Ferroptosis-Related Genes in Neurodevelopment and Central Nervous System
Abstract
Simple Summary
Abstract
1. Introduction
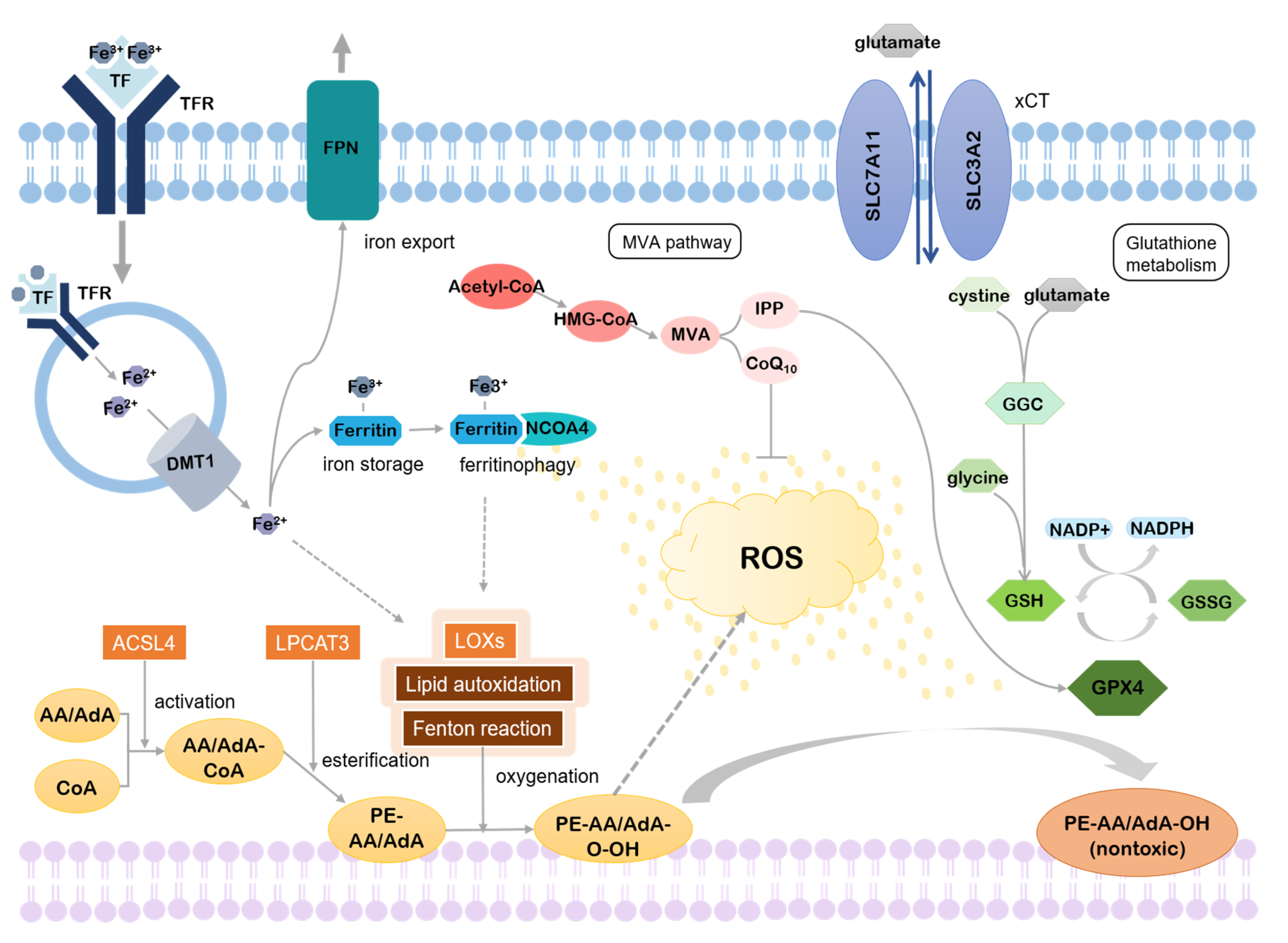
2. Mechanism of Ferroptosis
2.1. Lipid Metabolism
2.1.1. Accumulation of Lipid Peroxides
2.1.2. Inhibition of Lipid Peroxides Reduction
2.2. Iron Metabolism
2.3. Others
3. Association with Neurological Diseases
4. Brain Development
5. Ferroptosis in Brain Development
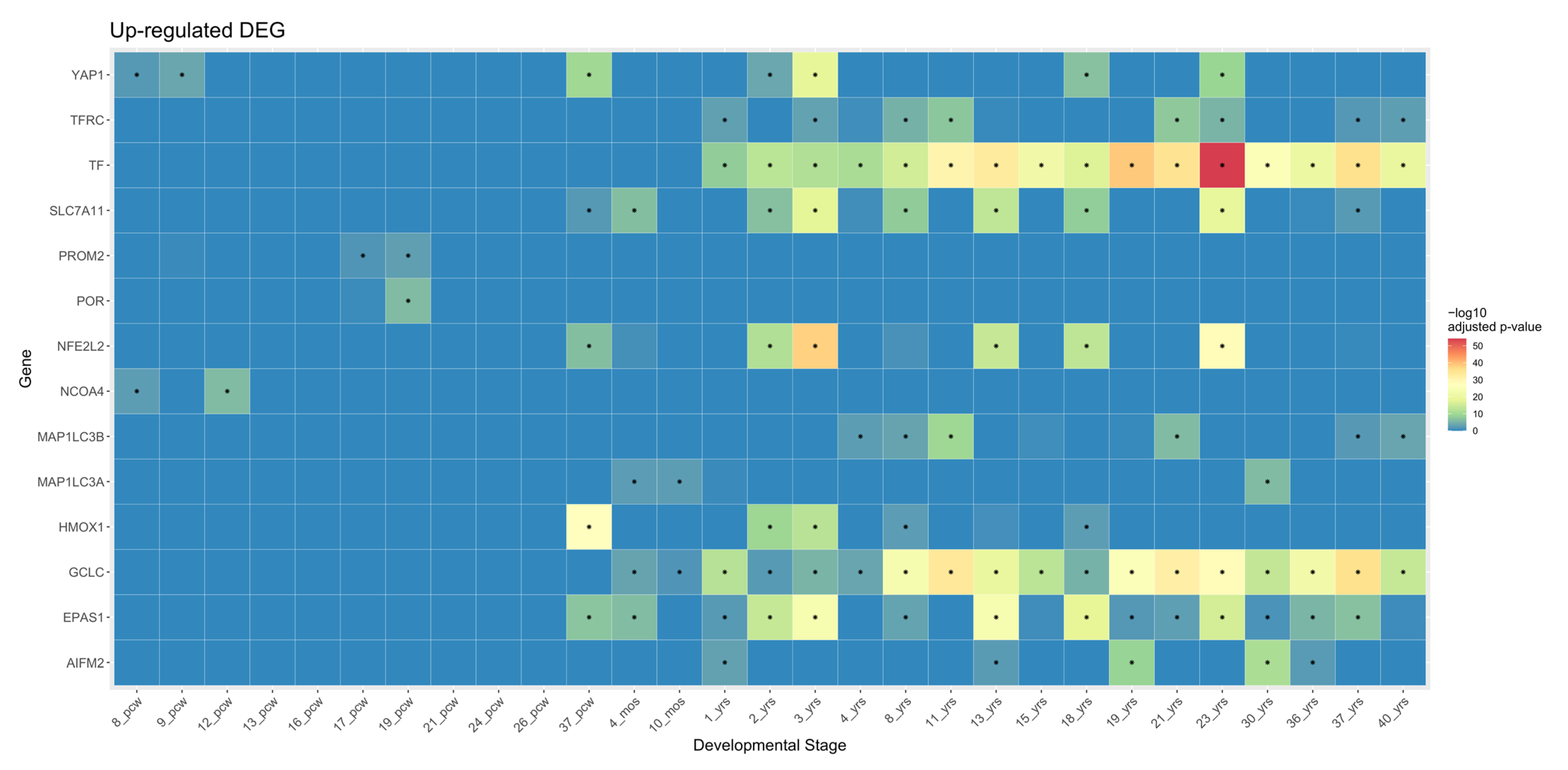
5.1. Ferroptosis Genes in Cortical Development
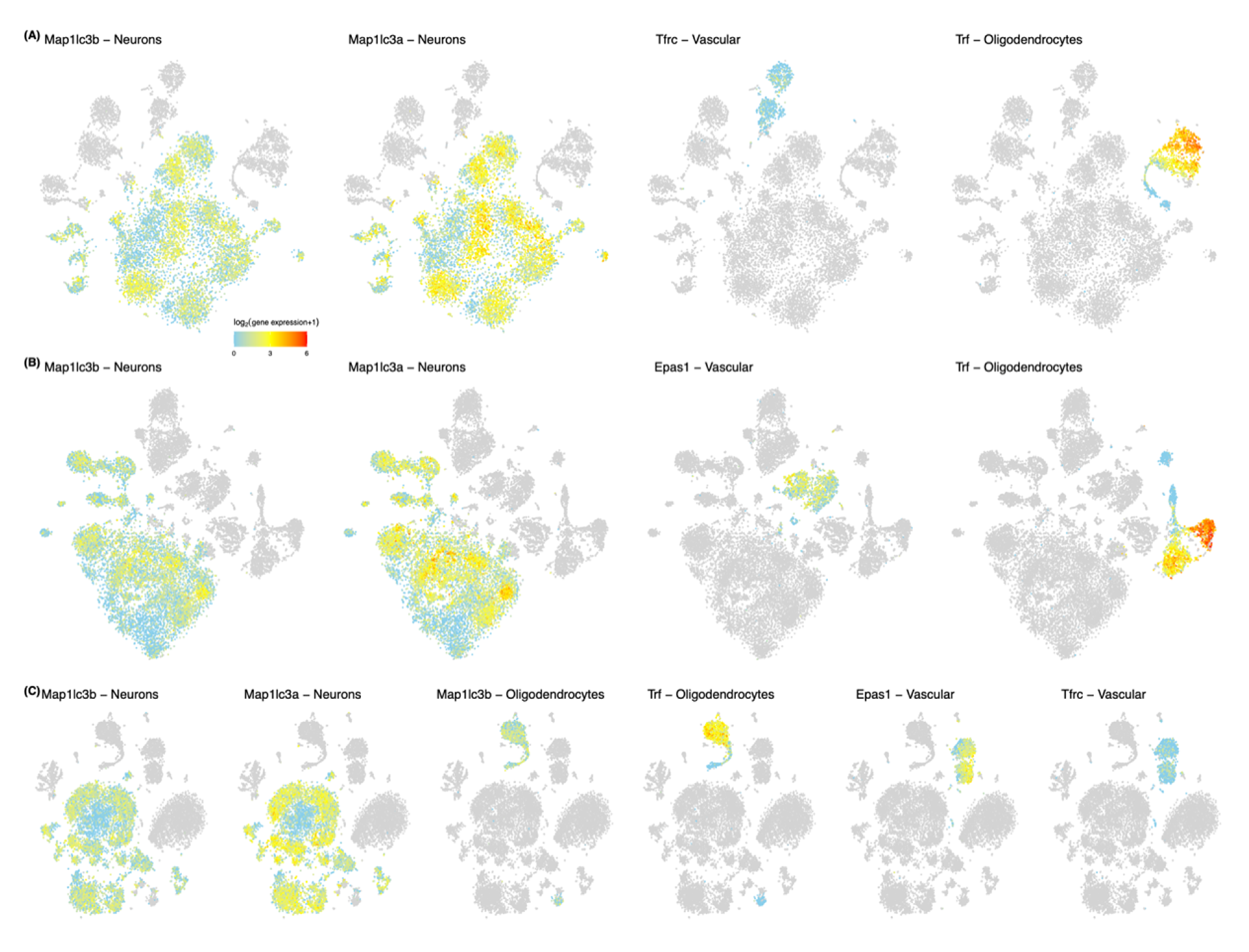
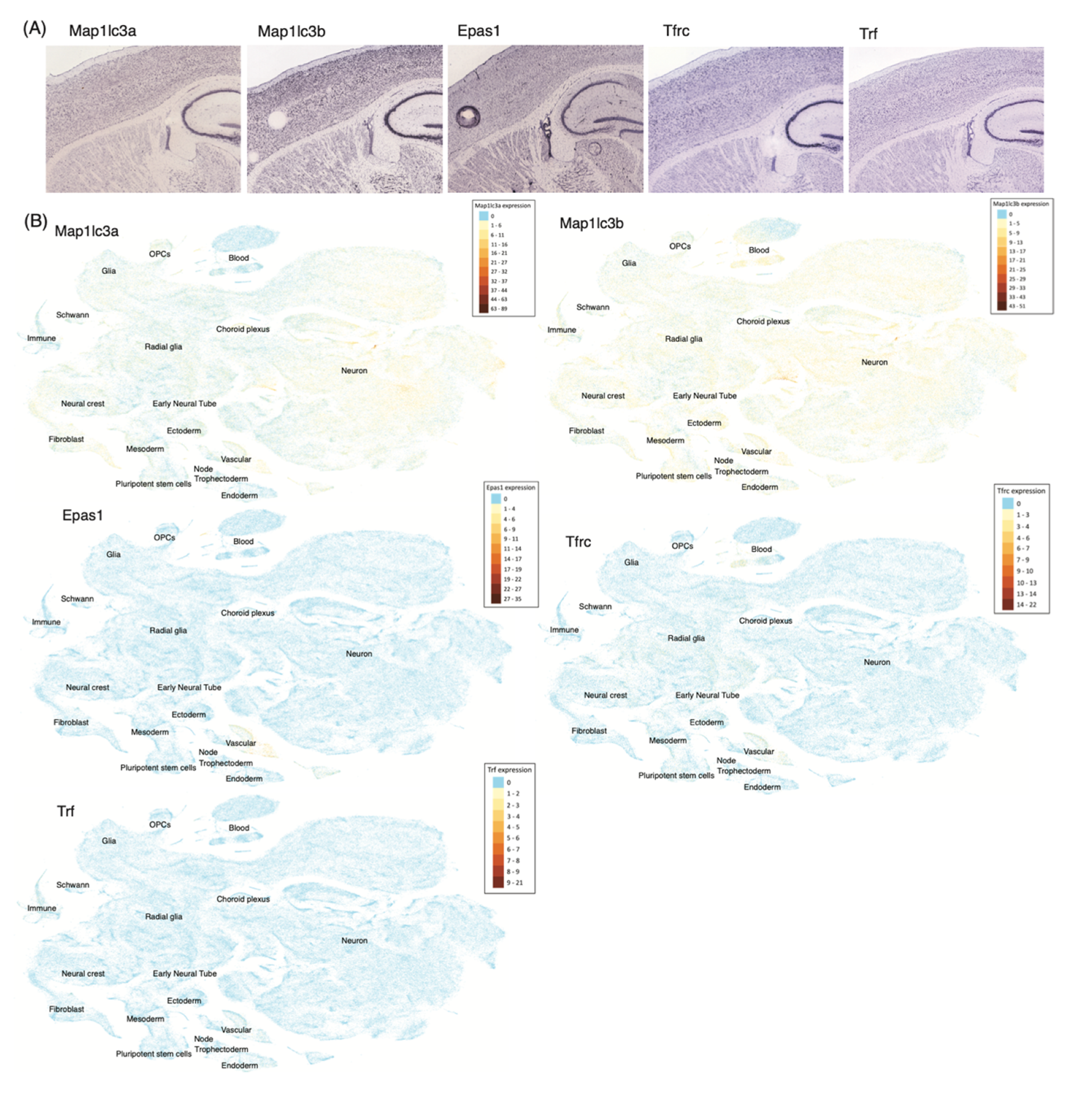
5.2. Ferroptosis Genes in Neurons and Non-Neuronal Cells
5.3. Functional Annotation of Ferroptosis Genes in Brain Development
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Yang, W.S.; Sriramaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Lewerenz, J.; Ates, G.; Methner, A.; Conrad, M.; Maher, P. Oxytosis/ferroptosis-(Re-) emerging roles for oxidative stress-dependent non-apoptotic cell death in diseases of the central nervous system. Front. Neurosci. 2018, 12, 214. [Google Scholar] [CrossRef] [PubMed]
- Praticò, D.; Sung, S. Lipid Peroxidation and Oxidative imbalance: Early functional events in Alzheimer’s disease. J. Alzheimer’s Dis. 2004, 6, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Castellani, R.J.; Moreira, P.I.; Liu, G.; Dobson, J.; Perry, G.; Smith, M.A.; Zhu, X. Iron: The redox-active center of oxidative stress in Alzheimer disease. Neurochem. Res. 2007, 32, 1640–1645. [Google Scholar] [CrossRef] [PubMed]
- Ayton, S.; Lei, P.; Duce, J.A.; Wong, B.X.W.; Sedjahtera, A.; Adlard, P.A.; Bush, A.I.; Finkelstein, D.I. Ceruloplasmin dysfunction and therapeutic potential for Parkinson disease. Ann. Neurol. 2013, 73, 554–559. [Google Scholar] [CrossRef]
- Lei, P.; Ayton, S.; Appukuttan, A.T.; Volitakis, I.; Adlard, P.A.; Finkelstein, D.I.; Bush, A.I. Clioquinol rescues Parkinsonism and dementia phenotypes of the tau knockout mouse. Neurobiol. Dis. 2015, 81, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.D.; Sbodio, J.I.; Xu, R.; Vandiver, M.S.; Cha, J.Y.; Snowman, A.M.; Snyder, S.H. Cystathionine γ-lyase deficiency mediates neurodegeneration in Huntington’s disease. Nature 2014, 509, 96–100. [Google Scholar] [CrossRef]
- Skouta, R.; Dixon, S.J.; Wang, J.; Dunn, D.E.; Orman, M.; Shimada, K.; Rosenberg, P.A.; Lo, D.C.; Weinberg, J.M.; Linkermann, A.; et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J. Am. Chem. Soc. 2014, 136, 4551–4556. [Google Scholar] [CrossRef]
- Kenny, E.M.; Fidan, E.; Yang, Q.; Anthonymuthu, T.S.; New, L.A.; Meyer, E.A.; Wang, H.; Kochanek, P.M.; Dixon, C.E.; Kagan, V.E.; et al. Ferroptosis Contributes to Neuronal Death and Functional Outcome after Traumatic Brain Injury. Crit. Care Med. 2019, 47, 410–418. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Li, Q.; Han, X.; Lan, X.; Gao, Y.; Wan, J.; Durham, F.; Cheng, T.; Yang, J.; Wang, Z.; Jiang, C.; et al. Inhibition of neuronal ferroptosis protects hemorrhagic brain. JCI Insight 2017, 2, e90777. [Google Scholar] [CrossRef] [PubMed]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Porto Freitas, F.; Seibt, T.; et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 2018, 172, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Lei, P.; Bai, T.; Sun, Y. Mechanisms of ferroptosis and relations with regulated cell death: A review. Front. Physiol. 2019, 10, 139. [Google Scholar] [CrossRef]
- Chu, B.; Kon, N.; Chen, D.; Li, T.; Liu, T.; Jiang, L.; Song, S.; Tavana, O.; Gu, W. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat. Cell Biol. 2019, 21, 579–591. [Google Scholar] [CrossRef]
- Ou, Y.; Wang, S.J.; Li, D.; Chu, B.; Gu, W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. USA 2016, 113, E6806–E6812. [Google Scholar] [CrossRef]
- Feng, H.; Schorpp, K.; Jin, J.; Yozwiak, C.E.; Hoffstrom, B.G.; Decker, A.M.; Rajbhandari, P.; Stokes, M.E.; Bender, H.G.; Csuka, J.M.; et al. Transferrin Receptor Is a Specific Ferroptosis Marker. Cell Rep. 2020, 30, 3411–3423. [Google Scholar] [CrossRef]
