Role of Transportome in the Gills of Chinese Mitten Crabs in Response to Salinity Change: A Meta-Analysis of RNA-Seq Datasets
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. De Novo Transcriptome Assembly
2.3. Assembly Statistics and Completeness
2.4. Functional Annotation of the Transcripts and Enrichment Analysis
2.5. Differential Expression and Transcriptome Meta-Analysis
2.6. Network Analysis and Community Detection
3. Results
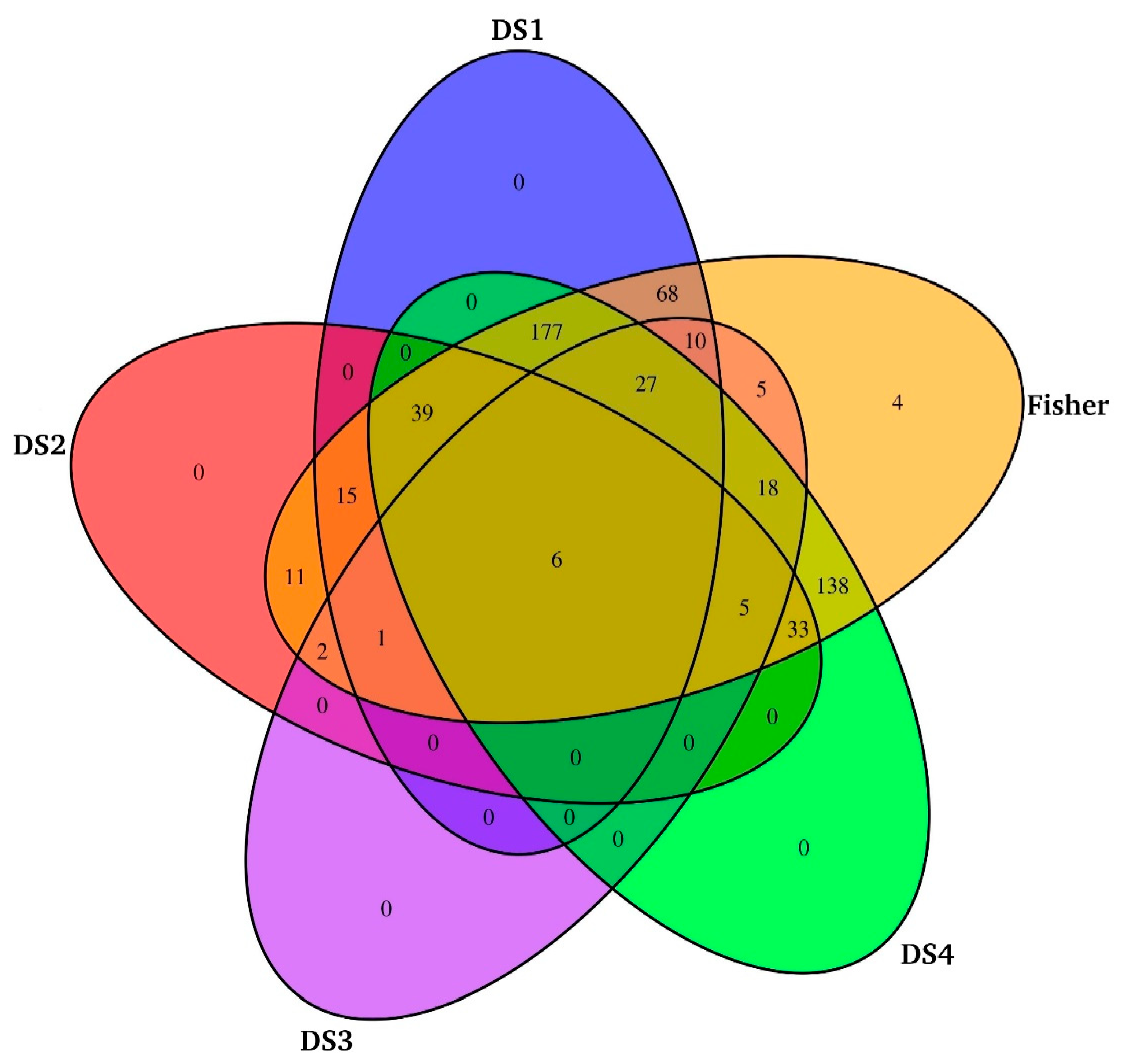
3.1. Datasets and De Novo Transcriptome Assembly
3.2. Identification of Coding Regions and Functional Annotation
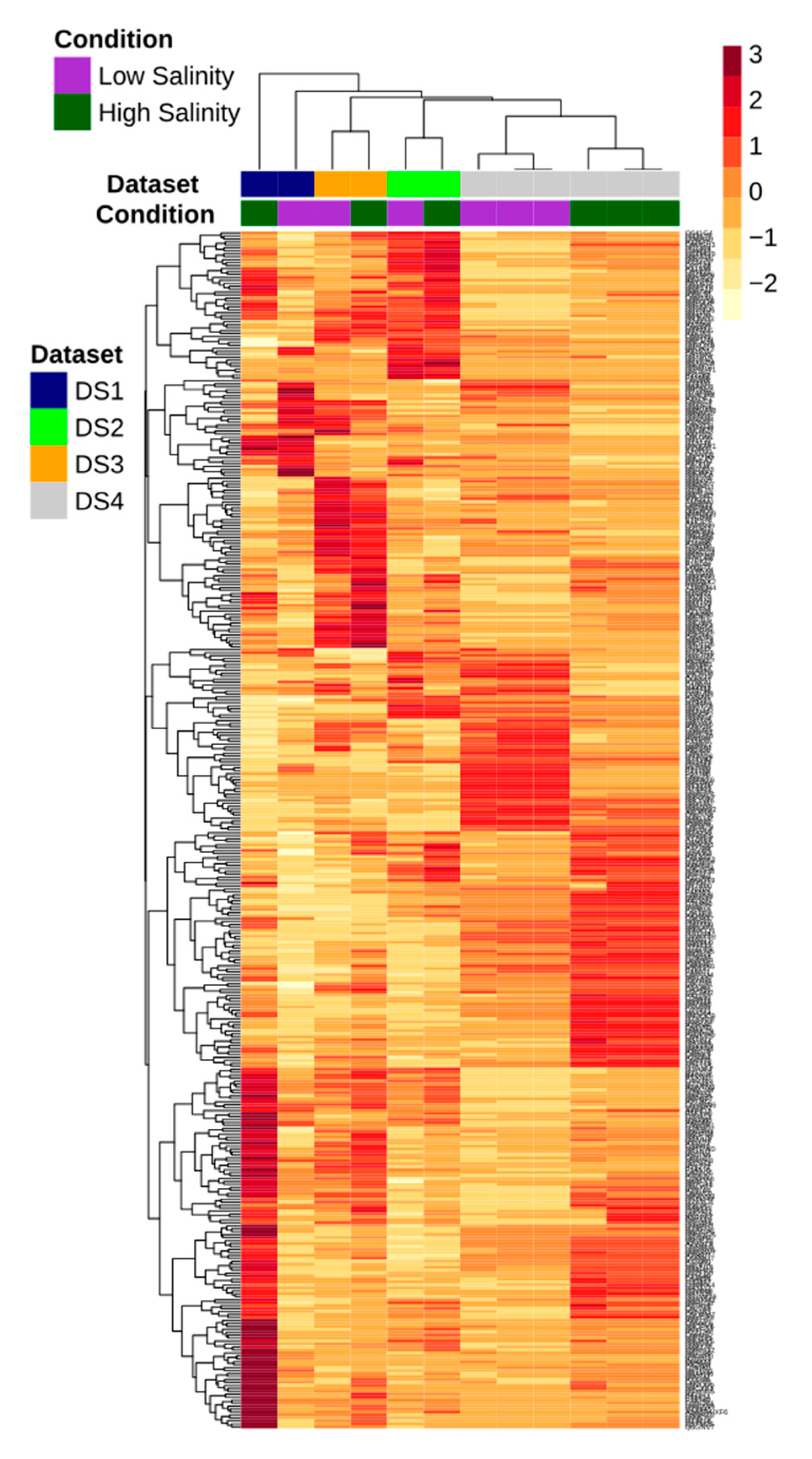
3.3. Meta-Analysis of CMC Gills Transcriptome
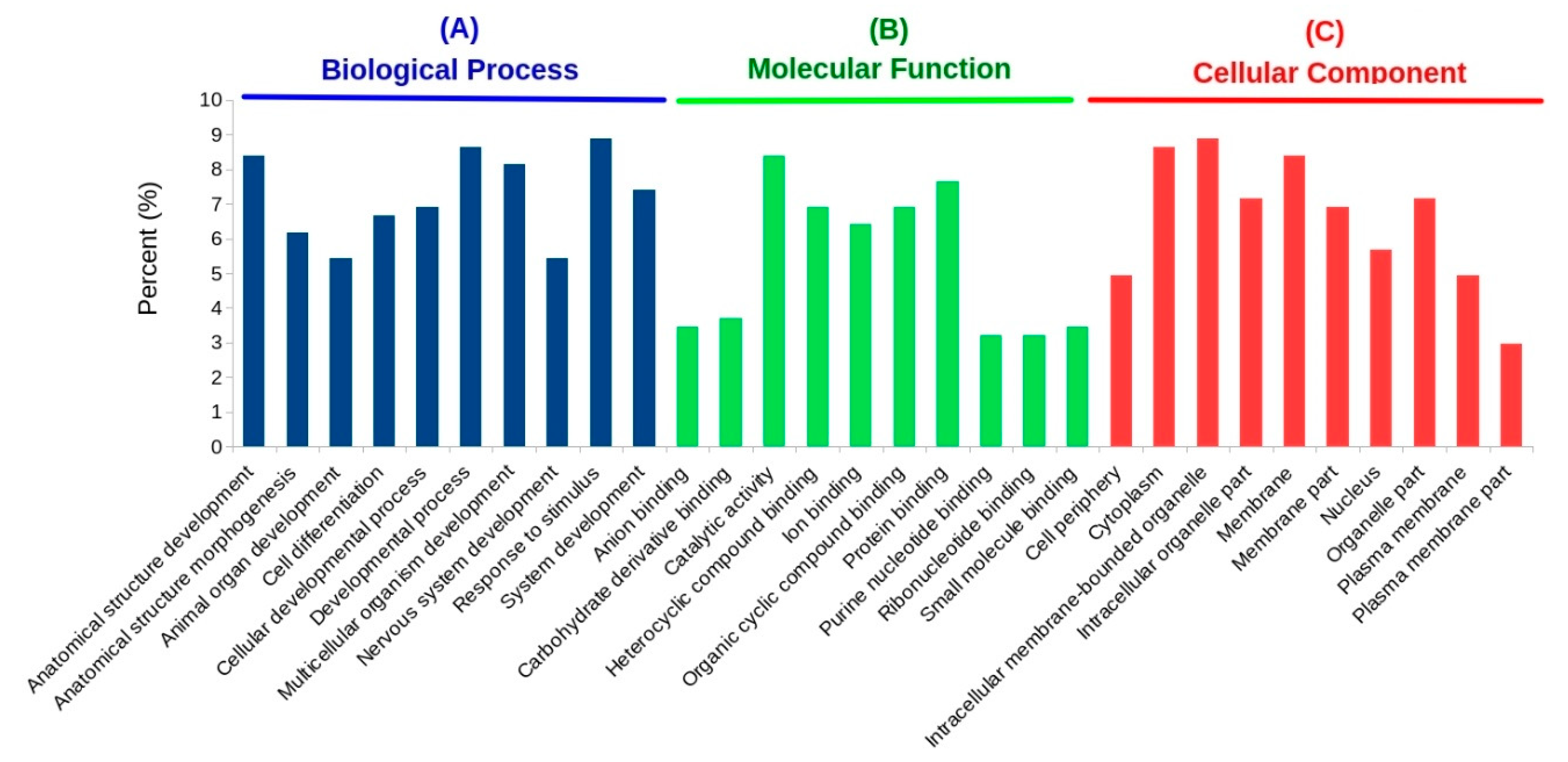
3.4. GO and KEGG Enrichment Analyses of DETs
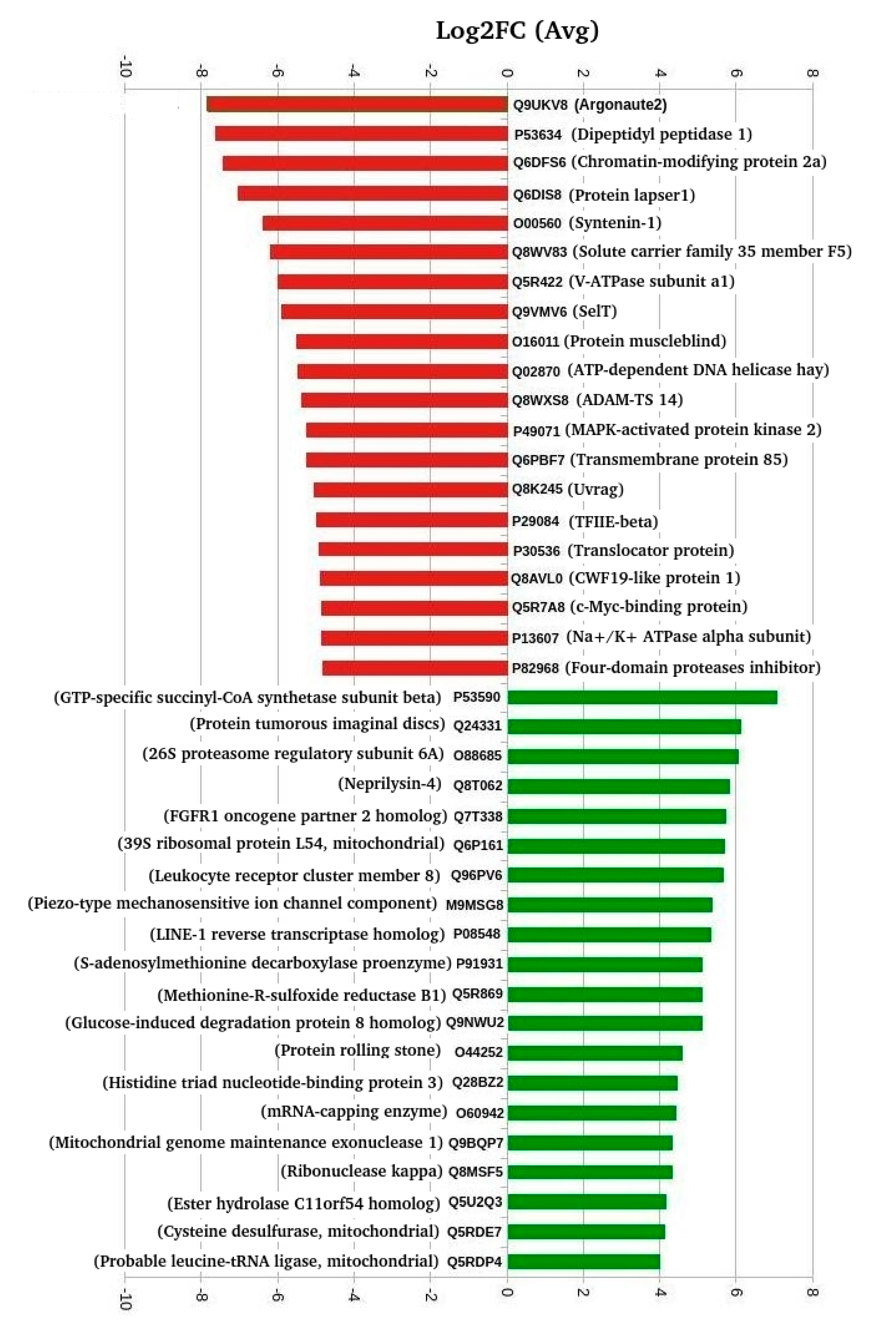
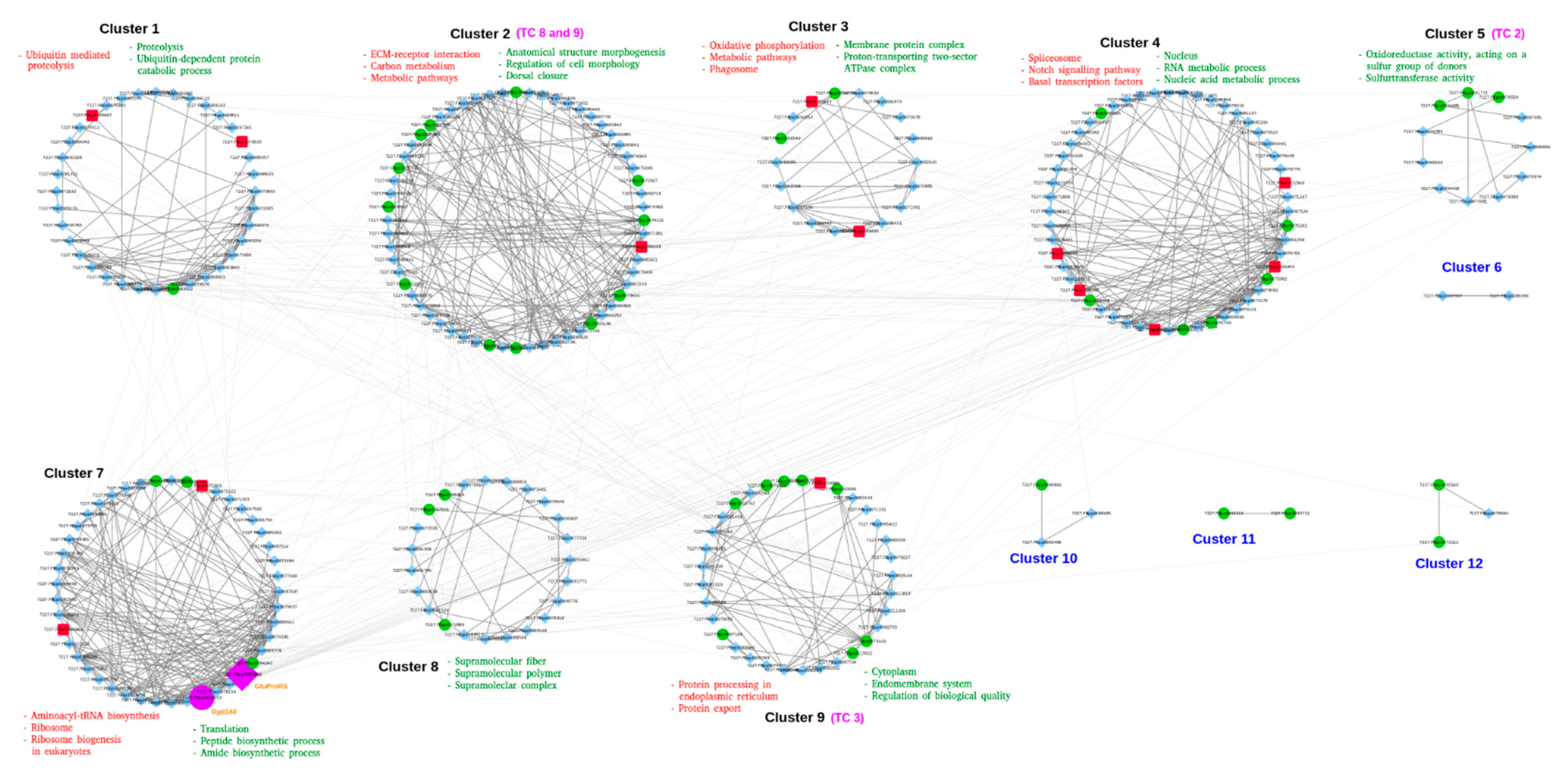
3.5. Interaction Network of DETs
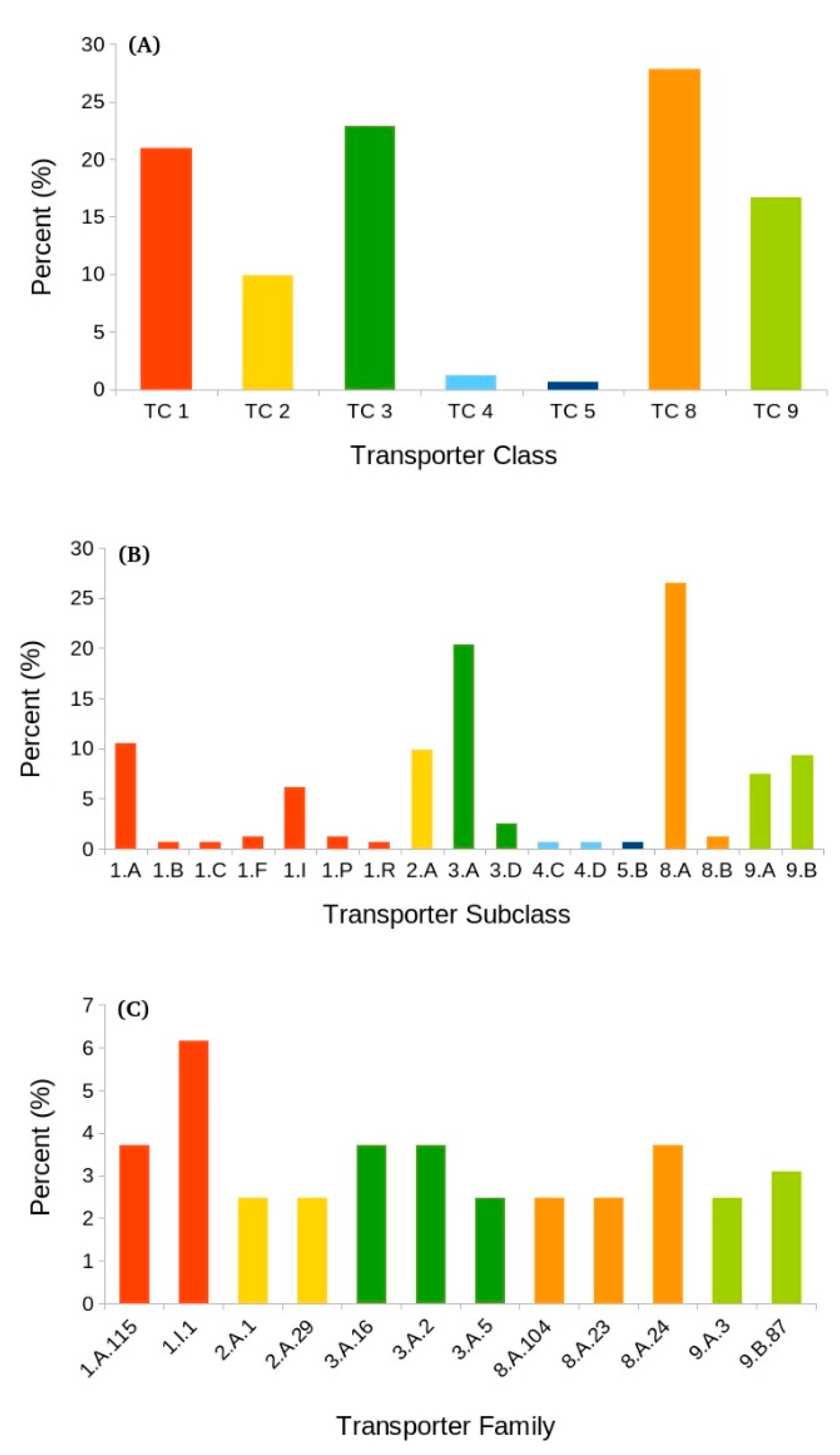
3.6. Transporters Implicated in Salinity Change
3.6.1. Accessory Factors Involved in Transport (TC 8)
Auxiliary Transport Proteins (TC 8.A)
Ribosomally Synthesized Protein/Peptide Toxins/Agonists that Target Channels and Carriers (TC 8.B)
3.6.2. Primary Active Transporters (TC 3)
P-P-Bond Hydrolysis-Driven Transporters (TC 3.A)
Oxidoreduction-Driven Transporters (TC 3.D)
3.6.3. Channels/Pores (TC 1)
α-Type Channels (TC 1.A)
Membrane-Bound Channels (TC 1.I)
Miscellaneous Channels/Pore Families
3.6.4. Incompletely Characterized Transport Systems (TC 9)
Recognized Transporters of Unknown Biochemical Mechanism (TC 9.A)
Putative Transport Proteins (TC 9.B)
3.6.5. Electrochemical Potential-Driven Transporters (TC 2)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anger, K. Effects of temperature and salinity on the larval development of the Chinese mitten crab Eriocheir sinensis (Decapoda: Grapsidae). Mar. Ecol. Prog. 1991, 72, 103–110. [Google Scholar] [CrossRef]
- Sui, L.; Zhang, F.; Wang, X.; Bossier, P.; Sorgeloos, P.; Hänfling, B. Genetic diversity and population structure of the Chinese mitten crab Eriocheir sinensis in its native range. Mar. Biol. 2009, 156, 1573–1583. [Google Scholar] [CrossRef]
- Zhang, D.; Qi, T.; Liu, J.; Liu, Q.; Jiang, S.; Zhang, H.; Wang, Z.; Ding, G.; Tang, B. Adaptively differential expression analysis in gill of Chinese mitten crabs (Eriocheir japonica sinensis) associated with salinity changes. Int. J. Biol. Macromol. 2018, 120, 2242–2246. [Google Scholar] [CrossRef] [PubMed]
- Dittel, A.I.; Epifanio, C.E. Invasion biology of the Chinese mitten crab Eriochier sinensis: A brief review. J. Exp. Mar. Biol. Ecol. 2009, 374, 79–92. [Google Scholar] [CrossRef]
- Kültz, D.; Fiol, D.; Valkova, N.; Gomez-Jimenez, S.; Chan, S.Y.; Lee, J. Functional genomics and proteomics of the cellular osmotic stress response in ‘non-model’ organisms. J. Exp. Biol. 2007, 210, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.H. Cell signaling and ion transport across the fish gill epithelium. J. Exp. Zool. 2002, 293, 336–347. [Google Scholar] [CrossRef]
- Evans, T.G.; Somero, G.N. A microarray-based transcriptomic time-course of hyper- and hypo-osmotic stress signaling events in the euryhaline fish Gillichthys mirabilis: Osmosensors to effectors. J. Exp. Biol. 2008, 211, 3636–3649. [Google Scholar] [CrossRef]
- Fiol, D.F.; Kültz, D. Osmotic stress sensing and signaling in fishes. Febs J. 2007, 274, 5790–5798. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, J.; Wei, B.; Cheng, Y.; Zhang, L.; Zhen, X. Comparative transcriptome analysis reveals osmotic-regulated genes in the gill of Chinese mitten crab (Eriocheir sinensis). PLoS ONE 2019, 14, e0210469. [Google Scholar] [CrossRef]
- Li, E.; Wang, S.; Li, C.; Wang, X.; Chen, K.; Chen, L. Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis. Physiol. Genom. 2014, 46, 177–190. [Google Scholar] [CrossRef]
