Promoting Human Nutrition and Health through Plant Metabolomics: Current Status and Challenges
Abstract
Simple Summary
Abstract
1. Introduction
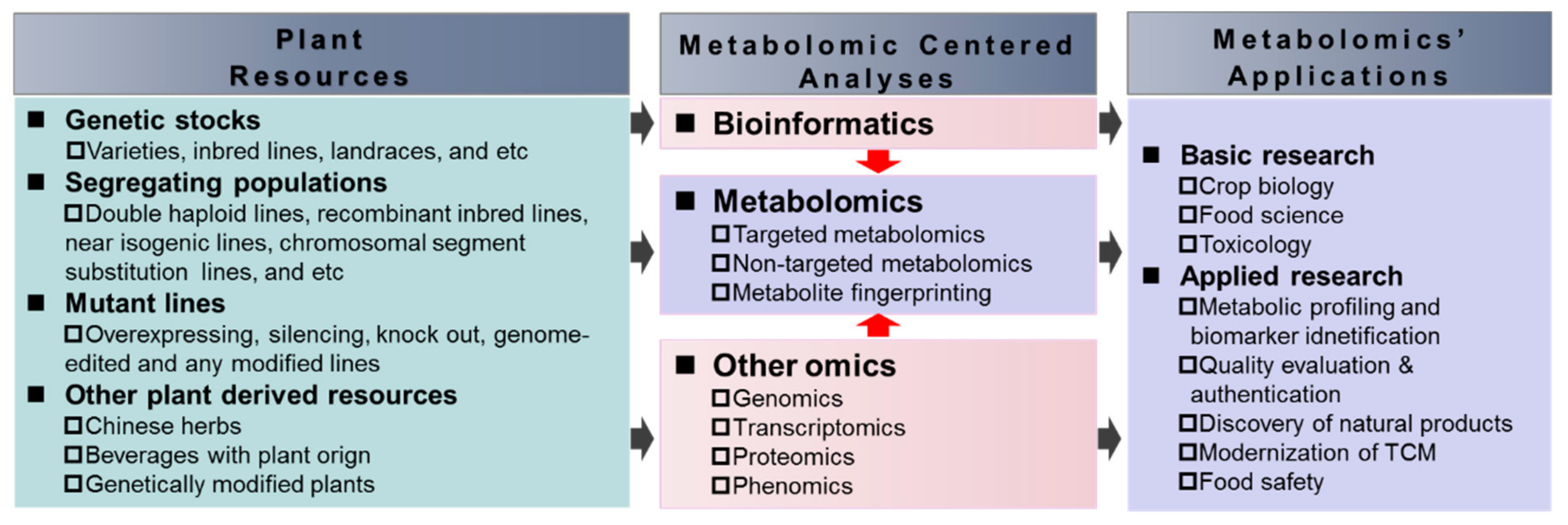
2. Approaches of Metabolomics
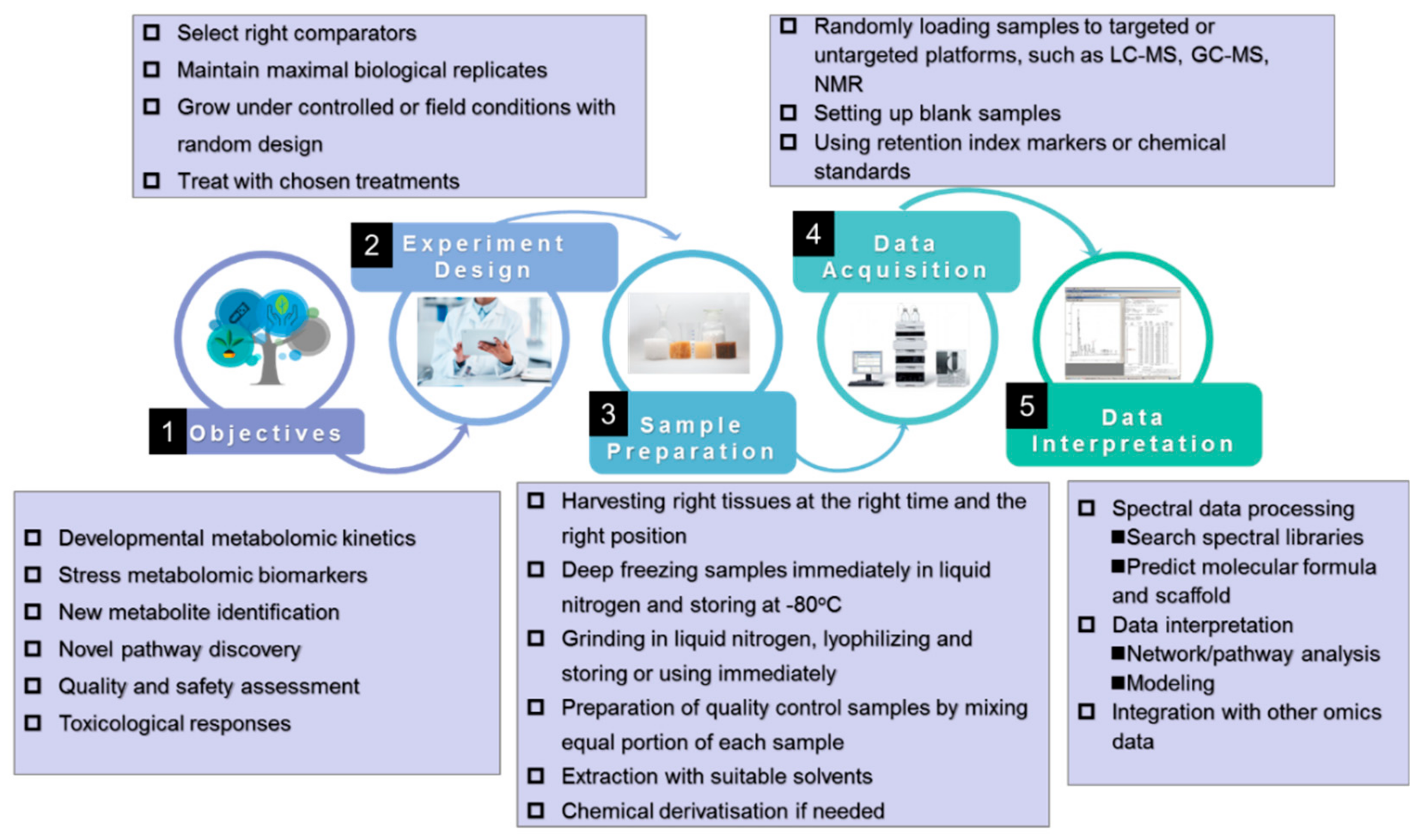
3. Standardization of Plant Metabolomics Studies
4. The Applications of Plant Metabolomics in Crop Improvement for Human Nutrition and Human Health
4.1. Plant Metabolomics and Natural Variations of Nutritional and Quality Relevant Metabolites and Underlying Genetic Mechanisms
4.1.1. Rice
4.1.2. Maize
4.1.3. Soybean
4.1.4. Wheat
4.1.5. Other Crops
4.1.6. Fruits and Vegetables
4.2. Plant Metabolomics and Quality Evaluation and Authentication of Plant-Derived Beverages
4.2.1. Tea
4.2.2. Coffee
4.2.3. Wine
4.3. Plant Metabolomics and the Discovery of Natural Plant Products and the Modernization of TCM
4.4. Plant Metabolomics and GM Food Safety
4.5. Plant Metabolomics and Plant-Derived Food Safety
5. Challenges and Future Perspective
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saito, K.; Matsuda, F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant. Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, C.; Gallage, N.; Hansen, C.I.C.; Møller, B.L.; Laursen, T. Dynamic metabolic solutions to the sessile life style of plants. Nat. Prod. Rep. 2018, 35, 1140–1155. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 2016, 17, 767. [Google Scholar] [CrossRef] [PubMed]
- Tenenboim, H.; Brotman, Y. Omic relief for the biotically stressed: Metabolomics of plant biotic interactions. Trends Plant. Sci. 2016, 21, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D.; Brouwer, I.D.; Fitzgerald, M.A. Plant metabolomics and its potential application for human nutrition. Physiol. Plant. 2007, 132, 162–175. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, J.; Yun, E.J.; Kim, K.H. Food metabolomics: From farm to human. Curr. Opin. Biotechnol. 2016, 37, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mosa, K.A.; Ji, L.; Kage, U.; Dhokane, D.; Karre, S.; Madalageri, D.; Pathania, N. Metabolomics-assisted biotechnological interventions for developing plant-based functional foods and nutraceuticals. Crit. Rev. Food Sci. Nutr. 2017, 58, 1791–1807. [Google Scholar] [CrossRef]
- Fernie, A.; Trethewey, R.N.; Krotzky, A.J.; Willmitzer, L. Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 2004, 5, 763–769. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Saito, K. Metabolomics for unknown plant metabolites. Anal. Bioanal. Chem. 2013, 405, 5005–5011. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Saito, K. Ultrahigh resolution metabolomics for S-containing metabolites. Curr. Opin. Biotechnol. 2017, 43, 8–16. [Google Scholar] [CrossRef]
- Panda, A.; Parida, A.K.; Rangani, J. Advancement of metabolomics techniques and their applications in plant science. In Plant Metabolites and Regulation Under Environmental Stress; Elsevier B.V.: London, UK, 2018; pp. 1–36. [Google Scholar]
- Tsugawa, H. Advances in computational metabolomics and databases deepen the understanding of metabolisms. Curr. Opin. Biotechnol. 2018, 54, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Willett, D.S.; Rering, C.C.; Ardura, D.A.; Beck, J.J. Application of mathematical models and computation in plant metabolomics. In Computational Phytochemistry; Sarker, S.D., Naha, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 231–254. [Google Scholar]
- Töpfer, N.; Seaver, S.M.D.; Aharoni, A. Integration of plant metabolomics data with metabolic networks: Progresses and challenges. Methods Mol. Biol. 2018, 1778, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.; Saito, K. Role of metabolomics in crop improvement. J. Plant. Biochem. Biotechnol. 2012, 21, 24–31. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Recent advances of metabolomics in plant biotechnology. Plant. Biotechnol. Rep. 2011, 6, 1–15. [Google Scholar] [CrossRef]
- Razzaq, A.; Sadia, B.; Raza, A.; Hameed, M.K.; Saleem, F. Metabolomics: A way forward for crop improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef]
- Alseekh, S.; Bermudez, L.; De Haro, L.A.; Fernie, A.R.; Carrari, F. Crop metabolomics: From diagnostics to assisted breeding. Metabolomics 2018, 14, 148. [Google Scholar] [CrossRef]
- Alawiye, T.T.; Babalola, O.O. Metabolomics: Current application and prospects in crop production. Biologia 2020, 1–13. [Google Scholar] [CrossRef]
- Zarei, I.; Brown, D.G.; Nealon, N.J.; Ryan, E.P. Rice bran metabolome contains amino acids, vitamins & cofactors, and phytochemicals with medicinal and nutritional properties. Rice 2017, 10, 24. [Google Scholar] [CrossRef]
- Mukherjee, P.K. Plant metabolomics and quality evaluation of herbal drugs. In Quality Control and Evaluation of Herbal Drugs-Evaluating Natural Products and Traditional Medicine; Mukherjee, P.K., Ed.; Elsevier: Amsterdam, The Netherland, 2019; pp. 629–653. [Google Scholar]