- Xie, Y.; Zhu, S.; Song, X.; Sun, X.; Fan, Y.; Liu, J.; Zhong, M.; Yuan, H.; Zhang, L.; Billiar, T.R.; et al. The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep. 2017, 20, 1692–1704. [Google Scholar] [CrossRef]
- Kwon, M.Y.; Park, E.; Lee, S.J.; Chung, S.W. Heme oxygenase-1 accelerates erastin-induced ferroptotic cell death. Oncotarget 2015, 6, 24393–24403. [Google Scholar] [CrossRef]
- Yang, Y.; Luo, M.; Zhang, K.; Zhang, J.; Gao, T.; Connell, D.O.; Yao, F.; Mu, C.; Cai, B.; Shang, Y.; et al. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Luo, M.; Yao, F.; Wang, S.; Yuan, Z.; Yang, Y. Ceruloplasmin suppresses ferroptosis by regulating iron homeostasis in hepatocellular carcinoma cells. Cell. Signal. 2020, 72, 109633. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Chang, S.Y.; Wu, Q.; Gou, Y.J.; Jia, L.; Cui, Y.M.; Yu, P.; Shi, Z.H.; Wu, W.S.; Gao, G.; et al. The protective role of mitochondrial ferritin on erastin-induced ferroptosis. Front. Aging Neurosci. 2016, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Kagan, V.E.; Mao, G.; Qu, F.; Angeli, J.P.F.; Doll, S.; Croix, C.S.; Dar, H.H.; Liu, B.; Tyurin, V.A.; Ritov, V.B.; et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017, 13, 81–90. [Google Scholar] [CrossRef]
- Toyokuni, S.; Ito, F.; Yamashita, K.; Okazaki, Y.; Akatsuka, S. Iron and thiol redox signaling in cancer: An exquisite balance to escape ferroptosis. Free Radic. Biol. Med. 2017, 108, 610–626. [Google Scholar] [CrossRef] [PubMed]
- Barelli, H.; Antonny, B. Lipid unsaturation and organelle dynamics. Curr. Opin. Cell Biol. 2016, 41, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Kim, K.J.; Gaschler, M.M.; Patel, M.; Shchepinov, M.S.; Stockwell, B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. USA 2016, 113, E4966–E4975. [Google Scholar] [CrossRef]
- Shah, R.; Shchepinov, M.S.; Pratt, D.A. Resolving the Role of Lipoxygenases in the Initiation and Execution of Ferroptosis. ACS Cent. Sci. 2018, 4, 387–396. [Google Scholar] [CrossRef]
- Soupene, E.; Fyrst, H.; Kuypers, F.A. Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Natl. Acad. Sci. USA 2008, 105, 88–93. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016, 26, 165–176. [Google Scholar] [CrossRef]
- Doll, S.; Proneth, B.; Tyurina, Y.Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, J.; Aichler, M.; Walch, A.; et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Winter, G.E.; Musavi, L.S.; Lee, E.D.; Snijder, B.; Rebsamen, M.; Superti-Furga, G.; Stockwell, B.R. Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem. Biol. 2015, 10, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Maillard, B.; Ingold, K.U.; Scaiano, J.C. Rate Constants for the Reactions of Free Radicals with Oxygen in Solution. J. Am. Chem. Soc. 1983, 105, 5095–5099. [Google Scholar] [CrossRef]
- Frankel, E.N. Free radical oxidation. In Lipid Oxidation; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Raefsky, S.M.; Furman, R.; Milne, G.; Pollock, E.; Axelsen, P.; Mattson, M.P.; Shchepinov, M.S. Deuterated polyunsaturated fatty acids reduce brain lipid peroxidation and hippocampal amyloid β-peptide levels, without discernable behavioral effects in an APP/PS1 mutant transgenic mouse model of Alzheimer’s disease. Neurobiol. Aging 2018, 66, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohé, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Tamba, M.; Ishii, T.; Bannai, S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 1999, 274, 11455–11458. [Google Scholar] [CrossRef]
- Shimada, K.; Stockwell, B.R. tRNA synthase suppression activates de novo cysteine synthesis to compensate for cystine and glutathione deprivation during ferroptosis. Mol. Cell. Oncol. 2016, 3, e1091059. [Google Scholar] [CrossRef][Green Version]