- Tseng, Y.C.; Hwang, P.P. Some insights into energy metabolism for osmoregulation in fish. Comp. Biochem. Physiol. Toxicol. Pharmacol. 2008, 148, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Anderle, P.; Bussey, K.J.; Barbacioru, C.; Shankavaram, U.; Dai, Z.; Reinhold, W.C.; Papp, A.; Weinstein, J.N.; Sadée, W. Membrane transporters and channels: Role of the transportome in cancer chemosensitivity and chemoresistance. Cancer Res. 2004, 64, 4294–4301. [Google Scholar] [CrossRef] [PubMed]
- Castilho, P.C.; Martins, I.A.; Bianchini, A. Gill Na(+),K(+)-ATPase and osmoregulation in the estuarine crab, Chasmagnathus granulata Dana, 1851 (Decapoda, Grapsidae). J. Exp. Mar. Biol. Ecol. 2001, 256, 215–227. [Google Scholar] [CrossRef]
- Cutler, C.P.; Cramb, G. Molecular physiology of osmoregulation in eels and other teleosts: The role of transporter isoforms and gene duplication. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2001, 130, 551–564. [Google Scholar] [CrossRef]
- Evans, D.H.; Piermarini, P.M.; Potts, W.T.W. Ionic transport in the fish gill epithelium. J. Exp. Zool. 1999, 283, 641–652. [Google Scholar] [CrossRef]
- Chen, X.; Peng, Z.; Hou, X.; Wang, J.; Wang, C. The Molecular Basis of Osmoregulation and Physiological Processes Associated with Salinity Changes in the Chinese Mitten Crab Eriocheir sinensis. J. Shellfish Res. 2019, 38, 643–653. [Google Scholar] [CrossRef]
- Charmantier, G.; Charmantier-Daures, M.; Bouaricha, N.; Thuet, P.; Trilles, J.P.; Aiken, D.E. Ontogeny of Osmoregulation and Salinity Tolerance in Two Decapod Crustaceans: Homarus americanus and Penaeus japonicus. Biol. Bull. 1988, 175, 102–110. [Google Scholar] [CrossRef]
- Henry, R.P.; Lucu, C.; Onken, H.; Weihrauch, D. Multiple functions of the crustacean gill: Osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front. Physiol. 2012, 3, 431. [Google Scholar] [CrossRef]
- Saier, M.H., Jr.; Reddy, V.S.; Tsu, B.V.; Ahmed, M.S.; Li, C.; Moreno-Hagelsieb, G. The Transporter Classification Database (TCDB): Recent advances. Nucleic Acids Res. 2016, 44, D372–D379. [Google Scholar] [CrossRef]
- Allan, A.K.; Du, J.; Davies, S.A.; Dow, J.A. Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol. Genom. 2005, 22, 128–138. [Google Scholar] [CrossRef]
- Marot, G.; Jaffr’ezic, F.; Rau, A. metaRNASeq: Differential meta-analysis of RNA-seq data. dim (param) 2020, 1, 3. [Google Scholar]
- Toro-Domínguez, D.; Villatoro-García, J.A.; Martorell-Marugán, J.; Román-Montoya, Y.; Alarcón-Riquelme, M.E.; Carmona-Sáez, P. A survey of gene expression meta-analysis: Methods and applications. Brief. Bioinform. 2020. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kim, H.; Na, D.; Kim, S.Y.; Jo, D.; Lee, D. Meta-analysis method for discovering reliable biomarkers by integrating statistical and biological approaches: An application to liver toxicity. Biochem. Biophys. Res. Commun. 2016, 471, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Piras, I.S.; Manchia, M.; Huentelman, M.J.; Pinna, F.; Zai, C.C.; Kennedy, J.L.; Carpiniello, B. Peripheral Biomarkers in Schizophrenia: A Meta-Analysis of Microarray Gene Expression Datasets. Int. J. Neuropsychopharmacol. 2019, 22, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Bian, C.; Luo, Y.; Wang, L.; You, X.; Li, J.; Qiu, Y.; Ma, X.; Zhu, Z.; Ma, L.; et al. Draft genome of the Chinese mitten crab, Eriocheir sinensis. GigaScience 2016, 5, 5. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Rau, A.; Marot, G.; Jaffrézic, F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinform. 2014, 15, 91. [Google Scholar] [CrossRef]
- Fisher, R.A. Statistical Methods for Research Workers; Oliver and Boyd: Edinburgh, UK, 1925. [Google Scholar]