- Olivares-Vicente, M.; Barrajón-Catalán, E.; Herranz-López, M.; Segura-Carretero, A.; Joven, J.; Encinar, J.A.; Micol, V. Plant-derived polyphenols in human health: Biological activity, metabolites and putative molecular targets. Curr. Drug Metab. 2018, 19, 351–369. [Google Scholar] [CrossRef]
- Scossa, F.; Benina, M.; Alseekh, S.; Zhang, Y.; Fernie, A.R. The Integration of metabolomics and next-generation sequencing data to elucidate the pathways of natural product metabolism in medicinal plants. Planta Medica 2018, 84, 855–873. [Google Scholar] [CrossRef]
- Hrbek, V.; Krtkova, V.; Rubert, J.; Chmelarova, H.; Demnerova, K.; Ovesna, J.; Hajslova, J. Metabolomic strategies based on high-resolution mass spectrometry as a tool for recognition of GMO (MON 89788 Variety) and non-GMO soybean: A critical assessment of two complementary methods. Food Anal. Methods 2017, 10, 3723–3737. [Google Scholar] [CrossRef]
- Stewart, D.; Shepherd, L.V.T. Metabolomics for the safety assessment of genetically modified (GM) crops. Metab. Food Nutr. 2013, 192–216. [Google Scholar] [CrossRef]
- Corujo, M.; Pla, M.; Van Dijk, J.P.; Voorhuijzen-Harink, M.M.; Staats, M.; Slot, M.; Lommen, A.; Barros, E.; Nadal, A.; Puigdomènech, P.; et al. Use of omics analytical methods in the study of genetically modified maize varieties tested in 90 days feeding trials. Food Chem. 2019, 292, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Hanhineva, K.; Aharoni, A. Metabolomics in fruit development. In Molecular Techniques in Crop Improvement; Jain, S.M., Brar, D.S., Eds.; Springer Science Business Media B.V.: Heidelberg, Germany, 2010; pp. 675–693. [Google Scholar]
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; Racchapannavar, V.; Vikram, A.; Jindal, S.K.; Kardile, H.; Kumar, R.; et al. Advances in omics approaches for abiotic stress tolerance in tomato. Biology 2019, 8, 90. [Google Scholar] [CrossRef] [PubMed]
- Zarei, I.; Luna, E.; Leach, J.E.; McClung, A.; Vilchez, S.; Koita, O.A.; Ryan, E.P. Comparative rice bran metabolomics across diverse cultivars and functional rice gene–bran metabolite relationships. Metabolites 2018, 8, 63. [Google Scholar] [CrossRef]
- Verma, A.; Kumar, S.; Hemansi; Kumar, G.; Saini, J.K.; Agrawal, R.; Satlewal, A.; Ansari, M.W. Rhizosphere metabolite profiling: An opportunity to understand plant-microbe interactions for crop improvement. In Crop Improvement through Microbial Biotechnology; Prasad, R., Ed.; Elsevier B.V.: Singapore, 2018; pp. 343–361. [Google Scholar]
- Calumpang, C.L.; Saigo, T.; Watanabe, M.; Tohge, T. Cross-species comparison of fruit-metabolomics to elucidate metabolic regulation of fruit polyphenolics among solanaceous crops. Metabolites 2020, 10, 209. [Google Scholar] [CrossRef]
- Vu, D.C.; Lei, Z.; Sumner, L.W.; Coggeshall, M.V.; Lin, C.-H. Identification and quantification of phytosterols in black walnut kernels. J. Food Compos. Anal. 2019, 75, 61–69. [Google Scholar] [CrossRef]
- Martin, C. The interface between plant metabolic engineering and human health. Curr. Opin. Biotechnol. 2013, 24, 344–353. [Google Scholar] [CrossRef]
- Li, X.; Pang, W.; Piao, Z. Omics meets phytonutrients in vegetable brassicas: For nutritional quality breeding. Hortic. Plant. J. 2017, 3, 247–254. [Google Scholar] [CrossRef]
- Piasecka, A.; Kachlicki, P.; Stobiecki, M. Analytical methods for detection of plant metabolomes changes in response to biotic and abiotic stresses. Int. J. Mol. Sci. 2019, 20, 379. [Google Scholar] [CrossRef]
- Beger, R.D.; Dunn, W.B.; Bandukwala, A.; Bethan, B.; Broadhurst, D.; Clish, C.B.; Dasari, S.; Derr, L.; Evans, A.; Fischer, S.; et al. Towards quality assurance and quality control in untargeted metabolomics studies. Metabolomics 2019, 15, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.C.M.; Caldana, C.; Wolf, L.D.; De Abreu, L.G.F. The importance of experimental design, quality assurance, and control in plant metabolomics experiments. In Methods in Molecular Biology; Springer Science and Business Media LLC: New York City, NY, USA, 2018; pp. 3–17. [Google Scholar]
- Kim, H.K.; Verpoorte, R. Sample preparation for plant metabolomics. Phytochem. Anal. 2010, 21, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Aharoni, A.; Willmitzer, L.; Stitt, M.; Tohge, T.; Kopka, J.; Carroll, A.J.; Saito, K.; Fraser, P.D.; DeLuca, V. Recommendations for reporting metabolite data. Plant. Cell 2011, 23, 2477–2482. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, H.; Hardy, N.; Beckmann, M.; Draper, J.; Smith, A.R.; Taylor, J.; Fiehn, O.; Goodacre, R.; Bino, R.J.; Hall, R.D.; et al. A proposed framework for the description of plant metabolomics experiments and their results. Nat. Biotechnol. 2004, 22, 1601–1606. [Google Scholar] [CrossRef]