- Shimada, K.; Hayano, M.; Pagano, N.C.; Stockwell, B.R. Cell-Line Selectivity Improves the Predictive Power of Pharmacogenomic Analyses and Helps Identify NADPH as Biomarker for Ferroptosis Sensitivity. Cell Chem. Biol. 2016, 23, 225–235. [Google Scholar] [CrossRef]
- Warner, G.J.; Berry, M.J.; Moustafa, M.E.; Carlson, B.A.; Hatfield, D.L.; Faust, J.R. Inhibition of selenoprotein synthesis by selenocysteine tRNA([Ser]Sec) lacking isopentenyladenosine. J. Biol. Chem. 2000, 276, 28110–28119. [Google Scholar] [CrossRef]
- Cardoso, B.R.; Hare, D.J.; Bush, A.I.; Roberts, B.R. Glutathione peroxidase 4: A new player in neurodegeneration? Mol. Psychiatry 2017, 22, 328–335. [Google Scholar] [CrossRef]
- Hayano, M.; Yang, W.S.; Corn, C.K.; Pagano, N.C.; Stockwell, B.R. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016, 23, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Kryukov, G.V.; Castellano, S.; Novoselov, S.V.; Lobanov, A.V.; Zehtab, O.; Guigó, R.; Gladyshev, V.N. Characterization of mammalian selenoproteomes. Science 2003, 300, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Skouta, R.; Kaplan, A.; Yang, W.S.; Hayano, M.; Dixon, S.J.; Brown, L.M.; Valenzuela, C.A.; Wolpaw, A.J.; Stockwell, B.R. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 2016, 12, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Kasai, H. Analysis of a form of oxidative DNA damage, 8-hydroxy-2’-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat. Res. Rev. Mutat. Res. 1997, 387, 147–163. [Google Scholar] [CrossRef]
- Toyokuni, S. Iron and carcinogenesis: From Fenton reaction to target genes. Redox Rep. 2002, 7, 189–197. [Google Scholar] [CrossRef]
- Gao, M.; Monian, P.; Quadri, N.; Ramasamy, R.; Jiang, X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol. Cell 2015, 59, 298–308. [Google Scholar] [CrossRef]
- Yang, W.S.; Stockwell, B.R. Synthetic Lethal Screening Identifies Compounds Activating Iron-Dependent, Nonapoptotic Cell Death in Oncogenic-RAS-Harboring Cancer Cells. Chem. Biol. 2008, 15, 234–245. [Google Scholar] [CrossRef]
- Gao, M.; Monian, P.; Pan, Q.; Zhang, W.; Xiang, J.; Jiang, X. Ferroptosis is an autophagic cell death process. Cell Res. 2016, 26, 1021–1032. [Google Scholar] [CrossRef]
- Hou, W.; Xie, Y.; Song, X.; Sun, X.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 2016, 12, 1425–1428. [Google Scholar] [CrossRef]
- Mancias, J.D.; Wang, X.; Gygi, S.P.; Harper, J.W.; Kimmelman, A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014, 509, 105–109. [Google Scholar] [CrossRef]
- Yuan, H.; Li, X.; Zhang, X.; Kang, R.; Tang, D. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem. Biophys. Res. Commun. 2016, 478, 838–844. [Google Scholar] [CrossRef]
- Fan, Z.; Wirth, A.-K.; Chen, D.; Wruck, C.J.; Rauh, M.; Buchfelder, M.; Savaskan, N. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis 2017, 6, e371. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Bersuker, K.; Hendricks, J.M.; Li, Z.; Magtanong, L.; Ford, B.; Tang, P.H.; Roberts, M.A.; Tong, B.; Maimone, T.J.; Zoncu, R.; et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019, 575, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Doll, S.; Freitas, F.P.; Shah, R.; Aldrovandi, M.; da Silva, M.C.; Ingold, I.; Grocin, A.G.; Xavier da Silva, T.N.; Panzilius, E.; Scheel, C.H.; et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 2019, 575, 693–698. [Google Scholar] [CrossRef]
- Grazia Cotticelli, M.; Xia, S.; Lin, D.; Lee, T.; Terrab, L.; Wipf, P.; Huryn, D.M.; Wilson, R.B. Ferroptosis as a novel therapeutic target for Friedreich’s ataxia. J. Pharmacol. Exp. Ther. 2019, 369, 47–54. [Google Scholar] [CrossRef]
- Johnson, W.M.; Wilson-Delfosse, A.L.; Mieyal, J.J. Dysregulation of glutathione homeostasis in neurodegenerative diseases. Nutrients 2012, 4, 1399–1440. [Google Scholar] [CrossRef]
- Hancock, D.B.; Markunas, C.A.; Bierut, L.J.; Johnson, E.O. Human Genetics of Addiction: New Insights and Future Directions. Curr. Psychiatry Rep. 2018, 20, 8. [Google Scholar] [CrossRef]