- Panahi, B.; Frahadian, M.; Dums, J.T.; Hejazi, M.A. Integration of Cross Species RNA-seq Meta-Analysis and Machine-Learning Models Identifies the Most Important Salt Stress-Responsive Pathways in Microalga Dunaliella. Front. Genet. 2019, 10, 752. [Google Scholar] [CrossRef]
- Chan, M.Y.; Efthymios, M.; Tan, S.H.; Pickering, J.W.; Troughton, R.; Pemberton, C.; Ho, H.H.; Prabath, J.F.; Drum, C.L.; Ling, L.H.; et al. Prioritizing Candidates of Post-Myocardial Infarction Heart Failure Using Plasma Proteomics and Single-Cell Transcriptomics. Circulation 2020, 142, 1408–1421. [Google Scholar] [CrossRef] [PubMed]
- Alimadadi, A.; Munroe, P.B.; Joe, B.; Cheng, X. Meta-Analysis of Dilated Cardiomyopathy Using Cardiac RNA-Seq Transcriptomic Datasets. Genes 2020, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Su, G.; Kuchinsky, A.; Morris, J.H.; States, D.J.; Meng, F. GLay: Community structure analysis of biological networks. Bioinformatics 2010, 26, 3135–3137. [Google Scholar] [CrossRef]
- Boube, M.; Martin-Bermudo, M.D.; Brown, N.H.; Casanova, J. Specific tracheal migration is mediated by complementary expression of cell surface proteins. Genes Dev. 2001, 15, 1554–1562. [Google Scholar] [CrossRef]
- Hartman, M.A.; Finan, D.; Sivaramakrishnan, S.; Spudich, J.A. Principles of unconventional myosin function and targeting. Annu. Rev. Cell Dev. Biol. 2011, 27, 133–155. [Google Scholar] [CrossRef]
- Peck, J.W.; Bowden, E.T.; Burbelo, P.D. Structure and function of human Vps20 and Snf7 proteins. Biochem. J. 2004, 377, 693–700. [Google Scholar] [CrossRef]
- Phillips, D.; Covian, R.; Aponte, A.M.; Glancy, B.; Taylor, J.F.; Chess, D.; Balaban, R.S. Regulation of oxidative phosphorylation complex activity: Effects of tissue-specific metabolic stress within an allometric series and acute changes in workload. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012, 302, R1034–R1048. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Lee, E.J.; Jan, A.T.; Ahmad, S.; Cho, K.H.; Kim, J.; Choi, I. Network Analysis for the Identification of Differentially Expressed Hub Genes Using Myogenin Knock-down Muscle Satellite Cells. PLoS ONE 2015, 10, e0133597. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Lee, J.; Lee, J. Community-based network study of protein-carbohydrate interactions in plant lectins using glycan array data. PLoS ONE 2014, 9, e95480. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.; Girvan, M. Finding and evaluating community structure in networks. Phys. Rev. E 2004, 69, 026113. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef]
- Koh, G.C.; Porras, P.; Aranda, B.; Hermjakob, H.; Orchard, S.E. Analyzing protein-protein interaction networks. J. Proteome Res. 2012, 11, 2014–2031. [Google Scholar] [CrossRef]
- Li, J.; Dai, Z.; Jana, D.; Callaway, D.J.; Bu, Z. Ezrin controls the macromolecular complexes formed between an adapter protein Na+/H+ exchanger regulatory factor and the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2005, 280, 37634–37643. [Google Scholar] [CrossRef]
- Ponting, C.P.; Phillips, C.; Davies, K.E.; Blake, D.J. PDZ domains: Targeting signalling molecules to sub-membranous sites. Bioessays 1997, 19, 469–479. [Google Scholar] [CrossRef]
- Macias, M.J.; Wiesner, S.; Sudol, M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002, 513, 30–37. [Google Scholar] [CrossRef]
- Muramatsu, T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J. Biochem. 2016, 159, 481–490. [Google Scholar] [CrossRef]
- Egan, D.F.; Shackelford, D.B.; Mihaylova, M.M.; Gelino, S.; Kohnz, R.A.; Mair, W.; Vasquez, D.S.; Joshi, A.; Gwinn, D.M.; Taylor, R.; et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011, 331, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Cotton, J.; Crest, M.; Bouet, F.; Alessandri, N.; Gola, M.; Forest, E.; Karlsson, E.; Castañeda, O.; Harvey, A.L.; Vita, C.; et al. A potassium-channel toxin from the sea anemone Bunodosoma granulifera, an inhibitor for Kv1 channels. Revision of the amino acid sequence, disulfide-bridge assignment, chemical synthesis, and biological activity. Eur. J. Biochem. 1997, 244, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Meusser, B.; Hirsch, C.; Jarosch, E.; Sommer, T. ERAD: The long road to destruction. Nat. Cell Biol. 2005, 7, 766–772. [Google Scholar] [CrossRef] [PubMed]
