C-type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors
Abstract
:Simple Summary
Abstract
1. Introduction
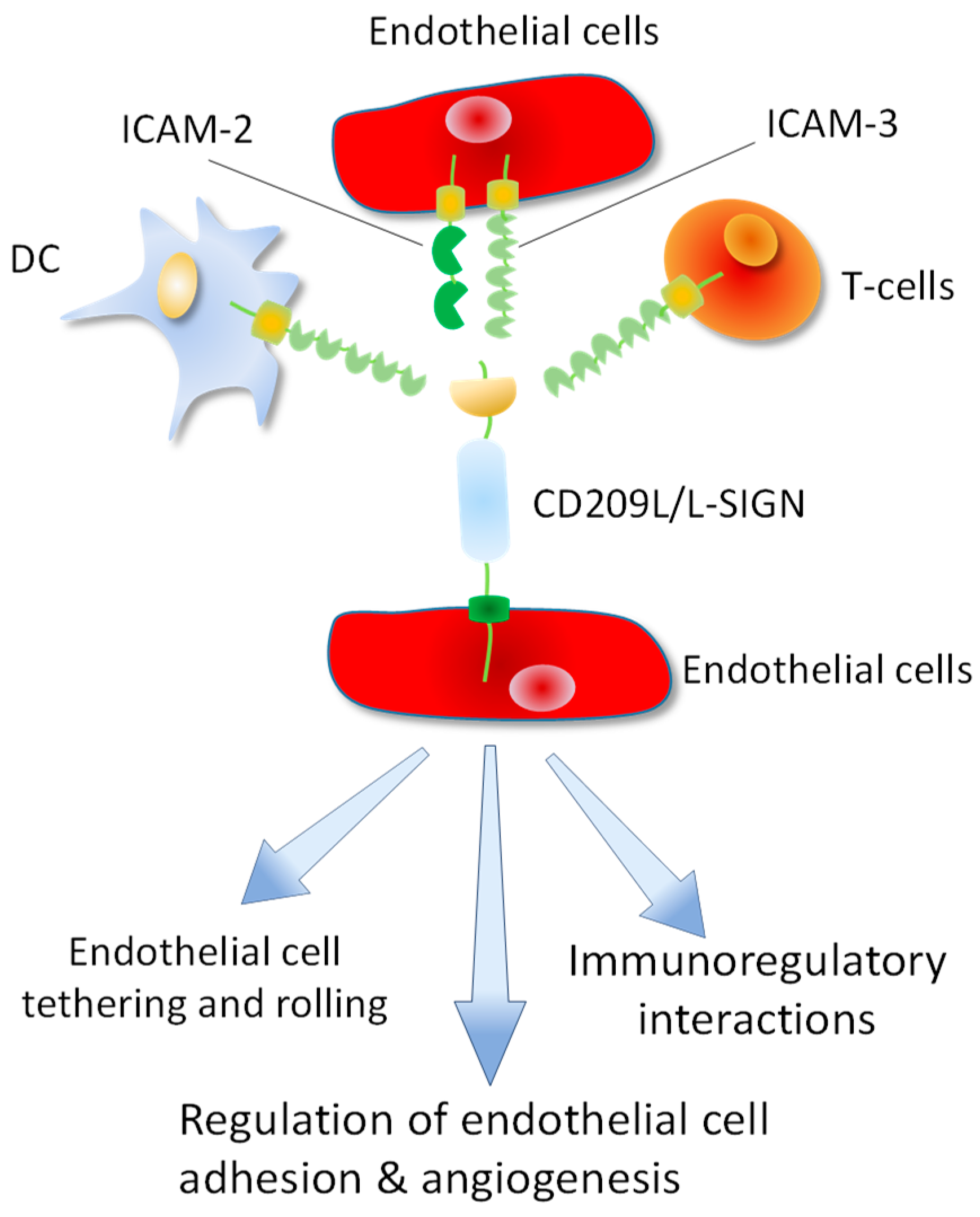
2. CD209/DC-SIGN and CD209L/L-SIGN Family Proteins: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors:
3. CD209/DC-SIGN and CD209L/L-SIGN Family Proteins and Coronaviruses:
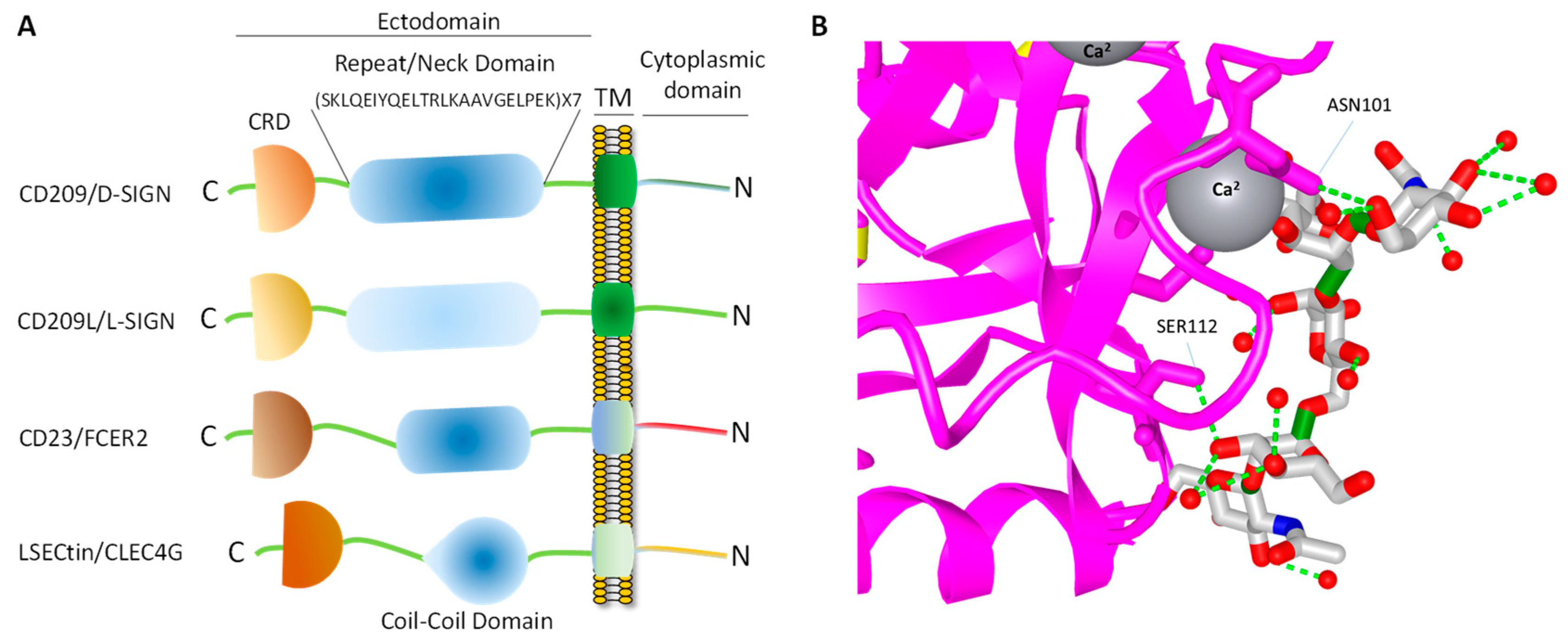
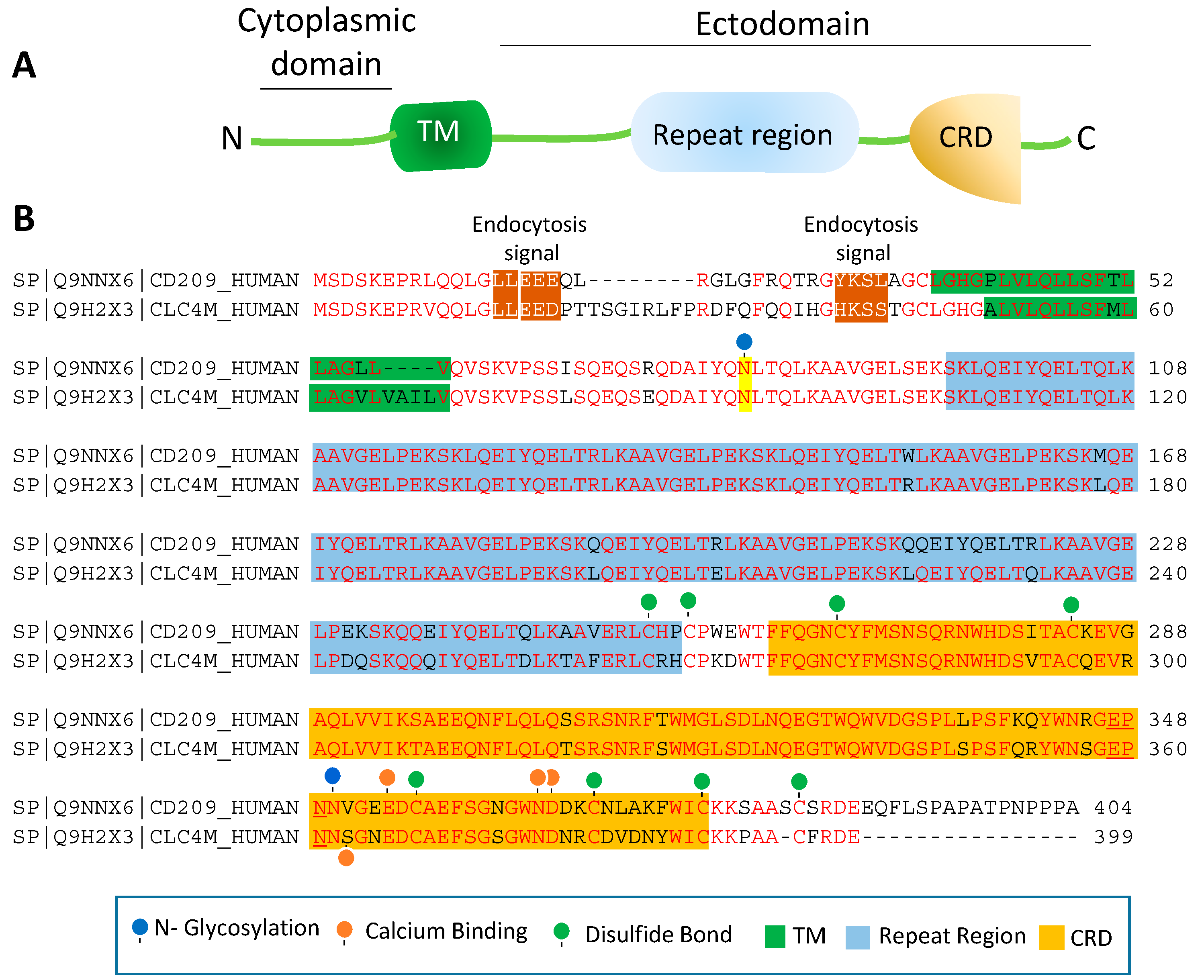
4. Topology of CD209L/L-SIGN and CD209/D-SIGN
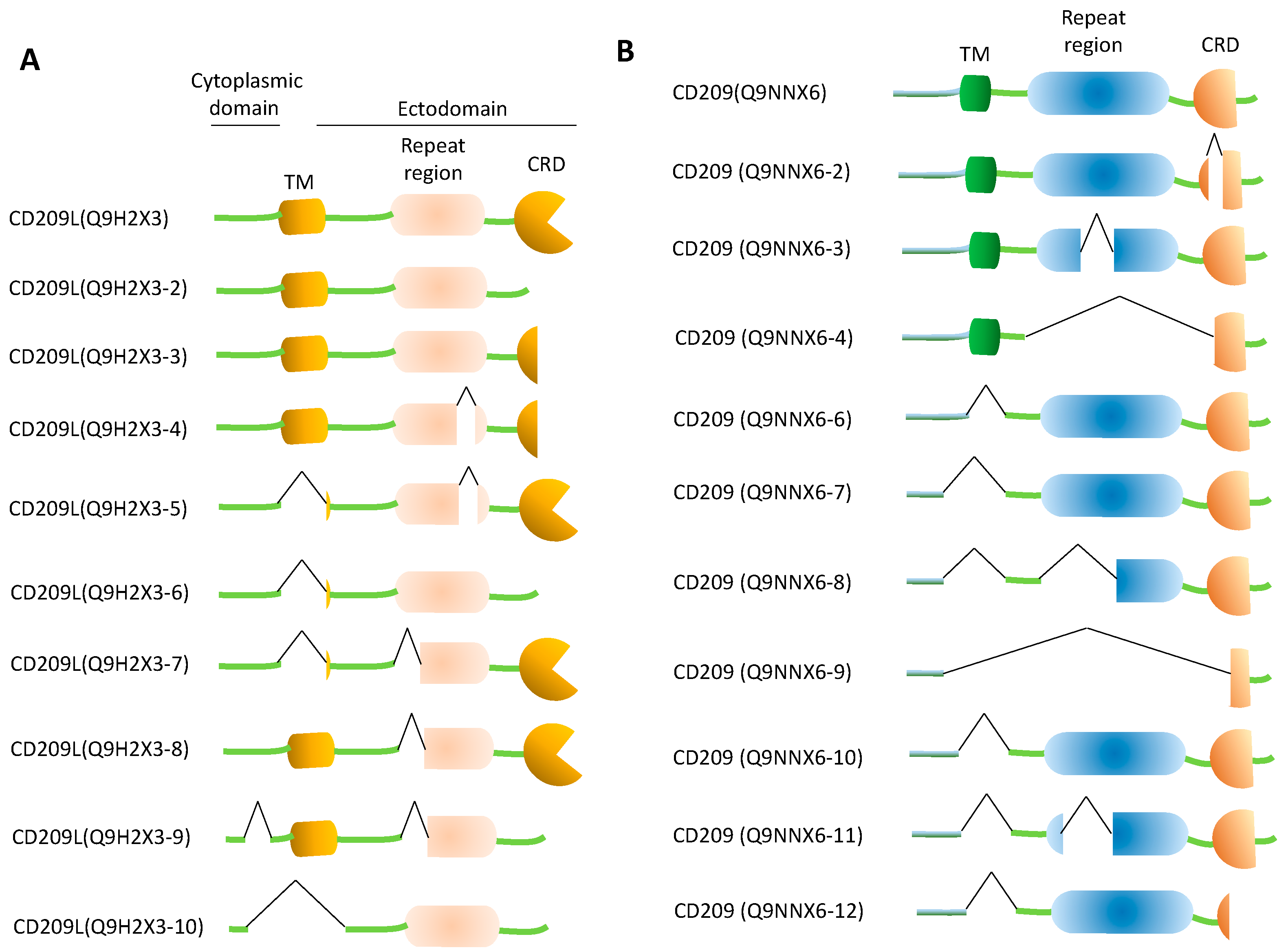
5. Decoy CD209L and CD209 Proteins:
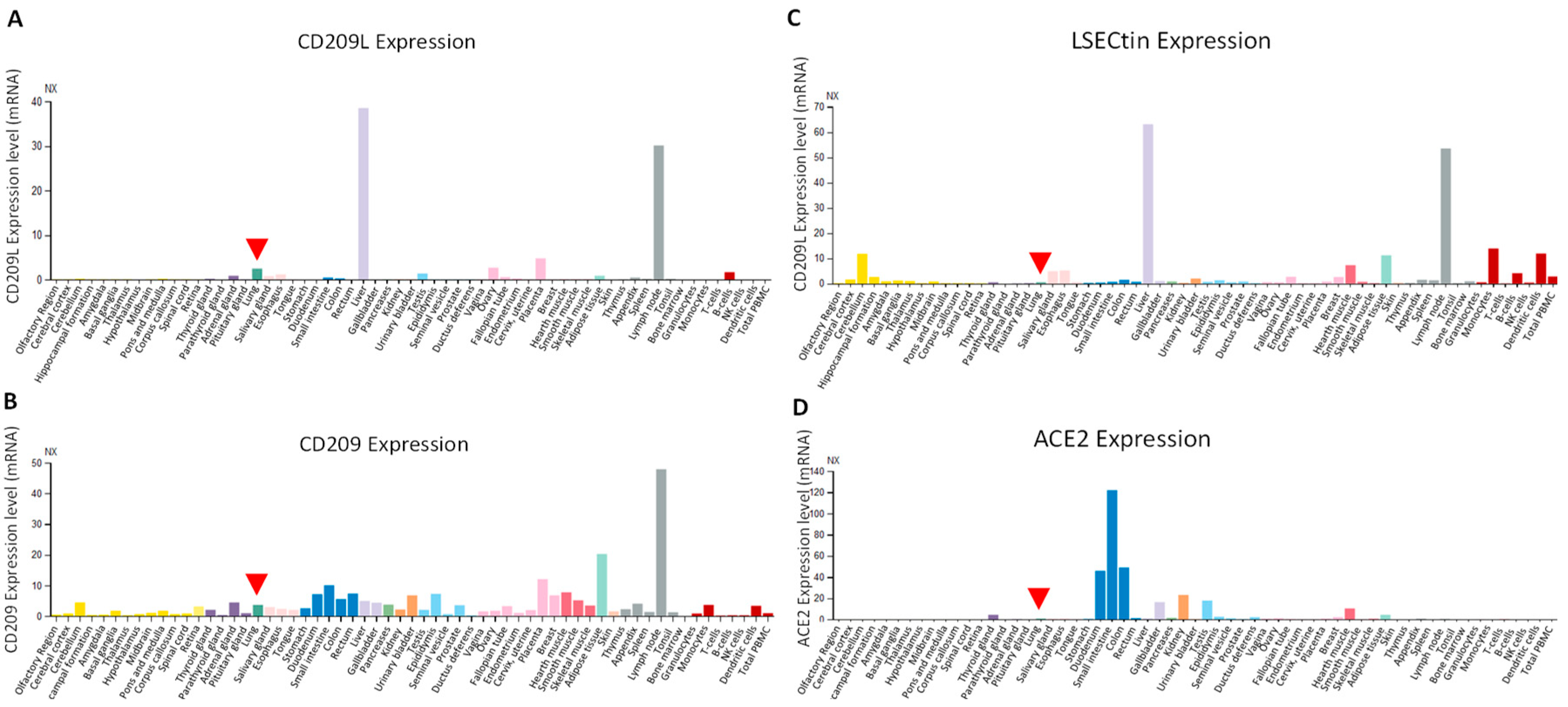
6. Expression Profile of CD209L Family Proteins in Human Tissues and Cells:
7. SARS-CoV-2, CD290 Family Proteins and Endothelial Cells:
8. Conclusions and Perspective:
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mulloy, B.; Linhardt, R.J. Order out of complexity—Protein structures that interact with heparin. Curr. Opin. Struct. Biol. 2001, 11, 623–628. [Google Scholar] [CrossRef]
- Esko, J.D.; Selleck, S.B. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Ann. Rev. Biochem. 2002, 71, 435–471. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, D.C. Animal lectins: A historical introduction and overview. Biochim. Biophys. Acta 2002, 1572, 187–197. [Google Scholar] [CrossRef]
- Van Breedam, W.; Pohlmann, S.; Favoreel, H.W.; de Groot, R.J.; Nauwynck, H.J. Bitter-sweet symphony: Glycan-lectin interactions in virus biology. FEMS Microbiol. Rev. 2014, 38, 598–632. [Google Scholar] [CrossRef] [Green Version]
- Sharon, N.; Lis, H. History of lectins: From hemagglutinins to biological recognition molecules. Glycobiology 2004, 14, 53R–62R. [Google Scholar] [CrossRef] [Green Version]
- Drickamer, K.; Fadden, A.J. Genomic analysis of C-type lectins. Biochem. Soc. Symp. 2002, 69, 59–72. [Google Scholar] [CrossRef]
- Zelensky, A.N.; Gready, J.E. The C-type lectin-like domain superfamily. FEBS J. 2005, 272, 6179–6217. [Google Scholar] [CrossRef]
- Sharon, N.; Lis, H. The structural basis for carbohydrate recognition by lectins. Adv. Exp. Med. Biol. 2001, 491, 1–16. [Google Scholar] [CrossRef]
- Ebner, S.; Sharon, N.; Ben-Tal, N. Evolutionary analysis reveals collective properties and specificity in the C-type lectin and lectin-like domain superfamily. Proteins 2003, 53, 44–55. [Google Scholar] [CrossRef]
- Drickamer, K.; Taylor, M.E. Recent insights into structures and functions of C-type lectins in the immune system. Curr. Opin. Struct. Biol. 2015, 34, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Soilleux, E.J.; Barten, R.; Trowsdale, J. DC-SIGN; a related gene, DC-SIGNR and CD23 form a cluster on 19p13. J. Immunol. 2000, 165, 2937–2942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Tang, L.; Zhang, G.; Wei, H.; Cui, Y.; Guo, L.; Gou, Z.; Chen, X.; Jiang, D.; Zhu, Y.; et al. Characterization of a novel C-type lectin-like gene, LSECtin: Demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J. Biol. Chem. 2004, 279, 18748–18758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, C.G.; Takahara, K.; Umemoto, E.; Yashima, Y.; Matsubara, K.; Matsuda, Y.; Clausen, B.E.; Inaba, K.; Steinman, R.M. Five mouse homologues of the human dendritic cell C-type lectin, DC-SIGN. Int. Immunol. 2001, 13, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Crocker, P.R.; Clark, E.A.; Filbin, M.; Gordon, S.; Jones, Y.; Kehrl, J.H.; Kelm, S.; Le Douarin, N.; Powell, L.; Roder, J.; et al. Siglecs: A family of sialic-acid binding lectins. Glycobiology 1998, 8. [Google Scholar] [CrossRef] [Green Version]