- Fiehn, O.; Robertson, D.; Griffin, J.; Van Der Werf, M.; Nikolau, B.; Morrison, N.; Sumner, L.W.; Goodacre, R.; Hardy, N.W.; Taylor, C.; et al. The metabolomics standards initiative (MSI). Metabolomics 2007, 3, 175–178. [Google Scholar] [CrossRef]
- Alseekh, S.; Wu, S.; Brotman, Y.; Fernie, A.R. Guidelines for sample normalization to minimize batch variation for large-scale metabolic profiling of plant natural genetic variance. In Methods in Molecular Biology; Springer Science and Business Media LLC: New York City, NY, USA, 2018; Volume 1778, pp. 33–46. [Google Scholar]
- Chen, W.; Wang, W.; Peng, M.; Gong, L.; Gao, Y.; Wan, J.; Wang, S.; Shi, L.; Zhou, B.; Li, Z.; et al. Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals. Nat. Commun. 2016, 7, 12767. [Google Scholar] [CrossRef]
- Sysi-Aho, M.; Katajamaa, M.; Yetukuri, L.; Orešič, M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinform. 2007, 8, 93. [Google Scholar] [CrossRef]
- Wehrens, R.; Hageman, J.A.; Van Eeuwijk, F.; Kooke, R.; Flood, P.J.; Wijnker, E.; Keurentjes, J.J.B.; Lommen, A.; Van Eekelen, H.D.L.M.; Hall, R.D.; et al. Improved batch correction in untargeted MS-based metabolomics. Metabolomics 2016, 12, 1–12. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Y.; Zeng, J.; Duan, L.; Xue, X.; Wang, H.; Lin, T.; Liu, Z.; Zeng, K.; Zhong, Y.; et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat. Plants 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Rastogi, S.; Shah, S.; Kumar, R.; Kumar, A.; Shasany, A.K. Comparative temporal metabolomics studies to investigate interspecies variation in three Ocimum species. Sci. Rep. 2020, 10, 5234. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Chen, W.; Sun, W.; Peng, M.; Yuan, Z.; Shen, S.; Xie, K.; Jin, C.; Sun, Y.; et al. Metabolome analysis of multi-connected biparental chromosome segment substitution line populations. Plant. Physiol. 2018, 178, 612–625. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Zhu, A.; Jia, J.; Hu, X.; Chen, J.; Liu, W.; Ren, X.; Sun, D.; Fernie, A.; Cui, F.; et al. Metabolomics analysis and metabolite-agronomic trait associations using kernels of wheat (Triticum aestivum) recombinant inbred lines. Plant. J. 2020, 103, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Luo, J. Metabolic GWAS-based dissection of genetic bases underlying the diversity of plant metabolism. Plant. J. 2019, 97, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Luo, J. Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant. Biol. 2015, 24, 31–38. [Google Scholar] [CrossRef]
- Chen, J.; Hu, X.; Shi, T.; Yin, H.; Sun, D.; Hao, Y.; Xia, X.; Luo, J.; Fernie, A.R.; He, Z.; et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant. Biotechnol. J. 2020, 18, 1722–1735. [Google Scholar] [CrossRef]
- Dong, X.; Chen, W.; Wang, W.-S.; Zhang, H.; Liu, X.; Luo, J. Comprehensive profiling and natural variation of flavonoids in rice. J. Integr. Plant. Biol. 2014, 56, 876–886. [Google Scholar] [CrossRef]
- Matsuda, F.; Nakabayashi, R.; Yang, Z.; Okazaki, Y.; Yonemaru, J.; Ebana, K.; Yano, M.; Saito, K. Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant. J. 2014, 81, 13–23. [Google Scholar] [CrossRef]
- Wen, W.; Li, D.; Li, X.; Gao, Y.; Li, W.; Li, H.; Liu, J.; Liu, H.; Chen, W.; Luo, J.; et al. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 2014, 5, 3438. [Google Scholar] [CrossRef]
- Wen, W.; Brotman, Y.; Willmitzer, L.; Yan, J.; Fernie, A. Broadening our portfolio in the genetic improvement of maize chemical composition. Trends Genet. 2016, 32, 459–469. [Google Scholar] [CrossRef]
- Beleggia, R.; Rau, D.; Laidò, G.; Platani, C.; Nigro, F.; Fragasso, M.; De Vita, P.; Scossa, F.; Fernie, A.R.; Nikoloski, Z.; et al. Evolutionary metabolomics reveals domestication-associated changes in tetraploid wheat kernels. Mol. Biol. Evol. 2016, 33, 1740–1753. [Google Scholar] [CrossRef]
- Ye, J.; Wang, X.; Hu, T.; Zhang, F.; Wang, B.; Li, C.; Yang, T.; Li, H.; Lu, Y.; Giovannoni, J.J.; et al. An InDel in the promoter of Al-Activated malate transporter9 selected during tomato domestication determines fruit malate contents and Aluminum Tolerance. Plant. Cell 2017, 29, 2249–2268. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Wang, S.; Huang, Z.; Zhang, S.; Liao, Q.; Zhang, C.; Lin, T.; Qin, M.; Peng, M.; Yang, C.; et al. Rewiring of the fruit metabolome in tomato breeding. Cell 2018, 172, 249–261.e12. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.Y.; Shi, J.; Quan, S.; Cui, B.; Kleessen, S.; Nikoloski, Z.; Tohge, T.; Alexander, D.; Guo, L.; Lin, H.; et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomicsd. Sci Rep. 2014, 4, 5067. [Google Scholar] [CrossRef] [PubMed]