- Kutalik, Z.; Benyamin, B.; Bergmann, S.; Mooser, V.; Waeber, G.; Montgomery, G.W.; Martin, N.G.; Madden, P.A.F.; Heath, A.C.; Beckmann, J.S.; et al. Genome-wide association study identifies two loci strongly affecting transferrin glycosylation. Hum. Mol. Genet. 2011, 20, 3710–3717. [Google Scholar] [CrossRef] [PubMed]
- Skonieczna-Zydecka, K.; Jamiol-Milc, D.; Borecki, K.; Stachowska, E.; Zabielska, P.; Kaminska, M.; Karakiewicz, B. The Prevalence of Insomnia and the Link between Iron Metabolism Genes Polymorphisms, TF rs1049296 C>T, TF rs3811647 G>A, TFR rs7385804 A>C, HAMP rs10421768 A>G and Sleep Disorders in Polish Individuals with ASD. Int. J. Environ. Res. Public Health 2020, 17, 400. [Google Scholar] [CrossRef] [PubMed]
- Mills, N.T.; Maier, R.; Whitfield, J.B.; Wright, M.J.; Colodro-Conde, L.; Byrne, E.M.; Scott, J.G.; Byrne, G.J.; Hansell, N.K.; Vinkhuyzen, A.A.E.; et al. Investigating the relationship between iron and depression. J. Psychiatr. Res. 2017, 94, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Grubman, A.; Chew, G.; Ouyang, J.F.; Sun, G.; Choo, X.Y.; McLean, C.; Simmons, R.; Buckberry, S.; Landin, D.V.; Pflueger, J.; et al. A single cell brain atlas in human Alzheimer’s disease. bioRxiv 2019. [Google Scholar] [CrossRef]
- Moradifard, S.; Hoseinbeyki, M.; Ganji, S.M.; Minuchehr, Z. Analysis of microRNA and Gene Expression Profiles in Alzheimer’s Disease: A Meta-Analysis Approach. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef]
- Ikiz, B.; Alvarez, M.J.; Ré, D.B.; Le Verche, V.; Politi, K.; Lotti, F.; Phani, S.; Pradhan, R.; Yu, C.; Croft, G.F.; et al. The Regulatory Machinery of Neurodegeneration in In Vitro Models of Amyotrophic Lateral Sclerosis. Cell Rep. 2015, 12, 335–345. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Daly, M.J.; Ripke, S.; Lewis, C.M.; Lin, D.Y.; Wray, N.R.; Neale, B.; Levinson, D.F.; Breen, G.; Byrne, E.M.; et al. A mega-Analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry 2013, 18, 497–511. [Google Scholar] [CrossRef]
- Vojinovic, D.; Adams, H.H.; Jian, X.; Yang, Q.; Smith, A.V.; Bis, J.C.; Teumer, A.; Scholz, M.; Armstrong, N.J.; Hofer, E.; et al. Genome-wide association study of 23,500 individuals identifies 7 loci associated with brain ventricular volume. Nat. Commun. 2018, 9, 3945. [Google Scholar] [CrossRef]
- Zuo, L.; Zhang, C.K.; Sayward, F.G.; Cheung, K.H.; Wang, K.; Krystal, J.H.; Zhao, H.; Luo, X. Gene-based and pathway-based genome-wide association study of alcohol dependence. Shanghai Arch. Psychiatry 2015, 27, 111–118. [Google Scholar]
- Vallerga, C.L.; Zhang, F.; Fowdar, J.; McRae, A.F.; Qi, T.; Nabais, M.F.; Zhang, Q.; Kassam, I.; Henders, A.K.; Wallace, L.; et al. Analysis of DNA methylation associates the cystine–glutamate antiporter SLC7A11 with risk of Parkinson’s disease. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Feng, X.; Bao, R.; Li, L.; Deisenhammer, F.; Arnason, B.G.W.; Reder, A.T. Interferon-β corrects massive gene dysregulation in multiple sclerosis: Short-term and long-term effects on immune regulation and neuroprotection: Short running title: Interferon-β corrects gene dysregulation in multiple sclerosis. EBioMedicine 2019, 49, 269–283. [Google Scholar] [CrossRef] [PubMed]
- Bishop, K.M.; Rubenstein, J.L.R.; O’Leary, D.D.M. Distinct actions of Emx1, Emx2, and Pax6 in regulating the specification of areas in the developing neocortex. J. Neurosci. 2002, 22, 7627–7638. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, T.; Leingärtner, A.; Ringstedt, T.; O’Leary, D.D.M. EMX2 regulates sizes and positioning of the primary sensory and motor areas in neocortex by direct specification of cortical progenitors. Neuron 2004, 43, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Rakic, P. Evolution of the neocortex: A perspective from developmental biology. Nat. Rev. Neurosci. 2009, 10, 724–735. [Google Scholar] [CrossRef]