- Varki, A.; Angata, T. Siglecs—The major subfamily of I-type lectins. Glycobiology 2006, 16, 1R–27R. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amraie, R.; Napoleon, M.A.; Yin, W.; Berrigan, J.; Suder, E.; Zhao, G.; Olejnik, J.; Gummuluru, S.; Muhlberger, E.; Chitalia, V.; et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Bashirova, A.A.; Geijtenbeek, T.B.; van Duijnhoven, G.C.; van Vliet, S.J.; Eilering, J.B.; Martin, M.P.; Wu, L.; Martin, T.D.; Viebig, N.; Knolle, P.A.; et al. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J. Exp. Med. 2001, 193, 671–678. [Google Scholar] [CrossRef]
- Hibbert, R.G.; Teriete, P.; Grundy, G.J.; Beavil, R.L.; Reljic, R.; Holers, V.M.; Hannan, J.P.; Sutton, B.J.; Gould, H.J.; McDonnell, J.M. The structure of human CD23 and its interactions with IgE and CD21. J. Exp. Med. 2005, 202, 751–760. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Yang, J.; Tang, X.; Ying, W.; Qian, X.; He, F. The DC-SIGN family member LSECtin is a novel ligand of CD44 on activated T cells. Eur. J. Immunol. 2010, 40, 1185–1191. [Google Scholar] [CrossRef]
- Kerr, J.R. Cell adhesion molecules in the pathogenesis of and host defence against microbial infection. Mol. Pathol. 1999, 52, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amraei, R.; Rahimi, N. COVID-19, Renin-Angiotensin System and Endothelial Dysfunction. Cells 2020, 9, 1652. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.P.; Lasala, F.; Carrillo, J.; Muniz, O.; Corbi, A.L.; Delgado, R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002, 76, 6841–6844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cormier, E.G.; Durso, R.J.; Tsamis, F.; Boussemart, L.; Manix, C.; Olson, W.C.; Gardner, J.P.; Dragic, T. L-SIGN (CD209L) and DC-SIGN (CD209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. USA 2004, 101, 14067–14072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffers, S.A.; Hemmila, E.M.; Holmes, K.V. Human coronavirus 229E can use CD209L (L-SIGN) to enter cells. Adv. Exp. Med. Biol. 2006, 581, 265–269. [Google Scholar] [CrossRef] [Green Version]
- Marzi, A.; Gramberg, T.; Simmons, G.; Moller, P.; Rennekamp, A.J.; Krumbiegel, M.; Geier, M.; Eisemann, J.; Turza, N.; Saunier, B.; et al. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004, 78, 12090–12095. [Google Scholar] [CrossRef] [Green Version]
- Londrigan, S.L.; Turville, S.G.; Tate, M.D.; Deng, Y.M.; Brooks, A.G.; Reading, P.C. N-linked glycosylation facilitates sialic acid-independent attachment and entry of influenza A viruses into cells expressing DC-SIGN or L-SIGN. J. Virol. 2011, 85, 2990–3000. [Google Scholar] [CrossRef] [Green Version]
- Tassaneetrithep, B.; Burgess, T.H.; Granelli-Piperno, A.; Trumpfheller, C.; Finke, J.; Sun, W.; Eller, M.A.; Pattanapanyasat, K.; Sarasombath, S.; Birx, D.L.; et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 2003, 197, 823–829. [Google Scholar] [CrossRef] [Green Version]
- Shimojima, M.; Takenouchi, A.; Shimoda, H.; Kimura, N.; Maeda, K. Distinct usage of three C-type lectins by Japanese encephalitis virus: DC-SIGN, DC-SIGNR, and LSECtin. Arch. Virol. 2014, 159, 2023–2031. [Google Scholar] [CrossRef]
- Yang, Z.Y.; Huang, Y.; Ganesh, L.; Leung, K.; Kong, W.P.; Schwartz, O.; Subbarao, K.; Nabel, G.J. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 2004, 78, 5642–5650. [Google Scholar] [CrossRef] [Green Version]