- Farré, G.; Twyman, R.M.; Christou, P.; Capell, T.; Zhu, C. Knowledge-driven approaches for engineering complex metabolic pathways in plants. Curr. Opin. Biotechnol. 2015, 32, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Tohge, T.; Chan, S.-A.; Song, Y.; Rao, J.; Cui, B.; Lin, H.; Wang, L.; Fernie, A.R.; Zhang, D.; et al. Identification of conserved and diverse metabolic shifts during rice grain development. Sci. Rep. 2016, 6, 20942. [Google Scholar] [CrossRef]
- Hu, C.; Li, Q.; Shen, X.; Quan, S.; Lin, H.; Duan, L.; Wang, Y.; Luo, Q.; Qu, G.; Han, Q.; et al. Characterization of factors underlying the metabolic shifts in developing kernels of colored maize. Sci. Rep. 2016, 6, 35479. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, Y.-X.; Sun, S.-Y.; Xie, M.-X.; Hu, C.; Shi, Y.-Q.; Shi, J.; Li, J.-Y. Identification of the biochemical characteristics of developing giant embryo rice grains using non-targeted metabolomics. J. Cereal Sci. 2019, 85, 70–76. [Google Scholar] [CrossRef]
- Daygon, V.D.; Calingacion, M.; Forster, L.C.; De Voss, J.J.; Schwartz, B.D.; Ovenden, B.; Alonso, D.E.; McCouch, S.R.; Garson, M.J.; Fitzgerald, M. Metabolomics and genomics combine to unravel the pathway for the presence of fragrance in rice. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Song, E.-H.; Jeong, J.; Park, C.Y.; Kim, H.-Y.; Kim, E.-H.; Bang, E.; Hong, Y.-S. Metabotyping of rice (Oryza sativa L.) for understanding its intrinsic physiology and potential eating quality. Food Res. Int. 2018, 111, 20–30. [Google Scholar] [CrossRef]
- Cocuron, J.-C.; Koubaa, M.; Kimmelfield, R.; Ross, Z.; Alonso, A.P. A combined metabolomics and fluxomics analysis identifies steps limiting oil synthesis in maize embryos. Plant. Physiol. 2019, 181, 961–975. [Google Scholar] [CrossRef]
- Shewry, P.R.; Rakszegi, M.; Lovegrove, A.; Amos, D.; Corol, D.-I.; Tawfike, A.; Miko, P.; Ward, J.L. Effects of organic and conventional crop nutrition on profiles of polar metabolites in grain of wheat. J. Agric. Food Chem. 2018, 66, 5346–5351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cao, S.; Zhang, Z.; Meng, X.; Hsiaoping, C.; Yin, C.; Jiang, H.; Wang, S. Nutritional quality and health risks of wheat grains from organic and conventional cropping systems. Food Chem. 2020, 308, 125584. [Google Scholar] [CrossRef]
- Drapal, M.; Lindqvist-Kreuze, H.; Mihovilovich, E.; Aponte, M.; Bonierbale, M.; Fraser, P.D. Cooking dependent loss of metabolites in potato breeding lines and their wild and landrace relatives. J. Food Compos. Anal. 2020, 88, 103432. [Google Scholar] [CrossRef]
- Galland, M.; He, D.; Lounifi, I.; Marc, G.; Clément, G.; Balzergue, S.; Huguet, S.; Cueff, G.; Godin, B.; Collet, B.; et al. An integrated “multi-omics” comparison of embryo and endosperm tissue-specific features and their impact on rice seed quality. Front. Plant. Sci. 2017, 8, 1984. [Google Scholar] [CrossRef] [PubMed]
- Bough, R.A.; Holm, D.G.; Jayanty, S.S. Evaluation of cooked flavor for fifteen potato genotypes and the correlation of sensory analysis to instrumental methods. Am. J. Potato Res. 2020, 97, 63–77. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Holm, D.G.; Broeckling, C.D.; Prenni, J.E.; Heuberger, A.L. Metabolomics and ionomics of potato tuber reveals an influence of cultivar and market class on human nutrients and bioactive compounds. Front. Nutr. 2018, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Mounet, F.; Lemaire-Chamley, M.; Maucourt, M.; Cabasson, C.; Giraudel, J.-L.; Deborde, C.; Lessire, R.; Gallusci, P.; Bertrand, A.; Gaudillère, M.; et al. Quantitative metabolic profiles of tomato flesh and seeds during fruit development: Complementary analysis with ANN and PCA. Metabolomics 2007, 3, 273–288. [Google Scholar] [CrossRef]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant. Physiol. 2006, 142, 839–854. [Google Scholar] [CrossRef]
- Fang, C.; Luo, J.; Wang, S. The diversity of nutritional metabolites: Origin, dissection, and application in crop breeding. Front. Plant. Sci. 2019, 10, 1028. [Google Scholar] [CrossRef]
- Hu, C.; Rao, J.; Song, Y.; Chan, S.-A.; Tohge, T.; Cui, B.; Lin, H.; Fernie, A.R.; Zhang, D.; Shi, J. Dissection of flag leaf metabolic shifts and their relationship with those occurring simultaneously in developing seed by application of non-targeted metabolomics. PLoS ONE 2020, 15, e0227577. [Google Scholar] [CrossRef]
- Ming, M.; Cui, S.W.; Zhang, T.; Jin, Z. Slowly digestible starch-A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1642–1657. [Google Scholar]
- Guzman, M.K.; Parween, S.; Butardo, V.M.; Alhambra, C.M.; Anacleto, R.; Seiler, C.; Bird, A.R.; Chow, C.-P.; Sreenivasulu, N. Investigating glycemic potential of rice by unraveling compositional variations in mature grain and starch mobilization patterns during seed germination. Sci Rep. 2017, 7, 5854. [Google Scholar] [CrossRef] [PubMed]
- Concepcion, J.C.T.; Calingacion, M.; Garson, M.J.; Fitzgerald, M.A. Lipidomics reveals associations between rice quality traits. Metabolomics 2020, 16, 2–16. [Google Scholar] [CrossRef] [PubMed]