- Rajagopalan, V.; Scott, J.; Habas, P.A.; Kim, K.; Corbett-Detig, J.; Rousseau, F.; Barkovich, A.J.; Glenn, O.A.; Studholme, C. Local tissue growth patterns underlying normal fetal human brain gyrification quantified in utero. J. Neurosci. 2011, 31, 2878–2887. [Google Scholar] [CrossRef]
- Voytek, B.; Kayser, A.S.; Badre, D.; Fegen, D.; Chang, E.F.; Crone, N.E.; Parvizi, J.; Knight, R.T.; D’Esposito, M. Oscillatory dynamics coordinating human frontal networks in support of goal maintenance. Nat. Neurosci. 2015, 18, 1318–1324. [Google Scholar] [CrossRef]
- Gazzaley, A.; Nobre, A.C. Top-down modulation: Bridging selective attention and working memory. Trends Cogn. Sci. 2012, 16, 129–135. [Google Scholar] [CrossRef]
- Feredoes, E.; Heinen, K.; Weiskopf, N.; Ruff, C.; Driver, J. Causal evidence for frontal involvement in memory target maintenance by posterior brain areas during distracter interference of visual working memory. Proc. Natl. Acad. Sci. USA 2011, 108, 17510–17515. [Google Scholar] [CrossRef]
- Jung, R.E.; Haier, R.J. The Parieto-Frontal Integration Theory (P-FIT) of intelligence: Converging neuroimaging evidence. Behav. Brain Sci. 2007, 30, 135–154. [Google Scholar] [CrossRef]
- Curtis, C.E.; D’Esposito, M. Persistent activity in the prefrontal cortex during working memory. Trends Cogn. Sci. 2003, 7, 415–423. [Google Scholar] [CrossRef]
- García-Cabezas, M.Á.; Zikopoulos, B.; Barbas, H. The Structural Model: A theory linking connections, plasticity, pathology, development and evolution of the cerebral cortex. Brain Struct. Funct. 2019, 224, 985–1008. [Google Scholar] [CrossRef]
- Cooper, J.A. A mechanism for inside-out lamination in the neocortex. Trends Neurosci. 2008, 31, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, B.; Parnavelas, J.G. Modes of neuronal migration in the developing cerebral cortex. Nat. Rev. Neurosci. 2002, 3, 423–432. [Google Scholar] [CrossRef]
- Rakic, P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 1972, 145, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Noctor, S.C.; Flint, A.C.; Weissman, T.A.; Dammerman, R.S.; Kriegstein, A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 2001, 409, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.A.; Ding, S.L.; Sunkin, S.M.; Smith, K.A.; Ng, L.; Szafer, A.; Ebbert, A.; Riley, Z.L.; Royall, J.J.; Aiona, K.; et al. Transcriptional landscape of the prenatal human brain. Nature 2014, 508, 199–206. [Google Scholar] [CrossRef]
- Kang, H.J.; Kawasawa, Y.I.; Cheng, F.; Zhu, Y.; Xu, X.; Li, M.; Sousa, A.M.M.; Pletikos, M.; Meyer, K.A.; Sedmak, G.; et al. Spatio-temporal transcriptome of the human brain. Nature 2011, 478, 483–489. [Google Scholar] [CrossRef]
- Werling, D.M.; Pochareddy, S.; Choi, J.; An, J.Y.; Sheppard, B.; Peng, M.; Li, Z.; Dastmalchi, C.; Santpere, G.; Sousa, A.M.M.; et al. Whole-Genome and RNA Sequencing Reveal Variation and Transcriptomic Coordination in the Developing Human Prefrontal Cortex. Cell Rep. 2020, 31, 107489. [Google Scholar] [CrossRef]
- Uchida, M.; Sugaya, M.; Kanamaru, T.; Hisatomi, H. Alternative RNA splicing in expression of the glutathione synthetase gene in human cells. Mol. Biol. Rep. 2010, 37, 2105–2109. [Google Scholar] [CrossRef]
- Bloch, B.; Popovici, T.; Levin, M.J.; Tuil, D.; Kahn, A. Transferrin gene expression visualized in oligodendrocytes of the rat brain by using in situ hybridization and immunohistochemistry. Proc. Natl. Acad. Sci. USA 1985, 82, 6706–6710. [Google Scholar] [CrossRef]
- Connor, J.R.; Fine, R.E. The distribution of transferrin immunoreactivity in the rat central nervous system. Brain Res. 1986, 368, 319–328. [Google Scholar] [CrossRef]
- Burdo, J.R.; Antonetti, D.A.; Wolpert, E.B.; Connor, J.R. Mechanisms and regulation of transferrin and iron transport in a model blood-brain barrier system. Neuroscience 2003, 121, 883–890. [Google Scholar] [CrossRef]