- Jeffers, S.A.; Tusell, S.M.; Gillim-Ross, L.; Hemmila, E.M.; Achenbach, J.E.; Babcock, G.J.; Thomas, W.D., Jr.; Thackray, L.B.; Young, M.D.; Mason, R.J.; et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 2004, 101, 15748–15753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colmenares, M.; Puig-Kroger, A.; Pello, O.M.; Corbi, A.L.; Rivas, L. Dendritic cell (DC)-specific intercellular adhesion molecule 3 (ICAM-3)-grabbing nonintegrin (DC-SIGN, CD209), a C-type surface lectin in human DCs, is a receptor for Leishmania amastigotes. J. Biol. Chem. 2002, 277, 36766–36769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Skurnik, M.; Zhang, S.S.; Schwartz, O.; Kalyanasundaram, R.; Bulgheresi, S.; He, J.J.; Klena, J.D.; Hinnebusch, B.J.; Chen, T. Human dendritic cell-specific intercellular adhesion molecule-grabbing nonintegrin (CD209) is a receptor for Yersinia pestis that promotes phagocytosis by dendritic cells. Infect. Immun. 2008, 76, 2070–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtis, B.M.; Scharnowske, S.; Watson, A.J. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 1992, 89, 8356–8360. [Google Scholar] [CrossRef] [Green Version]
- Geijtenbeek, T.B.H.; van Duijnhoven, G.C.F.; van Vliet, S.J.; Krieger, E.; Vriend, G.; Figdor, C.G.; van Kooyk, Y. Identification of different binding sites in the dendritic cell-specific receptor DC-SIGN for intercellular adhesion molecule 3 and HIV-1. J. Biol. Chem. 2002, 277, 11314–11320. [Google Scholar] [CrossRef] [Green Version]
- Simmons, G.; Reeves, J.D.; Grogan, C.C.; Vandenberghe, L.H.; Baribaud, F.; Whitbeck, J.C.; Burke, E.; Buchmeier, M.J.; Soilleux, E.J.; Riley, J.L.; et al. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 2003, 305, 115–123. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.; Simmons, G.; Pohlmann, S.; Baribaud, F.; Ni, H.; Leslie, G.J.; Haggarty, B.S.; Bates, P.; Weissman, D.; Hoxie, J.A.; et al. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J. Virol. 2003, 77, 1337–1346. [Google Scholar] [CrossRef] [Green Version]
- Lai, W.K.; Sun, P.J.; Zhang, J.; Jennings, A.; Lalor, P.F.; Hubscher, S.; McKeating, J.A.; Adams, D.H. Expression of DC-SIGN and DC-SIGNR on human sinusoidal endothelium: A role for capturing hepatitis C virus particles. Am. J. Pathol. 2006, 169, 200–208. [Google Scholar] [CrossRef] [Green Version]
- de Witte, L.; Abt, M.; Schneider-Schaulies, S.; van Kooyk, Y.; Geijtenbeek, T.B.H. Measles virus targets DC-SIGN to enhance dendritic cell infection. J. Virol. 2006, 80, 3477–3486. [Google Scholar] [CrossRef] [Green Version]
- de Jong, M.A.W.P.; de Witte, L.; Bolmstedt, A.; van Kooyk, Y.; Geijtenbeek, T.B.H. Dendritic cells mediate herpes simplex virus infection and transmission through the C-type lectin DC-SIGN. J. Gen. Virol. 2008, 89, 2398–2409. [Google Scholar] [CrossRef]
- Mou, H.; Raj, V.S.; van Kuppeveld, F.J.M.; Rottier, P.J.M.; Haagmans, B.L.; Bosch, B.J. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J. Virol. 2013, 87, 9379–9383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goncalves, A.-R.; Moraz, M.-L.; Pasquato, A.; Helenius, A.; Lozach, P.-Y.; Kunz, S. Role of DC-SIGN in Lassa virus entry into human dendritic cells. J. Virol. 2013, 87, 11504–11515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, T.R.; McLellan, J.S.; Graham, B.S. Respiratory syncytial virus glycoprotein G interacts with DC-SIGN and L-SIGN to activate ERK1 and ERK2. J. Virol. 2012, 86, 1339–1347. [Google Scholar] [CrossRef] [Green Version]
- Lozach, P.-Y.; Kuhbacher, A.; Meier, R.; Mancini, R.; Bitto, D.; Bouloy, M.; Helenius, A. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 2011, 10, 75–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, C.W.; Nguyen, H.-Y.; Hanna, S.L.; Sanchez, M.D.; Doms, R.W.; Pierson, T.C. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J. Virol. 2006, 80, 1290–1301. [Google Scholar] [CrossRef] [Green Version]
- Carroll, M.V.; Sim, R.B.; Bigi, F.; Jakel, A.; Antrobus, R.; Mitchell, D.A. Identification of four novel DC-SIGN ligands on Mycobacterium bovis BCG. Protein Cell 2010, 1, 859–870. [Google Scholar] [CrossRef] [Green Version]