- Rao, J.; Cheng, F.; Hu, C.; Quan, S.; Lin, H.; Wang, J.; Chen, G.; Zhao, X.; Alexander, D.; Guo, L.; et al. Metabolic map of mature maize kernels. Metabolomics 2014, 10, 775–787. [Google Scholar] [CrossRef]
- Harjes, C.E.; Rocheford, T.R.; Bai, L.; Brutnell, T.P.; Kandianis, C.B.; Sowinski, S.G.; Stapleton, A.E.; Vallabhaneni, R.; Williams, M.; Wurtzel, E.T.; et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 2008, 319, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Lin, H.; Rao, J.; Shi, J.; Hu, C.; Cheng, F.; Wilson, Z.A.; Zhang, D.; Quan, S. Seed metabolomic study reveals significant metabolite variations and correlations among different soybean cultivars. J. Integr. Plant. Biol. 2014, 56, 826–836. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, P.; Shi, X.; Yang, N.; Yan, L.; Zhao, Q.; Yang, C.; Guan, Y. Primary metabolite contents are correlated with seed protein and oil traits in near-isogenic lines of soybean. Crop. J. 2019, 7, 651–659. [Google Scholar] [CrossRef]
- Natarajan, S. Natural variability in abundance of prevalent soybean proteins. Regul. Toxicol. Pharmacol. 2010, 58, S26–S29. [Google Scholar] [CrossRef]
- Medic, J.; Atkinson, C.; Hurburgh, C.R. Current knowledge in soybean composition. J. Am. Oil Chem. Soc. 2014, 91, 363–384. [Google Scholar] [CrossRef]
- Francki, M.G.; Hayton, S.; Gummer, J.P.A.; Rawlinson, C.; Trengove, R.D. Metabolomic profiling and genomic analysis of wheat aneuploid lines to identify genes controlling biochemical pathways in mature grain. Plant. Biotechnol. J. 2015, 14, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Gorzolka, K.; Lissel, M.; Kessler, N.; Loch-Ahring, S.; Niehaus, K. Metabolite fingerprinting of barley whole seeds, endosperms, and embryos during industrial malting. J. Biotechnol. 2012, 159, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, A.L.; Broeckling, C.D.; Kirkpatrick, K.R.; Prenni, J.E. Application of nontargeted metabolite profiling to discover novel markers of quality traits in an advanced population of malting barley. Plant. Biotechnol. J. 2013, 12, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Khakimov, B.; Rasmussen, M.A.; Kannangara, R.M.; Jespersen, B.M.; Munck, L.; Engelsen, S.B. From metabolome to phenotype: GC-MS metabolomics of developing mutant barley seeds reveals effects of growth, temperature and genotype. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Turner, M.F.; Heuberger, A.L.; Kirkwood, J.S.; Collins, C.C.; Wolfrum, E.J.; Broeckling, C.D.; Prenni, J.E.; Jahn, C.E. Non-targeted metabolomics in diverse sorghum breeding lines indicates primary and secondary metabolite profiles are associated with plant biomass accumulation and photosynthesis. Front. Plant. Sci. 2016, 7, 953. [Google Scholar] [CrossRef]
- Loskutov, I.G.; Shelenga, T.V.; Konarev, A.V.; Shavarda, A.L.; Blinova, E.V.; Dzubenko, N.I. The metabolomic approach to the comparative analysis of wild and cultivated species of oats (Avena L.). Russ. J. Genet. Appl. Res. 2017, 7, 501–508. [Google Scholar] [CrossRef]
- Khakimov, B.; Jespersen, B.M.; Engelsen, S.B. Comprehensive and comparative metabolomic profiling of wheat, barley, oat and rye using gas chromatography-mass spectrometry and advanced chemometrics. Foods 2014, 3, 569–585. [Google Scholar] [CrossRef]
- Moing, A.; Aharoni, A.; Biais, B.; Rogachev, I.; Meir, S.; Brodsky, L.; Allwood, J.W.; Erban, A.; Dunn, W.B.; Kay, L.; et al. Extensive metabolic cross-talk in melon fruit revealed by spatial and developmental combinatorial metabolomics. New Phytol. 2011, 190, 683–696. [Google Scholar] [CrossRef]
- Hu, B.; Gao, J.; Xu, S.; Zhu, J.; Fan, X.; Zhou, X. Quality evaluation of different varieties of dry red wine based on nuclear magnetic resonance metabolomics. Appl. Biol. Chem. 2020, 63, 1–8. [Google Scholar] [CrossRef]
- Pinu, F.R.; Tumanov, S.; Grose, C.; Raw, V.; Albright, A.; Stuart, L.; Villas-Boas, S.; Martin, D.; Harker, F.R.; Greven, M. Juice Index: An integrated Sauvignon blanc grape and wine metabolomics database shows mainly seasonal differences. Metabolomics 2019, 15, 3. [Google Scholar] [CrossRef]
- Cheng, L.; Yang, Q.-Q.; Chen, Z.; Zhang, J.-R.; Chen, Q.; Wang, Y.; Wei, X.-L. Distinct changes of metabolic profile and sensory quality during Qingzhuan tea processing revealed by LC-MS-based metabolomics. J. Agric. Food Chem. 2020, 68, 4955–4965. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.-K.; Ma, J.-Q.; Apostolides, Z.; Chen, L. Metabolomics for a millenniums-old crop: Tea plant (Camellia sinensis). J. Agric. Food Chem. 2019, 67, 6445–6457. [Google Scholar] [CrossRef] [PubMed]