- Gunshin, H.; Mackenzie, B.; Berger, U.V.; Gunshin, Y.; Romero, M.F.; Boron, W.F.; Nussberger, S.; Gollan, J.L.; Hediger, M.A. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 1997, 388, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Burdo, J.R.; Menzies, S.L.; Simpson, I.A.; Garrick, L.M.; Garrick, M.D.; Dolan, K.G.; Haile, D.J.; Beard, J.L.; Connor, J.R. Distribution of Divalent Metal Transporter 1 and Metal Transport Protein 1 in the normal and Belgrade rat. J. Neurosci. Res. 2001, 66, 1198–1207. [Google Scholar] [CrossRef]
- Moos, T. Immunohistochemical localization of intraneuronal transferrin receptor immunoreactivity in the adult mouse central nervous system. J. Comp. Neurol. 1996. [Google Scholar] [CrossRef]
- Connor, J.R.; Menzies, S.L. Cellular management of iron in the brain. J. Neurol. Sci. 1995, 134, 33–44. [Google Scholar] [CrossRef]
- Connor, J.R.; Ponnuru, P.; Wang, X.S.; Patton, S.M.; Allen, R.P.; Earley, C.J. Profile of altered brain iron acquisition in restless legs syndrome. Brain 2011, 134, 959–968. [Google Scholar] [CrossRef]
- Thomas, T.; Schreiber, G.; Jaworowski, A. Developmental patterns of gene expression of secreted proteins in brain and choroid plexus. Dev. Biol. 1989, 134, 38–47. [Google Scholar] [CrossRef]
- Zhong, M.Q.; To, Y.; Pak, L.T.; You, M.F. Transferrin receptors on the plasma membrane of cultured rat astrocytes. Exp. Brain Res. 1999, 129, 0473–0476. [Google Scholar] [CrossRef]
- Hoepken, H.H.; Korten, T.; Robinsont, S.R.; Dringen, R. Iron accumulation, iron-mediated toxicity and altered levels of ferritin and transferrin receptor in cultured astrocytes during incubation with ferric ammonium citrate. J. Neurochem. 2004, 88, 1194–1202. [Google Scholar] [CrossRef]
- De los Monteros, A.E.; Peña, L.A.; de Vellis, J. Does transferrin have a special role in the nervous system? J. Neurosci. Res. 1989, 24, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Ling, E.A. Increased expression of transferrin receptors and iron in amoeboid microglial cells in postnatal rats following an exposure to hypoxia. Neurosci. Lett. 1999, 262, 183–186. [Google Scholar] [CrossRef]
- Moos, T.; Nielsen, T.R.; Skjørringe, T.; Morgan, E.H. Iron trafficking inside the brain. J. Neurochem. 2007, 103, 1730–1740. [Google Scholar] [CrossRef] [PubMed]
- Rouault, T.A.; Cooperman, S. Brain Iron Metabolism. Semin. Pediatr. Neurol. 2006, 13, 142–148. [Google Scholar] [CrossRef]
- Madsen, E.; Gitlin, J.D. Copper and iron disorders of the brain. Annu. Rev. Neurosci. 2007, 30, 317–337. [Google Scholar] [CrossRef]
- Tian, H.; McKnight, S.L.; Russell, D.W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997, 11, 72–82. [Google Scholar] [CrossRef]
- Maxwell, P.H.; Wlesener, M.S.; Chang, G.W.; Clifford, S.C.; Vaux, E.C.; Cockman, M.E.; Wykoff, C.C.; Pugh, C.W.; Maher, E.R.; Ratcliffe, P.J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999, 399, 271–275. [Google Scholar] [CrossRef]
- Bruick, R.K.; McKnight, S.L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 2001, 294, 1337–1340. [Google Scholar] [CrossRef]
- Epstein, A.C.R.; Gleadle, J.M.; McNeill, L.A.; Hewitson, K.S.; O’Rourke, J.; Mole, D.R.; Mukherji, M.; Metzen, E.; Wilson, M.I.; Dhanda, A.; et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 2001, 107, 43–54. [Google Scholar] [CrossRef]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, J. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef]
- Jaakkola, P.; Mole, D.R.; Tian, Y.M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; Von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.Y.; Tsai, M.Y.; Tseng, W.F.; Cheng, C.H.; Huang, C.R.; Wu, J.S.; Chung, H.Y.; Hsieh, C.S.; Sun, C.K.; Hwang, S.P.L.; et al. Integration of CNS survival and differentiation by HIF2α. Cell Death Differ. 2011, 18, 1757–1770. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smeyne, M.; Sladen, P.; Jiao, Y.; Dragatsis, I.; Smeyne, R.J. HIF1α is necessary for exercise-induced neuroprotection while HIF2α is needed for dopaminergic neuron survival in the substantia nigra pars compacta. Neuroscience 2015, 295, 23–38. [Google Scholar] [CrossRef]