- Halary, F.; Amara, A.; Lortat-Jacob, H.; Messerle, M.; Delaunay, T.; Houles, C.; Fieschi, F.; Arenzana-Seisdedos, F.; Moreau, J.F.; Dechanet-Merville, J. Human cytomegalovirus binding to DC-SIGN is required for dendritic cell infection and target cell trans-infection. Immunity 2002, 17, 653–664. [Google Scholar] [CrossRef] [Green Version]
- Gramberg, T.; Hofmann, H.; Moller, P.; Lalor, P.F.; Marzi, A.; Geier, M.; Krumbiegel, M.; Winkler, T.; Kirchhoff, F.; Adams, D.H.; et al. LSECtin interacts with filovirus glycoproteins and the spike protein of SARS coronavirus. Virology 2005, 340, 224–236. [Google Scholar] [CrossRef] [Green Version]
- Shimojima, M.; Stroher, U.; Ebihara, H.; Feldmann, H.; Kawaoka, Y. Identification of cell surface molecules involved in dystroglycan-independent Lassa virus cell entry. J. Virol. 2012, 86, 2067–2078. [Google Scholar] [CrossRef] [Green Version]
- Geijtenbeek, T.B.H.; Engering, A.; Van Kooyk, Y. DC-SIGN, a C-type lectin on dendritic cells that unveils many aspects of dendritic cell biology. J. Leukoc. Biol. 2002, 71, 921–931. [Google Scholar]
- Geijtenbeek, T.B.; Kwon, D.S.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Middel, J.; Cornelissen, I.L.; Nottet, H.S.; KewalRamani, V.N.; Littman, D.R.; et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 2000, 100, 587–597. [Google Scholar] [CrossRef] [Green Version]
- Moris, A.; Nobile, C.; Buseyne, F.; Porrot, F.; Abastado, J.-P.; Schwartz, O. DC-SIGN promotes exogenous MHC-I-restricted HIV-1 antigen presentation. Blood 2004, 103, 2648–2654. [Google Scholar] [CrossRef] [PubMed]
- Turville, S.G.; Santos, J.J.; Frank, I.; Cameron, P.U.; Wilkinson, J.; Miranda-Saksena, M.; Dable, J.; Stossel, H.; Romani, N.; Piatak, M., Jr.; et al. Immunodeficiency virus uptake, turnover, and 2-phase transfer in human dendritic cells. Blood 2004, 103, 2170–2179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hijazi, K.; Wang, Y.; Scala, C.; Jeffs, S.; Longstaff, C.; Stieh, D.; Haggarty, B.; Vanham, G.; Schols, D.; Balzarini, J.; et al. DC-SIGN increases the affinity of HIV-1 envelope glycoprotein interaction with CD4. PLoS ONE 2011, 6, e28307. [Google Scholar] [CrossRef] [Green Version]
- Burleigh, L.; Lozach, P.-Y.; Schiffer, C.; Staropoli, I.; Pezo, V.; Porrot, F.; Canque, B.; Virelizier, J.-L.; Arenzana-Seisdedos, F.; Amara, A. Infection of dendritic cells (DCs), not DC-SIGN-mediated internalization of human immunodeficiency virus, is required for long-term transfer of virus to T cells. J. Virol. 2006, 80, 2949–2957. [Google Scholar] [CrossRef] [Green Version]
- Geijtenbeek, T.B.; Krooshoop, D.J.; Bleijs, D.A.; van Vliet, S.J.; van Duijnhoven, G.C.; Grabovsky, V.; Alon, R.; Figdor, C.G.; van Kooyk, Y. DC-SIGN-ICAM-2 interaction mediates dendritic cell trafficking. Nat. Immunol. 2000, 1, 353–357. [Google Scholar] [CrossRef] [Green Version]
- Kwon, D.S.; Gregorio, G.; Bitton, N.; Hendrickson, W.A.; Littman, D.R. DC-SIGN-mediated internalization of HIV is required for trans-enhancement of T cell infection. Immunity 2002, 16, 135–144. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Wu, L.; Bohks, S.M.; KewalRamani, V.N.; Unutmaz, D.; Hope, T.J. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 2003, 300, 1295–1297. [Google Scholar] [CrossRef] [Green Version]
- Cameron, P.U.; Freudenthal, P.S.; Barker, J.M.; Gezelter, S.; Inaba, K.; Steinman, R.M. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 1992, 257, 383–387. [Google Scholar] [CrossRef]