- Gamboa-Becerra, R.; Hernández-Hernández, M.C.; González-Ríos, Ó.; Suárez-Quiroz, M.L.; Gálvez-Ponce, E.; Ordaz-Ortiz, J.J.; Winkler, R. Metabolomic markers for the early selection of coffea canephora plants with desirable cup quality traits. Metabolites 2019, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, S.; Yonetani, T.; Yuki, T.; Tomio, A.; Bamba, T.; Fukusaki, E. Quality evaluation of green tea leaf cultured under artificial light condition using gas chromatography/mass spectrometry. J. Biosci. Bioeng. 2017, 123, 197–202. [Google Scholar] [CrossRef]
- Lee, J.-E.; Lee, B.-J.; Chung, J.-O.; Kim, H.-N.; Kim, E.-H.; Jung, S.; Lee, H.; Lee, S.-J.; Hong, Y.-S. Metabolomic unveiling of a diverse range of green tea (Camellia sinensis) metabolites dependent on geography. Food Chem. 2015, 174, 452–459. [Google Scholar] [CrossRef]
- Jing, J.; Shi, Y.; Zhang, Q.; Wang, J.; Ruan, J. Prediction of Chinese green tea ranking by metabolite profiling using ultra-performance liquid chromatography–quadrupole time-of-flight mass spectrometry (UPLC–Q-TOF/MS). Food Chem. 2017, 221, 311–316. [Google Scholar] [CrossRef]
- Xu, Q.; He, Y.; Yan, X.; Zhao, S.; Zhu, J.; Wei, C. Unraveling a crosstalk regulatory network of temporal aroma accumulation in tea plant (Camellia sinensis) leaves by integration of metabolomics and transcriptomics. Environ. Exp. Bot. 2018, 149, 81–94. [Google Scholar] [CrossRef]
- Zhang, Q.; Tang, D.; Liu, M.; Ruan, J. Integrated analyses of the transcriptome and metabolome of the leaves of albino tea cultivars reveal coordinated regulation of the carbon and nitrogen metabolism. Sci. Hortic. 2018, 231, 272–281. [Google Scholar] [CrossRef]
- Yu, X.; Xiao, J.; Chen, S.; Yu, Y.; Ma, J.; Lin, Y.; Li, R.; Lin, J.; Fu, Z.; Zhou, Q.; et al. Metabolite signatures of diverse Camellia sinensis tea populations. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Souard, F.; Delporte, C.; Stoffelen, P.P.; Thévenot, E.E.; Noret, N.; Dauvergne, B.; Kauffmann, J.-M.; Van Antwerpen, P.; Stévigny, C. Metabolomics fingerprint of coffee species determined by untargeted-profiling study using LC-HRMS. Food Chem. 2018, 245, 603–612. [Google Scholar] [CrossRef]
- Pérez-Míguez, R.; Castro-Puyana, M.; Sánchez-López, E.; Plaza, M.; Marina, M.L. Untargeted HILIC-MS-based metabolomics approach to evaluate coffee roasting process: Contributing to an integrated metabolomics multiplatform. Molecules 2020, 25, 887. [Google Scholar] [CrossRef] [PubMed]
- Okaru, A.O.; Scharinger, A.; De Rezende, T.R.; Teipel, J.C.; Kuballa, T.; Walch, S.G.; Lachenmeier, D.W. Validation of a quantitative proton nuclear magnetic resonance spectroscopic screening method for coffee quality and authenticity (NMR coffee screener). Foods 2020, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Hang, D.; Zeleznik, O.A.; He, X.; Guasch-Ferre, M.; Jiang, X.; Li, J.; Liang, L.; Eliassen, A.H.; Clish, C.B.; Chan, A.T.; et al. Metabolomic signatures of long-term coffee consumption and risk of type 2 diabetes in women. Diabetes Care 2020, 43, 2588–2596. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, M.C.; Erlund, I.; Michelotti, G.A.; Herder, C.; Westerhuis, J.A.; Tuomilehto, J. Metabolomic response to coffee consumption: Application to a three-stage clinical trial. J. Intern. Med. 2018, 283, 544–557. [Google Scholar] [CrossRef]
- Kuang, A.; Erlund, I.; Herder, C.; Westerhuis, J.A.; Tuomilehto, J.; Cornelis, M.C. Lipidomic response to coffee consumption. Nutrients 2018, 10, 1851. [Google Scholar] [CrossRef]
- Seow, W.J.; Low, D.Y.; Pan, W.-C.; Gunther, S.H.; Sim, X.; Torta, F.; Herr, D.R.; Kovalik, J.-P.; Ching, J.; Khoo, C.M.; et al. Coffee, black tea, and green tea consumption in relation to plasma metabolites in an Asian population. Mol. Nutr Food Res. 2020, e2000527. [Google Scholar] [CrossRef]
- Arapitsas, P.; Ugliano, M.; Marangon, M.; Piombino, P.; Rolle, L.; Gerbi, V.; Versari, A.; Mattivi, F. Use of untargeted liquid chromatography-Mass spectrometry metabolome to discriminate Italian monovarietal red wines, produced in their different terroirs. J. Agric. Food Chem. 2020, 68, 13353–13366. [Google Scholar] [CrossRef]
- Springer, A.; Riedl, J.; Esslinger, S.; Roth, T.; Glomb, M.A.; Fauhl-Hassek, C. Validated modeling for german white wine varietal authentication based on headspace solid-phase microextraction online coupled with gas chromatography mass spectrometry fingerprinting. J. Agric. Food Chem. 2014, 62, 6844–6851. [Google Scholar] [CrossRef]
- Magdas, D.A.; Cozar, B.I.; Feher, I.; Guyon, F.; Dehelean, A.; Pinzaru, S.C. Testing the limits of FT-Raman spectroscopy for wine authentication: Cultivar, geographical origin, vintage and terroir effect influence. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef]
- Diamantidou, D.; Zotou, A.; Theodoridis, G. Wine and grape marc spirits metabolomics. Metabolomics 2018, 14, 159. [Google Scholar] [CrossRef]