- Collinson, I.; Breyton, C.; Duong, F.; Tziatzios, C.; Schubert, D.; Or, E.; Rapoport, T.; Kühlbrandt, W. Projection structure and oligomeric properties of a bacterial core protein translocase. Embo J. 2001, 20, 2462–2471. [Google Scholar] [CrossRef]
- Brandt, U. Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 2006, 75, 69–92. [Google Scholar] [CrossRef]
- Gemperli, A.C.; Schaffitzel, C.; Jakob, C.; Steuber, J. Transport of Na(+) and K (+) by an antiporter-related subunit from the Escherichia coli NADH dehydrogenase I produced in Saccharomyces cerevisiae. Arch. Microbiol. 2007, 188, 509–521. [Google Scholar] [CrossRef][Green Version]
- Cardol, P.; Vanrobaeys, F.; Devreese, B.; Van Beeumen, J.; Matagne, R.F.; Remacle, C. Higher plant-like subunit composition of mitochondrial complex I from Chlamydomonas reinhardtii: 31 conserved components among eukaryotes. Biochim. Et Biophys. Acta 2004, 1658, 212–224. [Google Scholar] [CrossRef]
- Lu, H.; Cao, X. GRIM-19 is essential for maintenance of mitochondrial membrane potential. Mol. Biol. Cell 2008, 19, 1893–1902. [Google Scholar] [CrossRef]
- Krüger, V.; Becker, T.; Becker, L.; Montilla-Martinez, M.; Ellenrieder, L.; Vögtle, F.N.; Meyer, H.E.; Ryan, M.T.; Wiedemann, N.; Warscheid, B.; et al. Identification of new channels by systematic analysis of the mitochondrial outer membrane. J. Cell Biol. 2017, 216, 3485–3495. [Google Scholar] [CrossRef]
- Adams, J.M.; Cory, S. The Bcl-2 protein family: Arbiters of cell survival. Science 1998, 281, 1322–1326. [Google Scholar] [CrossRef]
- Coste, B.; Mathur, J.; Schmidt, M.; Earley, T.J.; Ranade, S.; Petrus, M.J.; Dubin, A.E.; Patapoutian, A. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science 2010, 330, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Jahed, Z.; Soheilypour, M.; Peyro, M.; Mofrad, M.R. The LINC and NPC relationship—It’s complicated! J. Cell Sci. 2016, 129, 3219–3229. [Google Scholar] [CrossRef] [PubMed]
- Burington, B.; Barlogie, B.; Zhan, F.; Crowley, J.; Shaughnessy, J.D., Jr. Tumor cell gene expression changes following short-term in vivo exposure to single agent chemotherapeutics are related to survival in multiple myeloma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 4821–4829. [Google Scholar] [CrossRef] [PubMed]
- Gurgis, F.M.; Ziaziaris, W.; Munoz, L. Mitogen-activated protein kinase-activated protein kinase 2 in neuroinflammation, heat shock protein 27 phosphorylation, and cell cycle: Role and targeting. Mol. Pharmacol. 2014, 85, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, J.D.; Bibbs, L.; Ulevitch, R.J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994, 265, 808–811. [Google Scholar] [CrossRef] [PubMed]
- Dérijard, B.; Raingeaud, J.; Barrett, T.; Wu, I.H.; Han, J.; Ulevitch, R.J.; Davis, R.J. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science 1995, 267, 682–685. [Google Scholar] [CrossRef]
- Raingeaud, J.; Whitmarsh, A.J.; Barrett, T.; Dérijard, B.; Davis, R.J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 1996, 16, 1247–1255. [Google Scholar] [CrossRef]
- Rizo, J.; Xu, J. Synaptic vesicle fusion without SNARE transmembrane regions. Dev. Cell 2013, 27, 124–126. [Google Scholar] [CrossRef]
- Inoue, T.; Tsai, B. Regulated Erlin-dependent release of the B12 transmembrane J-protein promotes ER membrane penetration of a non-enveloped virus. PloS Pathog. 2017, 13, e1006439. [Google Scholar] [CrossRef]