- Cavrois, M.; Neidleman, J.; Greene, W.C. The achilles heel of the trojan horse model of HIV-1 trans-infection. PLoS Pathog. 2008, 4, e1000051. [Google Scholar] [CrossRef] [Green Version]
- Boukour, S.; Masse, J.M.; Benit, L.; Dubart-Kupperschmitt, A.; Cramer, E.M. Lentivirus degradation and DC-SIGN expression by human platelets and megakaryocytes. J. Thrombosis Haemost. JTH 2006, 4, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Chaipan, C.; Soilleux, E.J.; Simpson, P.; Hofmann, H.; Gramberg, T.; Marzi, A.; Geier, M.; Stewart, E.A.; Eisemann, J.; Steinkasserer, A.; et al. DC-SIGN and CLEC-2 mediate human immunodeficiency virus type 1 capture by platelets. J. Virol. 2006, 80, 8951–8960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rappocciolo, G.; Piazza, P.; Fuller, C.L.; Reinhart, T.A.; Watkins, S.C.; Rowe, D.T.; Jais, M.; Gupta, P.; Rinaldo, C.R. DC-SIGN on B lymphocytes is required for transmission of HIV-1 to T lymphocytes. PLoS Pathog. 2006, 2, e70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thépaut, M.; Luczkowiak, J.; Vivès, C.; Labiod, N.; Bally, I.; Lasala, F.; Grimoire, Y.; Fenel, D.; Sattin, S.; Thielens, N.; et al. DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. bioRxiv 2020. [Google Scholar] [CrossRef]
- Hodges, A.; Sharrocks, K.; Edelmann, M.; Baban, D.; Moris, A.; Schwartz, O.; Drakesmith, H.; Davies, K.; Kessler, B.; McMichael, A.; et al. Activation of the lectin DC-SIGN induces an immature dendritic cell phenotype triggering Rho-GTPase activity required for HIV-1 replication. Nat. Immunol. 2007, 8, 569–577. [Google Scholar] [CrossRef]
- Gringhuis, S.I.; den Dunnen, J.; Litjens, M.; van Het Hof, B.; van Kooyk, Y.; Geijtenbeek, T.B.H. C-type lectin DC-SIGN modulates Toll-like receptor signaling via Raf-1 kinase-dependent acetylation of transcription factor NF-kappaB. Immunity 2007, 26, 605–616. [Google Scholar] [CrossRef] [Green Version]
- Geijtenbeek, T.B.H.; Gringhuis, S.I. Signalling through C-type lectin receptors: Shaping immune responses. Nat. Rev. Immunol. 2009, 9, 465–479. [Google Scholar] [CrossRef]
- Delmas, B.; Gelfi, J.; L’Haridon, R.; Vogel, L.K.; Sjostrom, H.; Noren, O.; Laude, H. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature 1992, 357, 417–420. [Google Scholar] [CrossRef] [Green Version]
- Yeager, C.L.; Ashmun, R.A.; Williams, R.K.; Cardellichio, C.B.; Shapiro, L.H.; Look, A.T.; Holmes, K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 1992, 357, 420–422. [Google Scholar] [CrossRef] [Green Version]
- Matrosovich, M.; Herrler, G.; Klenk, H.D. Sialic Acid Receptors of Viruses. Top Curr. Chem. 2015, 367, 1–28. [Google Scholar]
- Huang, X.; Dong, W.; Milewska, A.; Golda, A.; Qi, Y.; Zhu, Q.K.; Marasco, W.A.; Baric, R.S.; Sims, A.C.; Pyrc, K.; et al. Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme. J. Virol. 2015, 89, 7202–7213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Muller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.-M.; Chu, H.; Wang, Y.; Wong, B.H.-Y.; Zhao, X.; Zhou, J.; Yang, D.; Leung, S.P.; Chan, J.F.-W.; Yeung, M.-L.; et al. Carcinoembryonic Antigen-Related Cell Adhesion Molecule 5 Is an Important Surface Attachment Factor That Facilitates Entry of Middle East Respiratory Syndrome Coronavirus. J. Virol. 2016, 90, 9114–9127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef]
- Hofmann, H.; Pyrc, K.; van der Hoek, L.; Geier, M.; Berkhout, B.; Pohlmann, S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Han, D.P.; Lohani, M.; Cho, M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007, 81, 12029–12039. [Google Scholar] [CrossRef] [Green Version]