- Hernández-Alonso, P.; Papandreou, C.; Bullo, M.; Ruiz-Canela, M.; Dennis, C.; Deik, A.; Wang, D.D.; Guasch-Ferré, M.; Yu, E.; Toledo, E.; et al. Plasma metabolites associated with frequent red wine consumption: A metabolomics approach within the PREDIMED study. Mol. Nutr. Food Res. 2019, 63, e1900140. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Fernández, A.; Ibañez, C.; Simó, C.; Bartolomé, B.; Moreno-Arribas, M.V. An ultrahigh-performance liquid chromatography-time-of-flight mass spectrometry metabolomic approach to studying the impact of moderate red-wine consumption on urinary metabolome. J. Proteome Res. 2018, 17, 1624–1635. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; De Souza, L.P.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the context of plant natural products research: From sample preparation to metabolite analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liang, H.; Cao, H.; Zhang, B.; Li, J.; Wang, W.; Qin, S.; Wang, Y.; Xuan, L.; Lai, L.; et al. Efficient ligand discovery from natural herbs by integrating virtual screening, affinity mass spectrometry and targeted metabolomics. Analyst 2019, 144, 2881–2890. [Google Scholar] [CrossRef] [PubMed]
- Gonulalan, E.M.; Nemutlu, E.; Bayazeid, O.; Koçak, E.; Yalçın, F.N.; Demirezer, O.L. Metabolomics and proteomics profiles of some medicinal plants and correlation with BDNF activity. Phytomedicine 2020, 74, 152920. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Nicholson, J.K.; Hylands, P.J.; Sampson, J.; Whitcombe, I.; Stewart, C.G.; Caiger, S.; Oru, I.; Holmes, E. Metabolomic strategy for the classification and quality control of phytomedicine: A case study of chamomile flower (Matricaria recutitaL.). Planta Medica 2004, 70, 250–255. [Google Scholar] [CrossRef]
- Choi, Y.H.; Kim, H.K.; Hazekamp, A.; Erkelens, C.; Lefeber, A.W.M.; Verpoorte, R. Metabolomic differentiation of Cannabis s ativa cultivars using 1H NMR spectroscopy and principal component analysis. J. Nat. Prod. 2004, 67, 953–957. [Google Scholar] [CrossRef]
- Kooy, F.V.D.; Verpoorte, R.; Meyer, M. Metabolomic quality control of claimed anti-malarial Artemisia afra herbal remedy and A. afra and A. annua plant extracts. S Afr. J. Bot. 2008, 74, 186–189. [Google Scholar] [CrossRef]
- Qiu, S.; Yang, W.; Yao, C.; Qiu, Z.-D.; Shi, X.-J.; Zhang, J.-X.; Hou, J.; Wang, Q.-R.; Wu, W.-Y.; Guo, D.-A. Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. J. Chromatogr. A 2016, 1453, 78–87. [Google Scholar] [CrossRef]
- Chang, X.; Zhang, J.; Li, D.; Zhou, D.; Zhang, Y.; Wang, J.; Hu, B.; Ju, A.; Ye, Z. Nontargeted metabolomics approach for the differentiation of cultivation ages of mountain cultivated ginseng leaves using UHPLC/QTOF-MS. J. Pharm. Biomed. Anal. 2017, 141, 108–122. [Google Scholar] [CrossRef]
- Booker, A.; Jalil, B.; Frommenwiler, D.A.; Reich, E.; Zhai, L.; Kulic, Z.; Heinrich, M. The authenticity and quality of Rhodiola rosea products. Phytomedicine 2016, 23, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Crighton, E.; Weisenseel, J.; Bunce, M.; Musgrave, I.F.; Trengove, R.; Maker, G.L. Exploring the application of the DSA-TOF, a direct, high-resolution time-of-flight mass spectrometry technique for the screening of potential adulterated and contaminated herbal medicines. J. Am. Soc. Mass Spectrom. 2019, 30, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Yao, C.; Yao, S.; Yang, W.; Wu, W.-Y.; Guo, D.-A. A metabolomics strategy for authentication of plant medicines with multiple botanical origins, a case study of Uncariae Rammulus Cum Uncis. J. Sep. Sci. 2020, 43, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Haq, F.U.; Ali, A.; Khan, M.N.; Shah, S.M.Z.; Adhikhari, A.; El-Seedi, H.R.; Musharraf, S.G. Combining untargeted and targeted metabolomics approaches for the standardization of polyherbal formulations through UPLC–MS/MS. Metabolomics 2019, 15, 116. [Google Scholar] [CrossRef]
- Liu, Y.-T.; Peng, J.-B.; Jia, H.-M.; Cai, D.; Zhang, H.-W.; Yu, C.-Y.; Zou, Z.-M. UPLC-Q/TOF MS standardized Chinese formula Xin-Ke-Shu for the treatment of atherosclerosis in a rabbit model. Phytomedicine 2014, 21, 1364–1372. [Google Scholar] [CrossRef]
- Zuo, R.; Ren, W.; Bian, B.-L.; Wang, H.; Wang, Y.-N.; Hu, H.; Zhao, H.-Y.; Si, N. Metabolic fate analysis of Huang-Lian-Jie-Du Decoction in rat urine and feces by LC-IT-MS combining with LC–FT-ICR-MS: A feasible strategy for the metabolism study of Chinese medical formula. Xenobiotica 2015, 46, 65–81. [Google Scholar] [CrossRef]
- Tian, J.-S.; Peng, G.-J.; Wu, Y.-F.; Zhou, J.-J.; Xiang, H.; Gao, X.-X.; Zhou, Y.-Z.; Qin, X.; Du, G.-H. A GC-MS urinary quantitative metabolomics analysis in depressed patients treated with TCM formula of Xiaoyaosan. J. Chromatogr. B 2016, 1026, 227–235. [Google Scholar] [CrossRef]
- Ding, Z.; Zhong, R.; Yang, Y.; Xia, T.; Wang, W.; Wang, Y.; Xing, N.; Luo, Y.; Li, S.; Shang, L.; et al. Systems pharmacology reveals the mechanism of activity of Ge-Gen-Qin-Lian decoction against LPS-induced acute lung injury: A novel strategy for exploring active components and effective mechanism of TCM formulae. Pharmacol. Res. 2020, 156, 104759. [Google Scholar] [CrossRef]