- Arlt, H.; Reggiori, F.; Ungermann, C. Retromer and the dynamin Vps1 cooperate in the retrieval of transmembrane proteins from vacuoles. J. Cell Sci. 2015, 128, 645–655. [Google Scholar] [CrossRef]
- Lv, X.; Pu, X.; Qin, G.; Zhu, T.; Lin, H. The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis Int. J. Program. Cell Death 2014, 19, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Kozyraki, R. Cubilin, a multifunctional epithelial receptor: An overview. J. Mol. Med. 2001, 79, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Bouton, D.; Escriva, H.; de Mendonça, R.L.; Glineur, C.; Bertin, B.; Noël, C.; Robinson-Rechavi, M.; de Groot, A.; Cornette, J.; Laudet, V.; et al. A conserved retinoid X receptor (RXR) from the mollusk Biomphalaria glabrata transactivates transcription in the presence of retinoids. J. Mol. Endocrinol. 2005, 34, 567–582. [Google Scholar] [CrossRef]
- Song, Z. Roles of the nucleotide sugar transporters (SLC35 family) in health and disease. Mol. Asp. Med. 2013, 34, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Picco, C.; Scholz-Starke, J.; Festa, M.; Costa, A.; Sparla, F.; Trost, P.; Carpaneto, A. Direct Recording of Trans-Plasma Membrane Electron Currents Mediated by a Member of the Cytochrome b561 Family of Soybean. Plant Physiol. 2015, 169, 986–995. [Google Scholar] [CrossRef]
- Walker, E.; Hernandez, A.V.; Kattan, M.W. Meta-analysis: Its strengths and limitations. Cleve Clin. J. Med. 2008, 75, 431–439. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Huang, J.; Fraser, M.E. Structural basis for the binding of succinate to succinyl-CoA synthetase. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 912–921. [Google Scholar] [CrossRef]
- Lambeth, D.O.; Tews, K.N.; Adkins, S.; Frohlich, D.; Milavetz, B.I. Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J. Biol. Chem. 2004, 279, 36621–36624. [Google Scholar] [CrossRef]
- Araújo, S.J.; Cela, C.; Llimargas, M. Tramtrack regulates different morphogenetic events during Drosophila tracheal development. Development 2007, 134, 3665–3676. [Google Scholar] [CrossRef]
- Shirangi, S.A.; Kalbassi, M.R.; Khodabandeh, S.; Jafarian, H.; Lorin-Nebel, C.; Farcy, E.; Lignot, J.H. Salinity effects on osmoregulation and gill morphology in juvenile Persian sturgeon (Acipenser persicus). Fish Physiol. Biochem. 2016, 42, 1741–1754. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.D.; Regish, A.M.; Christensen, A.K. Distinct freshwater and seawater isoforms of Na+/K+-ATPase in gill chloride cells of Atlantic salmon. J. Exp. Biol. 2009, 212, 3994–4001. [Google Scholar] [CrossRef] [PubMed]
- Towle, D.W.; Weihrauch, D. Osmoregulation by Gills of Euryhaline Crabs: Molecular Analysis of Transporters1. Am. Zool. 2015, 41, 770–780. [Google Scholar] [CrossRef]
- Mitsou, I.; Multhaupt, H.A.B.; Couchman, J.R. Proteoglycans, ion channels and cell-matrix adhesion. Biochem. J. 2017, 474, 1965–1979. [Google Scholar] [CrossRef] [PubMed]
- Stogmann, E.; Reinthaler, E.; Eltawil, S.; El Etribi, M.A.; Hemeda, M.; El Nahhas, N.; Gaber, A.M.; Fouad, A.; Edris, S.; Benet-Pages, A.; et al. Autosomal recessive cortical myoclonic tremor and epilepsy: Association with a mutation in the potassium channel associated gene CNTN2. Brain 2013, 136, 1155–1160. [Google Scholar] [CrossRef]
- Giepmans, B.N.G. Role of connexin43-interacting proteins at gap junctions. Adv. Cardiol. 2006, 42, 41–56. [Google Scholar] [CrossRef]
- Ojala, V.K.; Knittle, A.M.; Kirjalainen, P.; Merilahti, J.A.M.; Kortesoja, M.; Tvorogov, D.; Vaparanta, K.; Lin, S.; Kast, J.; Pulliainen, A.T.; et al. The guanine nucleotide exchange factor VAV3 participates in ERBB4-mediated cancer cell migration. J. Biol. Chem. 2020, 295, 11559–11571. [Google Scholar] [CrossRef]