- Soh, W.T.; Liu, Y.; Nakayama, E.E.; Ono, C.; Torii, S.; Nakagami, H.; Matsuura, Y.; Shioda, T.; Arase, H. The N-terminal domain of spike glycoprotein mediates SARS-CoV-2 infection by associating with L-SIGN and DC-SIGN. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gao, C.; Zeng, J.; Jia, N.; Stavenhagen, K.; Matsumoto, Y.; Zhang, H.; Li, J.; Hume, A.J.; Muhlberger, E.; van Die, I.; et al. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chiodo, F.; Bruijns, S.C.M.; Rodriguez, E.; Li, R.J.E.; Molinaro, A.; Silipo, A.; Di Lorenzo, F.; Garcia-Rivera, D.; Valdes-Balbin, Y.; Verez-Bencomo, V.; et al. Novel ACE2-Independent Carbohydrate-Binding of SARS-CoV-2 Spike Protein to Host Lectins and Lung Microbiota. bioRxiv 2020. [Google Scholar] [CrossRef]
- Snyder, G.A.; Colonna, M.; Sun, P.D. The structure of DC-SIGNR with a portion of its repeat domain lends insights to modeling of the receptor tetramer. J. Mol. Biol. 2005, 347, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Sol-Foulon, N.; Moris, A.; Nobile, C.; Boccaccio, C.; Engering, A.; Abastado, J.P.; Heard, J.M.; van Kooyk, Y.; Schwartz, O. HIV-1 Nef-induced upregulation of DC-SIGN in dendritic cells promotes lymphocyte clustering and viral spread. Immunity 2002, 16, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, D.A.; Fadden, A.J.; Drickamer, K. A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 2001, 276, 28939–28945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cambi, A.; de Lange, F.; van Maarseveen, N.M.; Nijhuis, M.; Joosten, B.; van Dijk, E.M.; de Bakker, B.I.; Fransen, J.A.; Bovee-Geurts, P.H.; van Leeuwen, F.N.; et al. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J. Cell Biol. 2004, 164, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.D.; Oldring, A.P.; Powlesland, A.S.; Tso, C.K.; Yang, C.; Drickamer, K.; Taylor, M.E. Autonomous tetramerization domains in the glycan-binding receptors DC-SIGN and DC-SIGNR. J. Mol. Biol. 2009, 387, 1075–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feinberg, H.; Tso, C.K.; Taylor, M.E.; Drickamer, K.; Weis, W.I. Segmented helical structure of the neck region of the glycan-binding receptor DC-SIGNR. J. Mol. Biol. 2009, 394, 613–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drickamer, K. C-type lectin-like domains. Curr. Opin. Struct. Biol. 1999, 9, 585–590. [Google Scholar] [CrossRef]
- Van Liempt, E.; Imberty, A.; Bank, C.M.; Van Vliet, S.J.; Van Kooyk, Y.; Geijtenbeek, T.B.; Van Die, I. Molecular basis of the differences in binding properties of the highly related C-type lectins DC-SIGN and L-SIGN to Lewis X trisaccharide and Schistosoma mansoni egg antigens. J. Biol. Chem. 2004, 279, 33161–33167. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Chandler, K.B.; Leon, D.R.; Kuang, J.; Meyer, R.D.; Rahimi, N.; Costello, C.E. N-Glycosylation regulates ligand-dependent activation and signaling of vascular endothelial growth factor receptor 2 (VEGFR2). J. Biol. Chem. 2019, 294, 13117–13130. [Google Scholar] [CrossRef]
- Daeron, M.; Jaeger, S.; Du Pasquier, L.; Vivier, E. Immunoreceptor tyrosine-based inhibition motifs: A quest in the past and future. Immunol. Rev. 2008, 224, 11–43. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, N. The ubiquitin-proteasome system meets angiogenesis. Mol. Cancer Ther. 2012, 11, 538–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Zhu, T. Determination of DC-SIGN and DC-SIGNR repeat region variations. Methods Mol. Biol. 2005, 304, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Kawana-Tachikawa, A.; Iwamoto, A.; Kitamura, Y. Influence of polymorphism in dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin-related (DC-SIGNR) gene on HIV-1 trans-infection. Biochem. Biophys. Res. Commun. 