- Kusano, M.; Baxter, I.; Fukushima, A.; Oikawa, A.; Okazaki, Y.; Nakabayashi, R.; Bouvrette, D.J.; Achard, F.; Jakubowski, A.R.; Ballam, J.M.; et al. Assessing metabolomic and chemical diversity of a soybean lineage representing 35 years of breeding. Metabolomics 2014, 11, 261–270. [Google Scholar] [CrossRef]
- Kusano, M.; Redestig, H.; Hirai, T.; Oikawa, A.; Matsuda, F.; Fukushima, A.; Arita, M.; Watanabe, S.; Yano, M.; Hiwasa-Tanase, K.; et al. Covering chemical diversity of genetically-modified tomatoes using metabolomics for objective substantial equivalence assessment. PLoS ONE 2011, 6, e16989. [Google Scholar] [CrossRef]
- Rao, J.; Yang, L.; Guo, J.; Quan, S.; Chen, G.; Zhao, X.; Zhang, D.; Shi, J. Metabolic changes in transgenic maize mature seeds over-expressing the Aspergillus niger phyA2. Plant. Cell Rep. 2015, 35, 429–437. [Google Scholar] [CrossRef]
- Delaney, B.; Hazebroek, J.; Herman, R.; Juberg, D.; Storer, N.P. untargeted metabolomics are not useful in the risk assessment of GM crops. Trends Plant. Sci. 2019, 24, 383–384. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, G.G.; Lundry, D.; Drury, S.; Berman, K.; Riordan, S.G.; Nemeth, M.A.; Ridley, W.P.; Glenn, K.C. Natural variation in crop composition and the impact of transgenesis. Nat. Biotechnol. 2010, 28, 402–404. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Aharoni, A.; Hall, R.D.; Huang, S.; Giovannoni, J.J.; Sonnewald, U.; Fernie, A. Metabolomics should be deployed in the identification and characterization of gene-edited crops. Plant. J. 2020, 102, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Aliferis, K.A. Metabolomics in plant protection product research and development. In Environmental Metabolomics; Álvarez-Muñoz, D., Farré, M., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 163–194. [Google Scholar]
- Matich, E.K.; Soria, N.G.C.; Aga, D.S.; Atilla-Gokcumen, G.E. Applications of metabolomics in assessing ecological effects of emerging contaminants and pollutants on plants. J. Hazard. Mater. 2019, 373, 527–535. [Google Scholar] [CrossRef] [PubMed]
- de Souza, M.C.; Rosa, A.L.; Poschenrieder, C.; Tolra, R.; Da Costa, F.B. Fingerprinting metabolomics in tropical mistletoes: A case study with facultative aluminum-accumulating species. Phytochem. Let. 2018, 25, 90–94. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, Y.; Paglia, K.; Vaniya, A.; Wancewicz, B.; Keller, A.A. Metabolomics reveals the molecular mechanisms of copper induced cucumber leaf (Cucumis sativus) senescence. Environ. Sci. Technol. 2018, 52, 7092–7100. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Shen, H.; Wang, J.; Liu, W.; Zhu, X.; Wang, R.; Sun, X.; Liu, L. Metabolomic analysis with GC-MS to reveal potential metabolites and biological pathways involved in Pb & Cd stress response of radish roots. Sci. Rep. 2015, 5, 18296. [Google Scholar] [CrossRef]
- Youssef, R.; Colla, G.; Bernardo, L.; Kane, D.; Trevisan, M.; Lucini, L. Zinc excess triggered polyamines accumulation in lettuce root metabolome, as compared to osmotic stress under high salinity. Front. Plant. Sci. 2016, 7, 842. [Google Scholar]
- Alseekh, S.; Fernie, A. Metabolomics 20 years on: What have we learned and what hurdles remain? Plant. J. 2018, 94, 933–942. [Google Scholar] [CrossRef]
- Bayram, B.; González-Sarrías, A.; Istas, G.; Garcia-Aloy, M.; Morand, C.; Tuohy, K.; García-Villalba, R.; Mena, P. Breakthroughs in the health effects of plant food bioactives: A perspective on microbiomics, nutri(epi)genomics, and metabolomics. J. Agric. Food Chem. 2018, 66, 10686–10692. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Dudley, S.; Sun, C.; Schlenk, D.; Gan, J. Stable isotope labeling-assisted metabolite probing for emerging contaminants in plants. Anal. Chem. 2018, 90, 11040–11047. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Chen, Z.; Hong, J.; Shi, J. Promoting Human Nutrition and Health through Plant Metabolomics: Current Status and Challenges. Biology 2021, 10, 20. https://doi.org/10.3390/biology10010020
Sun W, Chen Z, Hong J, Shi J. Promoting Human Nutrition and Health through Plant Metabolomics: Current Status and Challenges. Biology. 2021; 10(1):20. https://doi.org/10.3390/biology10010020
Chicago/Turabian StyleSun, Wenli, Zican Chen, Jun Hong, and Jianxin Shi. 2021. "Promoting Human Nutrition and Health through Plant Metabolomics: Current Status and Challenges" Biology 10, no. 1: 20. https://doi.org/10.3390/biology10010020
APA StyleSun, W., Chen, Z., Hong, J., & Shi, J. (2021). Promoting Human Nutrition and Health through Plant Metabolomics: Current Status and Challenges. Biology, 10(1), 20. https://doi.org/10.3390/biology10010020