- Towle, D.W.; Palmer, G.; Harris, J. Role of gill Na+ + K+ -dependent ATPase in acclimation of blue crabs (Callinectes sapidus) to low salinity. J. Exp. Zool. 1976, 196, 315–321. [Google Scholar] [CrossRef]
- Perreira, J.M.; Aker, A.M.; Savidis, G.; Chin, C.R.; McDougall, W.M.; Portmann, J.M.; Meraner, P.; Smith, M.C.; Rahman, M.; Baker, R.E.; et al. RNASEK Is a V-ATPase-Associated Factor Required for Endocytosis and the Replication of Rhinovirus, Influenza A Virus, and Dengue Virus. Cell Rep. 2015, 12, 850–863. [Google Scholar] [CrossRef]
- Busch, W.; Saier, M.H., Jr. The IUBMB-endorsed transporter classification system. Mol. Biotechnol. 2004, 27, 253–262. [Google Scholar] [CrossRef]
- Anwar, T.; Samudrala, G. Bioinformatics Analysis and Functional Prediction of Transmembrane Proteins in Entamoeba histolytica. Genes 2018, 9, 499. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Tian, W.; Kleist, T.; He, K.; Garcia, V.; Bai, F.; Hao, Y.; Luan, S.; Li, L. DUF221 proteins are a family of osmosensitive calcium-permeable cation channels conserved across eukaryotes. Cell Res. 2014, 24, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, J.; Sun, L. Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2. Nat. Commun. 2018, 9, 5060. [Google Scholar] [CrossRef] [PubMed]
- Pang, C.; Yuan, H.; Ren, S.; Chen, Y.; An, H.; Zhan, Y. TMEM16A/B associated CaCC: Structural and functional insights. Protein Pept. Lett. 2014, 21, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Prole, D.L.; Taylor, C.W. Identification and analysis of putative homologues of mechanosensitive channels in pathogenic protozoa. PLoS ONE 2013, 8, e66068. [Google Scholar] [CrossRef]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. 1998, 62, 1–34. [Google Scholar] [CrossRef]
- Kumar, S.; Lekshmi, M.; Parvathi, A.; Ojha, M.; Wenzel, N.; Varela, M.F. Functional and Structural Roles of the Major Facilitator Superfamily Bacterial Multidrug Efflux Pumps. Microorganisms 2020, 8, 266. [Google Scholar] [CrossRef]
- Palmieri, F. The mitochondrial transporter family SLC25: Identification, properties and physiopathology. Mol. Asp. Med. 2013, 34, 465–484. [Google Scholar] [CrossRef]
| Dataset | BioProject Accession | Type * | #Raw Reads | #Trimmed Reads | Reads Retained Postfiltering (%) | Reference |
|---|---|---|---|---|---|---|
| DS1 | PRJNA481259 | PE | 49,648,084 | 47,331,397 | 95.33 | [3] |
| DS2 | PRJNA488907 | 17,817,553,2 | 14,353,948,4 | 80.42 | [16] | |
| DS3 | PRJNA80779 | 64,212,034 | 60,050,212 | 93.51 | [10] | |
| DS4 | PRJNA508867 | SE | 75,845,511 | 75,292,415 | 99.28 | [9] |
| P1 | P1 Description | P2 | P2 Description | Effect | Cor | Corrected p-Value |
|---|---|---|---|---|---|---|
| Q9D4P0 | ADP-ribosylation factor-like protein 5B | B2D0J5 | Venom carboxylesterase-6 | ↓ | 1 | 9.4355 × 10−7 |
| Q7ZV80 | Survival of motor neuron-related-splicing factor 30 | B0WTN3 | Eukaryotic translation initiation factor 3 subunit M | ↓ | 0.99 | 1.2953 × 10−6 |
| Q04164 | Putative epidermal cell surface receptor | Q9UQC9 | Calcium-activated chloride channel regulator 2 | ↓ | 0.99 | 1.5143 × 10−6 |
| Q8C0K5 | Graves disease carrier protein homolog (GDC) (Mitochondrial solute carrier protein homolog) | Q13336 | Urea transporter 1 | ↑ | 0.99 | 3.4321 × 10−6 |
| Q5REW1 | Iodotyrosine deiodinase 1 (IYD-1) | Q5I7G2 | Retinoic acid receptor RXR | ↑ | 0.99 | 3.4321 × 10−6 |
| Q9Y535 | DNA-directed RNA polymerase III subunit RPC8 | Q5VWG9 | Transcription initiation factor TFIID subunit 3 | ↓ | 0.99 | 3.4321 × 10−6 |
| Q04164 | Putative epidermal cell surface receptor | Q27421 | Protein outspread | ↓ | 0.99 | 3.4321 × 10−6 |
| Q8WXS8 | A disintegrin and metalloproteinase with thrombospondin motifs 14 | O16011 | Protein muscleblind | ↓ | 0.99 | 3.4321 × 10−6 |
| O74503 | Upstream activation factor subunit spp27 | Q53CF6 | Cytochrome c oxidase subunit 7A1, mitochondrial | ↑ | 0.99 | 3.7031 × 10−6 |