2010, 393, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Kashima, S.; Rodrigues, E.S.; Azevedo, R.; da Cruz Castelli, E.; Mendes-Junior, C.T.; Yoshioka, F.K.N.; da Silva, I.T.; Takayanagui, O.M.; Covas, D.T. DC-SIGN (CD209) gene promoter polymorphisms in a Brazilian population and their association with human T-cell lymphotropic virus type 1 infection. J. Gen. Virol. 2009, 90, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, L.B.; Neyrolles, O.; Babb, C.L.; Tailleux, L.; Quach, H.; McElreavey, K.; Helden, P.D.; Hoal, E.G.; Gicquel, B.; Quintana-Murci, L. Promoter variation in the DC-SIGN-encoding gene CD209 is associated with tuberculosis. PLoS Med. 2006, 3, e20. [Google Scholar] [CrossRef] [Green Version]
- Kagone, T.S.; Bisseye, C.; Meda, N.; Testa, J.; Pietra, V.; Kania, D.; Yonli, A.T.; Compaore, T.R.; Nikiema, J.B.; de Souza, C.; et al. A variant of DC-SIGN gene promoter associated with resistance to HIV-1 in serodiscordant couples in Burkina Faso. Asian Pac. J. Trop. Med. 2014, 7S1, S93–S96. [Google Scholar] [CrossRef] [Green Version]
- Tailleux, L.; Pham-Thi, N.; Bergeron-Lafaurie, A.; Herrmann, J.-L.; Charles, P.; Schwartz, O.; Scheinmann, P.; Lagrange, P.H.; de Blic, J.; Tazi, A.; et al. DC-SIGN induction in alveolar macrophages defines privileged target host cells for mycobacteria in patients with tuberculosis. PLoS Med. 2005, 2, e381. [Google Scholar] [CrossRef] [Green Version]
- Pohlmann, S.; Soilleux, E.J.; Baribaud, F.; Leslie, G.J.; Morris, L.S.; Trowsdale, J.; Lee, B.; Coleman, N.; Doms, R.W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 2001, 98, 2670–2675. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamming, I.; Timens, W.; Bulthuis, M.L.; Lely, A.T.; Navis, G.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. bioRxiv 2020. [Google Scholar] [CrossRef] [PubMed]
- Ganier, C.; Du-Harpur, X.; Harun, N.; Wan, B.; Arthurs, C.; Luscombe, N.; Watt, F.; Lynch, M. CD147 (BSG) but not ACE2 expression is detectable in vascular endothelial cells within single cell RNA sequencing datasets derived from multiple tissues in healthy individuals. bioRxiv 2020. [Google Scholar] [CrossRef]
- Engering, A.; van Vliet, S.J.; Hebeda, K.; Jackson, D.G.; Prevo, R.; Singh, S.K.; Geijtenbeek, T.B.; van Krieken, H.; van Kooyk, Y. Dynamic populations of dendritic cell-specific ICAM-3 grabbing nonintegrin-positive immature dendritic cells and liver/lymph node-specific ICAM-3 grabbing nonintegrin-positive endothelial cells in the outer zones of the paracortex of human lymph nodes. Am. J. Pathol. 2004, 164, 1587–1595. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, C.Y.; Wang, J.X.; Tang, N.L.; Xie, L.; Gong, Y.Y.; Yang, Z.; Xu, L.Y.; Kong, Q.P.; Zhang, Y.P. The neck-region polymorphism of DC-SIGNR in peri-centenarian from Han Chinese population. BMC Med. Genet. 2009, 10, 134. [Google Scholar] [CrossRef] [Green Version]
- Chan, V.S.; Chan, K.Y.; Chen, Y.; Poon, L.L.; Cheung, A.N.; Zheng, B.; Chan, K.H.; Mak, W.; Ngan, H.Y.; Xu, X.; et al. Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 2006, 38, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Fawcett, J.; Holness, C.L.; Needham, L.A.; Turley, H.; Gatter, K.C.; Mason, D.Y.; Simmons, D.L. Molecular cloning of ICAM-3, a third ligand for LFA-1, constitutively expressed on resting leukocytes. Nature 1992, 360, 481–484. [Google Scholar] [CrossRef]
- Kristof, E.; Zahuczky, G.; Katona, K.; Doro, Z.; Nagy, E.; Fesus, L. Novel role of ICAM3 and LFA-1 in the clearance of apoptotic neutrophils by human macrophages. Apoptosis 2013, 18, 1235–1251. [Google Scholar] [CrossRef]