- Sian, J.; Dexter, D.T.; Lees, A.J.; Daniel, S.; Jenner, P.; Marsden, C.D. Glutathione-related enzymes in brain in Parkinson’s disease. Ann. Neurol. 1994, 36, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Labak, M.; Foniok, T.; Kirk, D.; Rushforth, D.; Tomanek, B.; Jasiński, A.; Grieb, P. Metabolic changes in rat brain following intracerebroventricular injections of streptozotocin: A model of sporadic Alzheimer’s disease. In Proceedings of the Acta Neurochirurgica, Supplementum; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Lanius, R.A.; Shaw, C.A.; Wagey, R.; Krieger, C. Characterization, Distribution, and Protein Kinase C-Mediated Regulation of [35S]Glutathione Binding Sites in Mouse and Human Spinal Cord. J. Neurochem. 1994, 63, 155–160. [Google Scholar] [CrossRef]
- Chinta, S.J.; Kumar, M.J.; Hsu, M.; Rajagopalan, S.; Kaur, D.; Rane, A.; Nicholls, D.G.; Choi, J.; Andersen, J.K. Inducible alterations of glutathione levels in adult dopaminergic midbrain neurons result in nigrostriatal degeneration. J. Neurosci. 2007, 27, 13997–14006. [Google Scholar] [CrossRef]
- Garrido, M.; Tereshchenko, Y.; Zhevtsova, Z.; Taschenberger, G.; Bähr, M.; Kügler, S. Glutathione depletion and overproduction both initiate degeneration of nigral dopaminergic neurons. Acta Neuropathol. 2011, 121, 475–485. [Google Scholar] [CrossRef]
- Feng, W.; Rosca, M.; Fan, Y.; Hu, Y.; Feng, P.; Lee, H.G.; Monnier, V.M.; Fan, X. Gclc deficiency in mouse CNS causes mitochondrial damage and neurodegeneration. Hum. Mol. Genet. 2017, 26, 1376–1390. [Google Scholar] [CrossRef]
- Kuusisto, E.; Salminen, A.; Alafuzoff, I. Ubiquitin-binding protein p62 is present in neuronal and glial inclusions in human tauopathies and synucleinopathies. Neuroreport 2001, 12, 2085–2090. [Google Scholar] [CrossRef]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Higashi, S.; Moore, D.J.; Minegishi, M.; Kasanuki, K.; Fujishiro, H.; Kabuta, T.; Togo, T.; Katsuse, O.; Uchikado, H.; Furukawa, Y.; et al. Localization of MAP1-LC3 in vulnerable neurons and lewy bodies in brains of patients with dementia with lewy bodies. J. Neuropathol. Exp. Neurol. 2011, 70, 264–280. [Google Scholar] [CrossRef] [PubMed]
- Maines, M.D. The heme oxygenase system: A regulator of second messenger gases. Annu. Rev. Pharmacol. Toxicol. 1997, 37, 517–554. [Google Scholar] [CrossRef] [PubMed]
- Panahian, N.; Yoshiura, M.; Maines, M.D. Overexpression of heme oxygenase-1 is neuroprotective in a model of permanent middle cerebral artery occlusion in transgenic mice. J. Neurochem. 1999, 72, 1187–1203. [Google Scholar] [CrossRef] [PubMed]
- Poitelon, Y.; Lopez-Anido, C.; Catignas, K.; Berti, C.; Palmisano, M.; Williamson, C.; Ameroso, D.; Abiko, K.; Hwang, Y.; Gregorieff, A.; et al. YAP and TAZ control peripheral myelination and the expression of laminin receptors in Schwann cells. Nat. Neurosci. 2016, 19, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Hindley, C.J.; Condurat, A.L.; Menon, V.; Thomas, R.; Azmitia, L.M.; Davis, J.A.; Pruszak, J. The Hippo pathway member YAP enhances human neural crest cell fate and migration. Sci. Rep. 2016, 6, 23208. [Google Scholar] [CrossRef] [PubMed]
- Liddell, J.R. Are astrocytes the predominant cell type for activation of Nrf2 in aging and neurodegeneration? Antioxidants 2017, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Rojo, A.I.; Pajares, M.; Rada, P.; Nuñez, A.; Nevado-Holgado, A.J.; Killik, R.; Van Leuven, F.; Ribe, E.; Lovestone, S.; Yamamoto, M.; et al. NRF2 deficiency replicates transcriptomic changes in Alzheimer’s patients and worsens APP and TAU pathology. Redox Biol. 2017, 13, 444–451. [Google Scholar] [CrossRef]
- Calkins, M.J.; Jakel, R.J.; Johnson, D.A.; Chan, K.; Yuen, W.K.; Johnson, J.A. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc. Natl. Acad. Sci. USA 2005, 102, 244–249. [Google Scholar] [CrossRef]