| Q4R3Y4 | Long-chain fatty acid transport protein 4 (FATP-4) | Q7ZV80 | Survival of motor neuron-related-splicing factor 30 | ↓ | 0.99 | 3.9073 × 10−6 |
| Q68HB4 | Profilin | P56616 | Ubiquitin-conjugating enzyme E2 C | ↓ | 0.99 | 6.1125 × 10−6 |
| Q4R3Y4 | Long-chain fatty acid transport protein 4(FATP-4) | B0WTN3 | Eukaryotic translation initiation factor 3 subunit M | ↓ | 0.99 | 6.1125 × 10−6 |
| Q86WZ6 | Zinc finger protein 227 | Q8C0K5 | Graves disease carrier protein homolog | ↑ | 0.99 | 7.7076 × 10−6 |
| P53590 | Succinate–CoA ligase (GDP-forming) subunit beta, mitochondrial | P21158 | C-factor | ↑ | 0.99 | 7.7076 × 10−6 |
| Q9D4P0 | ADP-ribosylation factor-like protein 5B | P82968 | Four-domain proteases inhibitor | ↓ | 0.99 | 7.7076 × 10−6 |
| Q04833 | Low-density lipoprotein receptor-related protein | P82968 | Four-domain proteases inhibitor | ↓ | 0.99 | 8.8569 × 10−6 |
| Q9VB68 | Serine protease grass | Q5VWG9 | Transcription initiation factor TFIID subunit 3 | ↓ | 0.99 | 8.8569 × 10−6 |
| Q8K0U4 | Heat shock 70-kDa protein 12A | Q8N539 | Fibrinogen C domain-containing protein 1 | ↓ | 0.99 | 1.2159 × 10−5 |
| A6QP05 | Dehydrogenase/reductase SDR family member 12 | Q9CY58 | Plasminogen activator inhibitor 1 RNA-binding protein | ↑ | 0.99 | 1.5922 × 10−5 |
| P29844 | Endoplasmic reticulum chaperone BiP | Q91V92 | ATP-citrate synthase | ↓ | 0.99 | 1.6280 × 10−5 |
| UniProt ID | Gene Name | Organism | Protein Name | Gene Ontology (Biological Process) | Effect |
|---|---|---|---|---|---|
| P17789 | ttk | Drosophila melanogaster (Fruit fly) | Protein tramtrack, beta isoform | branch fusion, open tracheal system (GO:0035147) | ↓ |
| P53590 | SUCLG2 | Sus scrofa (Pig) | Succinate–CoA ligase (GDP-forming) subunit beta, mitochondrial | succinyl–CoA metabolic process (GO:0006104) | ↑ |
| Q28BZ2 | Hint3 | Xenopus tropicalis (Western clawed frog) | Histidine triad nucleotide-binding protein 3 | NA | ↑ |
| Q5U2Q3 | NA | Rattus norvegicus (Rat) | Ester hydrolase C11orf54 homolog | NA | ↑ |
| Q6DIS8 | Lzts2 | Xenopus tropicalis (Western clawed frog) | Leucine zipper putative tumor suppressor 2 homolog | microtubule severing (GO:0051013) | ↓ |
| Q8BTN6 | Leng9 | Mus musculus (Mouse) | Leukocyte receptor cluster member 9 | NA | ↓ |
| Pathway Name | Pathway ID | No. of Input Genes | Total No. of Genes | Corrected p-Value |
|---|---|---|---|---|
| Metabolic pathways | dme01100 | 52 | 1111 | 0.00001 |
| Apoptosis—fly | dme04214 | 9 | 63 | 0.00034 |
| Phagosome | dme04145 | 10 | 89 | 0.00067 |
| ECM-receptor interaction | dme04512 | 4 | 12 | 0.00316 |
| Oxidative phosphorylation | dme00190 | 11 | 144 | 0.00471 |
| Endocytosis | dme04144 | 10 | 122 | 0.00497 |
| Glycerophospholipid metabolism | dme00564 | 6 | 63 | 0.02182 |
| Pyruvate metabolism | dme00620 | 5 | 46 | 0.02654 |
| Sphingolipid metabolism | dme00600 | 4 | 28 | 0.02680 |
| Spliceosome | dme03040 | 8 | 128 | 0.04231 |
| Glycolysis/Gluconeogenesis | dme00010 | 5 | 55 | 0.04283 |
| Protein processing in endoplasmic reticulum | dme04141 | 8 | 133 | 0.04868 |
| Class | Class Description | Subclass | Subclass Description | Family | Family Description | Representative Transcript | Effect |
|---|---|---|---|---|---|---|---|
| TC 8 (45) | Accessory Factors Involved in Transport | TC 8.A (43) | Auxiliary transport proteins | TC 8.A.24 (6) | The Ezrin/Radixin/Moesin-binding Phosphoprotein 50 (EBP50) family | Syntenin-1 | ↓ |
| TC 8.A.23 (4) | The Basigin family | Tyrosine-protein kinase Abl | ↓ | ||||
| TC 8.A.104 (4) | The 5’-AMP-activated protein kinase (AMPK) family | Serine/threonine-protein kinase pim-3 | ↓ | ||||
| TC 8.B (2) | Ribosomally synthesized protein/peptide toxins/agonists that target channels and carriers | TC 8.B.14 (2) | The Sea Anemone Peptide Toxin, Class 1 (BgK) family | Matrix metalloproteinase-24 | ↓ | ||