- Patey, N.; Vazeux, R.; Canioni, D.; Potter, T.; Gallatin, W.M.; Brousse, N. Intercellular adhesion molecule-3 on endothelial cells. Expression in tumors but not in inflammatory responses. Am. J. Pathol. 1996, 148, 465–472. [Google Scholar] [PubMed]
- Huang, M.T.; Mason, J.C.; Birdsey, G.M.; Amsellem, V.; Gerwin, N.; Haskard, D.O.; Ridley, A.J.; Randi, A.M. Endothelial intercellular adhesion molecule (ICAM)-2 regulates angiogenesis. Blood 2005, 106, 1636–1643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, J.M.; Gonagle, D.M.; Ward, S.E.; Preston, R.J.S.; O’Donnell, J.S. Endothelial cells orchestrate COVID-19 coagulopathy. Lancet Haematol. 2020, 7, e553–e555. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Wiersinga, W.J.; Rhodes, A.; Cheng, A.C.; Peacock, S.J.; Prescott, H.C. Pathophysiology, Transmission, Diagnosis, and Treatment of Coronavirus Disease 2019 (COVID-19): A Review. JAMA 2020, 324, 782–793. [Google Scholar] [CrossRef]
- Fried, J.A.; Ramasubbu, K.; Bhatt, R.; Topkara, V.K.; Clerkin, K.J.; Horn, E.; Rabbani, L.; Brodie, D.; Jain, S.S.; Kirtane, A.J.; et al. The Variety of Cardiovascular Presentations of COVID-19. Circulation 2020, 141, 1930–1936. [Google Scholar] [CrossRef] [Green Version]
- Arentz, M.; Yim, E.; Klaff, L.; Lokhandwala, S.; Riedo, F.X.; Chong, M.; Lee, M. Characteristics and Outcomes of 21 Critically Ill Patients With COVID-19 in Washington State. JAMA 2020, 323, 1612–1614. [Google Scholar] [CrossRef] [Green Version]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W.; the Northwell, C.-R.C.; Barnaby, D.P.; Becker, L.B.; Chelico, J.D.; et al. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA 2020, 323, 2052–2059. [Google Scholar] [CrossRef]
- Xiang-Hua, Y.; Le-Min, W.; Ai-Bin, L.; Zhu, G.; Riquan, L.; Xu-You, Z.; Wei-Wei, R.; Ye-Nan, W. Severe acute respiratory syndrome and venous thromboembolism in multiple organs. Am. J. Respir. Crit. Care Med. 2010, 182, 436–437. [Google Scholar] [CrossRef]
- Ng, K.H.; Wu, A.K.; Cheng, V.C.; Tang, B.S.; Chan, C.Y.; Yung, C.Y.; Luk, S.H.; Lee, T.W.; Chow, L.; Yuen, K.Y. Pulmonary artery thrombosis in a patient with severe acute respiratory syndrome. Postgrad. Med. J. 2005, 81, e3. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
| Gene Name | Pathogen Name | References |
|---|---|---|
| CD209 | HIV-1 and HIV-2 | [34,35] |
| Ebolavirus | [36,37] | |
| Cytomegalovirus | [24,38] | |
| Hepatitis C virus | [24] | |
| Dengue virus | [28] | |
| Measles virus | [39] | |
| Herpes simplex virus | [40] | |
| Influenza virus A | [27] | |
| SARS-CoV-2 | [16] | |
| SARS-CoV | [30] | |
| MERS | [41] | |
| Japanese encephalitis virus | [29] | |
| Lassa virus | [42] | |
| Respiratory syncytial virus | [43] | |
| Rift valley fever virus | [44] | |
| Uukuniemi virus | [44] | |
| West-Nile virus | [45] | |
| CD209L | Ebolavirus | [23] |
| Hepatitis C virus | [24] | |
| HIV-1 | [37,46] | |
| Human coronavirus 229E | [25] | |
| Human cytomegalovirus/HHV-5 | [47] | |
| Influenza virus | [27] | |
| SARS-CoV | [26] | |
| SARS-CoV-2 | [16] | |
| West-Nile virus | [26] | |
| Japanese encephalitis virus | [29] | |
| Marburg virus | [26] | |
| LSECtin | Japanese encephalitis virus | [12] |
| Ebolavirus | [48] | |
| SARS-CoV | [48] | |
| Lassa virus | [49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahimi, N. C-type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors. Biology 2021, 10, 1. https://doi.org/10.3390/biology10010001
Rahimi N. C-type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors. Biology. 2021; 10(1):1. https://doi.org/10.3390/biology10010001
Chicago/Turabian StyleRahimi, Nader. 2021. "C-type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors" Biology 10, no. 1: 1. https://doi.org/10.3390/biology10010001