- Johnson, D.A.; Amirahmadi, S.; Ward, C.; Fabry, Z.; Johnson, J.A. The absence of the pro-antioxidant transcription factor Nrf2 exacerbates experimental autoimmune encephalomyelitis. Toxicol. Sci. 2009, 114, 237–246. [Google Scholar] [CrossRef]
- Sasaki, H.; Sato, H.; Kuriyama-Matsumura, K.; Sato, K.; Maebara, K.; Wang, H.; Tamba, M.; Itoh, K.; Yamamoto, M.; Bannai, S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002, 277, 44765–44771. [Google Scholar] [CrossRef]
- Dringen, R.; Gutterer, J.M.; Hirrlinger, J. Glutathione metabolism in brain: Metabolic interaction between astrocytes and neurons in the defense against reactive oxygen species. Eur. J. Biochem. 2000, 267, 4912–4916. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, A.; Hochgerner, H.; Lönnerberg, P.; Johnsson, A.; Memic, F.; van der Zwan, J.; Häring, M.; Braun, E.; Borm, L.E.; La Manno, G.; et al. Molecular Architecture of the Mouse Nervous System. Cell 2018, 174, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Leitner, D.F.; Connor, J.R. Functional roles of transferrin in the brain. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Takeda, N.; Maemura, K.; Imai, Y.; Harada, T.; Kawanami, D.; Nojiri, T.; Manabe, I.; Nagai, R. Endothelial PAS domain protein 1 gene promotes angiogenesis through the transactivation of both vascular endothelial growth factor and its receptor, Flt-1. Circ. Res. 2004, 95, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef] [PubMed]
- La Manno, G.; Siletti, K.; Furlan, A.; Gyllborg, D.; Vinsland, E.; Langseth, C.M.; Khven, I.; Johnsson, A.; Nilsson, M.; Lönnerberg, P.; et al. Molecular architecture of the developing mouse brain. bioRxiv 2020. [Google Scholar] [CrossRef]
- Cougnoux, A.; Yerger, J.C.; Fellmeth, M.; Serra-Vinardell, J.; Martin, K.; Navid, F.; Iben, J.; Wassif, C.A.; Cawley, N.X.; Porter, F.D. Single cell transcriptome analysis of niemann–pick disease, type c1 cerebella. Int. J. Mol. Sci. 2020, 21, 5368. [Google Scholar] [CrossRef]
| Gene Symbol | Trajectory Group | Module Assigned | Module Description | Cellular Group |
|---|---|---|---|---|
| FR-inducing genes | ||||
| NCOA4 | Non- transitional | M5 | RNA processing | No cellular specificity |
| ATF4 | M6 | RNA processing | ||
| VDAC2 | ||||
| VDAC3 | ||||
| POR | M7 | RNA processing | ||
| LPCAT3 | M9 | Chromosome organization | ||
| MAP1LC3B | Postnatal | M16 | Ribose phosphate metabolic process | Excitatory neuron |
| TFRC | ||||
| MAP1LC3A | M17 | Macroautophagy | ||
| HMOX1 | M18 | Immune effector process | Non-neuronal cell | |
| YAP1 | ||||
| EPAS1 | M19 | Synaptic signaling | Excitatory neuron, Non-neuronal cell | |
| TF | ||||
| FR-suppressing genes | ||||
| GSS | Non- transitional | M6 | RNA processing | No cellular specificity |
| PROM2 | M9 | Chromosome organization | ||
| AIFM2 | M12 | Regulation of ion transmembrane transport | ||
| FTL | M14 | Mitochondrion organization | ||
| GPX4 | ||||
| SLC3A2 | ||||
| NFE2L2 | Postnatal | M18 | Immune effector process | Non-neuronal cell |
| SLC7A11 | ||||
| GCLC | M19 | Synaptic signaling | Excitatory neuron, Non-neuronal cell | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-W.; Kim, Y.; Kim, S.E.; An, J.-Y. Ferroptosis-Related Genes in Neurodevelopment and Central Nervous System. Biology 2021, 10, 35. https://doi.org/10.3390/biology10010035
Kim S-W, Kim Y, Kim SE, An J-Y. Ferroptosis-Related Genes in Neurodevelopment and Central Nervous System. Biology. 2021; 10(1):35. https://doi.org/10.3390/biology10010035
Chicago/Turabian StyleKim, Soo-Whee, Yujin Kim, Sung Eun Kim, and Joon-Yong An. 2021. "Ferroptosis-Related Genes in Neurodevelopment and Central Nervous System" Biology 10, no. 1: 35. https://doi.org/10.3390/biology10010035
APA StyleKim, S.-W., Kim, Y., Kim, S. E., & An, J.-Y. (2021). Ferroptosis-Related Genes in Neurodevelopment and Central Nervous System. Biology, 10(1), 35. https://doi.org/10.3390/biology10010035






