superfamily of Transposons. Microbiol. Spectr. 2014, 3, MDNA3-0033-2014. [Google Scholar]
- Zhan, H.H.; Li, G.Y.; Xiong, X.M.; Ha, M.J.; Dai, F.Y. TRT, a Vertebrate and Protozoan Tc1-Like Transposon: Current Activity and Horizontal Transfer. Genome Biol. Evol. 2016, 8, 2994–3005. [Google Scholar] [CrossRef]
- Puzakov, M.; Puzakova, L.V.; Cheresiz, S.V. The Tc1-like elements with the spliceosomal introns in mollusk genomes. Mol. Genet. Genom. 2020, 295, 621–633. [Google Scholar] [CrossRef]
- Dupeyron, M.; Baril, T.; Bass, C.; Hayward, A. Phylogenetic analysis of the Tc1/mariner superfamily reveals the unexplored diversity of pogo-like elements. Mob. DNA 2020, 11, 21. [Google Scholar] [CrossRef]
- Gao, B.; Wang, Y.L.; Diaby, M.; Zong, W.C.; Shen, D.; Wang, S.; Chen, C.; Wang, X.; Song, C. Evolution of pogo, a separate superfamily of IS630-Tc1-mariner transposons, revealing recurrent domestication events in vertebrates. Mob. DNA 2020, 11, 25. [Google Scholar] [CrossRef]
- Coy, M.R.; Tu, Z.J. Gambol and Tc1 are two distinct families of DD34E transposons: Analysis of the Anopheles gambiae genome expands the diversity of the IS630-Tc1-mariner superfamily. Insect Mol. Biol. 2010, 14, 537–546. [Google Scholar] [CrossRef]
- Bao, W.; Jurka, M.G.; Kapitonov, V.V.; Jurka, J. New Superfamilies of Eukaryotic DNA Transposons and Their Internal Divisions. Mol. Biol. Evol. 2009, 26, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- McGuffin, L.J.; Bryson, K. The PSIPRED protein structure prediction server. Bioinformatics 2000, 16, 404–405. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772. [Google Scholar] [CrossRef]
- Harris, J.K.; Kelley, S.T.; Spiegelman, G.B.; Pace, N.R. The genetic core of the universal ancestor. Genome Res. 2003, 13, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Sudhir, K.; Glen, S.; Koichiro, T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar]
- Smith, T.C. A revised six-kingdom system of life. Biol. Rev. Camb. Philos. Soc. 1998, 73, 203–266. [Google Scholar] [CrossRef]
- Dupeyron, M.; Leclercq, S.; Cerveau, N.; Bouchon, D.; Gilbert, C. Horizontal transfer of transposons between and within crustaceans and insects. Mob. DNA 2014, 5, 4. [Google Scholar] [CrossRef]
- Wallau, G.L.; Vieira, C.; Loreto, É.L.S. Genetic exchange in eukaryotes through horizontal transfer: Connected by the mobilome. Mob. Dna 2018, 9, 6. [Google Scholar] [CrossRef]
- Hedges, S.B.; Marin, J.; Suleski, M.; Paymer, M.; Kumar, S. Tree of Life Reveals Clock-Like Speciation and Diversification. Mol. Biol. Evol. 2015, 32, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Schaack, S.; Gilbert, C.; Feschotte, C. Promiscuous DNA: Horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 2010, 25, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Blumenstiel, J.P. Birth, School, Work, Death, and Resurrection: The Life Stages and Dynamics of Transposable Element Proliferation. Genes 2019, 10, 335. [Google Scholar] [CrossRef]
- Jangam, D.; Feschotte, C.; Betrán, E. Transposable Element Domestication As an Adaptation to Evolutionary Conflicts. Trends Genet. 2017, 33, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Palmer, J.D. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 2008, 9, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Cordaux, R. Horizontal Transfer and Evolution of Prokaryote Transposable Elements in Eukaryotes. Genome Biol. Evol. 2013, 5, 822–832. [Google Scholar] [CrossRef]
- Hall, R.J.; Whelan, F.J.; McInerney, J.; Ou, Y.Q.; Sananes, M.R.D. Horizontal Gene Transfer as a Source of Conflict and Cooperation in Prokaryotes. Front. Microbiol. 2020, 11, 1569. [Google Scholar] [CrossRef]
- Gilbert, C.; Schaack, S.; Pace, J.K., II; Brindley, J.P.; Feschotte, C. A role for host-parasite interactions in the horizontal transfer of transposons across phyla. Nature 2010, 464, 1347. [Google Scholar] [CrossRef]
- Touchon, M.; Rocha, E.P.C. Causes of Insertion Sequences Abundance in Prokaryotic Genomes. Mol. Biol. Evol. 2007, 24, 969–981. [Google Scholar] [CrossRef]