| TC 3 (37) | Primary Active Transporters | TC 3.A (33) | P-P-bond hydrolysis-driven transporters | TC 3.A.2 (6) | The H+- or Na+-translocating F-type, V-type and A-type ATPase (F-ATPase) superfamily | V-type proton ATPase subunit a1 | ↓ |
| TC 3.A.16 (6) | The Endoplasmic Reticular Retrotranslocon (ER-RT) family | 26S proteasome regulatory subunit 6A | ↑ | ||||
| TC 3.A.5 (4) | The General Secretory Pathway (Sec) family | Putative U5 small nuclear ribonucleoprotein 200-kDa helicase | ↓ | ||||
| TC 3.D (4) | Oxidoreduction-driven transporters | TC 3.D.1 (3) | The H+ or Na+-translocating NADH Dehydrogenase (NDH) family | NADH dehydrogenase (ubiquinone) iron-sulfur protein 8, mitochondrial | ↑ | ||
| TC 3.D.4 (1) | The Proton-translocating Cytochrome Oxidase (COX) Superfamily | Cytochrome c oxidase subunit 7A1, mitochondrial | ↑ | ||||
| TC 1 (34) | Channels/Pores | TC 1.A (17) | α-Type Channels | TC 1.A.115 (6) | The Nonselective Cation Channel-2 (NSCC2) family | Dehydrogenase/reductase SDR family member 12 | ↑ |
| TC 1.A.17 (2) | The Calcium-dependent Chloride Channel (Ca-ClC) family | Transmembrane channel-like protein 7 | ↓ | ||||
| TC 1.I (10) | Membrane-bounded Channels | TC 1.I.1 (10) | The Nuclear Pore Complex (NPC) family | MAP kinase-activated protein kinase 2 | ↓ | ||
| TC 9 (27) | Incompletely Characterized Transport Systems | TC 9.A (12) | Recognized Transporters of Unknown Biochemical Mechanism | TC 9.A.3 (4) | The Sorting Nexin27 (SNX27)-Retromer Assembly Apparatus | Ras-related protein Rap-1b | ↓ |
| TC 9.A.63 (3) | The Retromer-dependent Vacuolar Protein Sorting (R-VPS) family | Cell division control protein 42 homolog | ↓ | ||||
| TC 9.B (15) | Putative uncharacterized transport proteins. | TC 9.B.87 (5) | The Selenoprotein P Receptor (SelP-Receptor) family | Cubilin | ↓ | ||
| TC 2 (16) | Electrochemical Potential-driven Transporters | TC 2.A (16) | Porters (uniporters, symporters, antiporters) | TC 2.A.1 (4) | The Major Facilitator Superfamily (MFS) | Solute carrier family 49 member 4 homolog | ↓ |
| TC 2.A.29 (4) | The Mitochondrial Carrier (MC) family | Mitochondrial coenzyme A transporter SLC25A42 | ↑ | ||||
| TC 2.A.7 (3) | The Drug/Metabolite Transporter (DMT) Superfamily | Solute carrier family 35 member F5 | ↓ | ||||
| TC 4 (2) | Group Translocators | TC 4.C (1) | Acyl CoA ligase-coupled transporters | TC 4.C.1 (1) | The Fatty Acid Transporter (FAT) Family | Long-chain fatty acid transport protein 4 | ↓ |
| TC 4.D (1) | Polysaccharide Synthase/Exporters | TC 4.D.1 (1) | The Putative Vectorial Glycosyl Polymerization (VGP) Family | Beta-1,4-mannosyltransferase egh | ↓ | ||
| TC 5 (1) | Transmembrane Electron Carriers | TC 5.B (1) | Transmembrane 1-Electron Transfer Carriers | TC 5.B.2 (1) | The Eukaryotic Cytochrome b561 (Cytb561) Family | Putative ferric-chelate reductase 1 homolog | ↓ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malik, A.; Kim, C.-B. Role of Transportome in the Gills of Chinese Mitten Crabs in Response to Salinity Change: A Meta-Analysis of RNA-Seq Datasets. Biology 2021, 10, 39. https://doi.org/10.3390/biology10010039
Malik A, Kim C-B. Role of Transportome in the Gills of Chinese Mitten Crabs in Response to Salinity Change: A Meta-Analysis of RNA-Seq Datasets. Biology. 2021; 10(1):39. https://doi.org/10.3390/biology10010039
Chicago/Turabian StyleMalik, Adeel, and Chang-Bae Kim. 2021. "Role of Transportome in the Gills of Chinese Mitten Crabs in Response to Salinity Change: A Meta-Analysis of RNA-Seq Datasets" Biology 10, no. 1: 39. https://doi.org/10.3390/biology10010039
APA StyleMalik, A., & Kim, C.-B. (2021). Role of Transportome in the Gills of Chinese Mitten Crabs in Response to Salinity Change: A Meta-Analysis of RNA-Seq Datasets. Biology, 10(1), 39. https://doi.org/10.3390/biology10010039