- Zhang, H.H.; Cédric, F.; Han, M.J.; Zhang, Z. Recurrent Horizontal Transfers of Chapaev Transposons in Diverse Invertebrate and Vertebrate Animals. Genome Biol. Evol. 2014, 6, 1375–1386. [Google Scholar] [CrossRef]
- Walsh, A.M.; Kortschak, R.D.; Gardner, M.G.; Bertozzi, T.; Adelson, D.L. Widespread horizontal transfer of retrotransposons. Proc. Natl. Acad. Sci. USA 2012, 110, 1012–1016. [Google Scholar] [CrossRef]
- Wallau, G.L.; Ortiz, M.F.; Loreto, E.L.S. Horizontal Transposon Transfer in Eukarya: Detection, Bias, and Perspectives. Genome Biol. Evol. 2012, 4, 801–811. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, H.H.; Huang, K.; Zhang, X.G.; Han, M.J.; Zhang, Z. Repeated horizontal transfers of four DNA transposons in invertebrates and bats. Mob. DNA 2015, 6, 3. [Google Scholar] [CrossRef]
- Dunning, H.J.C. Horizontal gene transfer between bacteria and animals. Trends Genet. 2011, 27, 157–163. [Google Scholar] [CrossRef]
- Novick, P.; Smith, J.; Ray, D.; Boissinot, S. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene 2010, 449, 85–94. [Google Scholar] [CrossRef]
- Gilbert, C.; Hernandez, S.S.; Flores-Benabib, J.; Smith, E.N.; Feschotte, C. Rampant horizontal transfer of SPIN transposons in squamate reptiles. Mol. Biol. Evol. 2012, 29, 503–515. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oliveira, S.G.; Bao, W.; Martins, C.; Jurka, J. Horizontal transfers of Mariner transposons between mammals and insects. Mob. DNA 2012, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Faridi, N.; Casola, C. An Ancient Transkingdom Horizontal Transfer of Penelope-Like Retroelements from Arthropods to Conifers. Genome Biol. Evol. 2016, 8, 1252–1266. [Google Scholar] [PubMed]
- Suh, A.; Witt, C.C.; Menger, J.; Sadanandan, K.R.; Podsiadlowski, L.; Gerth, M.; Weigert, A.; McGuire, J.A.; Mudge, J.; Edwards, S.V.; et al. Ancient horizontal transfers of retrotransposons between birds and ancestors of human pathogenic nematodes. Nat. Commun. 2016, 7, 11396. [Google Scholar] [CrossRef]

| Taxa Distribution | Number of Species Containing Sailor | Number of Species Containing FL Sailor | Number of Species Containing Intact Sailor | Length of FL Sailor | Length of Intact Sailor | Tpase Length of Intact Sailor | TIR Length of Intact Sailor |
|---|---|---|---|---|---|---|---|
| Bacteria | 9 | 4 | 4 | 1379–1437 | 1379–1437 | 376–422 | 23–57 |
| Red algae | 1 | 1 | – | 3034 | – | – | – |
| Land Plants | 1 | – | – | – | – | – | – |
| Stramenopiles | 23 | 8 | 7 | 1757–4441 | 1757–4441 | 323–507 | 24–57 |
| Amoebozoa | 2 | – | – | – | – | – | – |
| Ichthyosporea | 1 | – | – | – | – | – | – |
| Choanoflagellata | 1 | – | – | – | – | – | – |
| Fungi | 12 | 2 | 2 | 2427–6979 | 2427–6979 | 368–607 | 52–1362 |
| Porifera | 1 | 1 | – | 4115 | – | – | – |
| Mollusca | 17 | 4 | 2 | 2220–2465 | 2220–2465 | 338–432 | 34–237 |
| Annelida | 1 | 1 | 1 | 1927 | 1927 | 429 | 27 |
| Nematoda | 12 | 2 | 1 | 2754 | 2757 | 452 | 24 |
| Arthropoda | 174 | 65 | 41 | 1799–4408 | 1799–4408 | 334–676 | 18–61 |
| Echinodermata | 1 | 1 | 1 | 2763 | 2763 | 334 | 28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.; Puzakov, M.; Guan, Z.; Xiang, K.; Diaby, M.; Wang, Y.; Wang, S.; Song, C.; Gao, B. Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons. Biology 2021, 10, 1005. https://doi.org/10.3390/biology10101005
Shi S, Puzakov M, Guan Z, Xiang K, Diaby M, Wang Y, Wang S, Song C, Gao B. Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons. Biology. 2021; 10(10):1005. https://doi.org/10.3390/biology10101005
Chicago/Turabian StyleShi, Shasha, Mikhail Puzakov, Zhongxia Guan, Kuilin Xiang, Mohamed Diaby, Yali Wang, Saisai Wang, Chengyi Song, and Bo Gao. 2021. "Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons" Biology 10, no. 10: 1005. https://doi.org/10.3390/biology10101005
APA StyleShi, S., Puzakov, M., Guan, Z., Xiang, K., Diaby, M., Wang, Y., Wang, S., Song, C., & Gao, B. (2021). Prokaryotic and Eukaryotic Horizontal Transfer of Sailor (DD82E), a New Superfamily of IS630-Tc1-Mariner DNA Transposons. Biology, 10(10), 1005. https://doi.org/10.3390/biology10101005







