Abstract
In plants there is no universal protocol for RNA extraction, since optimizations are required depending on the species, tissues and developmental stages. Some plants/tissues are rich in secondary metabolites or synthesize thick cell walls, which hinder an efficient RNA extraction. One such example is bast fibres, long extraxylary cells characterized by a thick cellulosic cell wall. Given the economic importance of bast fibres, which are used in the textile sector, as well as in biocomposites as green substitutes of glass fibres, it is desirable to better understand their development from a molecular point of view. This knowledge favours the development of biotechnological strategies aimed at improving specific properties of bast fibres. To be able to perform high-throughput analyses, such as, for instance, transcriptomics of bast fibres, RNA extraction is a crucial and limiting step. We here detail a protocol enabling the rapid extraction of high quality RNA from the bast fibres of textile hemp, Cannabis sativa L., a multi-purpose fibre crop standing in the spotlight of research.
1. Introduction
The extraction of high quality ribonucleic acid (RNA) is a crucial step for the analysis of gene expression profiles using both targeted real-time PCR (RT-qPCR) and next generation sequencing (NGS). RT-qPCR enables the study of specific genes, by monitoring the dynamics in their expression following specific treatments, or in response to exogenous/endogenous factors; NGS is a high-throughput technology allowing the experimenter to get information on the dynamics of the whole set of expressed genes in a given organism (transcriptome). The rich compositional variety of plant tissues poses a challenge to the separation of RNA, because the presence of secondary metabolites, polysaccharides, as well as thick cell walls, can compromise the extraction. As reliable gene expression profiling depends not only on the purity of the extracted RNA, but also on its integrity [1,2], selective but not destructive extraction buffers should be used.
Bast fibres are cells of the sclerenchyma which elongate considerably and possess a thick cellulosic cell wall (known as the G-layer) [3,4,5]. Bast fibres are in the spotlight of research since they are widely used in industry [6,7]. Their importance has grown in the last years, because they represent an important bioresource enabling the development of a bio-economy: these natural fibres can indeed be used to substitute glass fibres and, besides lowering the C footprint, they raise no health-related issues [7] (as opposed to glass fibres).
To study the development of bast fibres using a molecular approach, it is necessary to isolate them from the surrounding tissues composed mainly of parenchyma/collenchyma and to extract pure and intact RNA.
We have here tested four different methods to extract RNA from isolated hemp bast fibres, i.e., (1) the RNeasy mini kit (Qiagen, Leusden, The Netherlands) with both buffers RLT and RLC; (2) a sodium dodecyl sulfate (SDS)-phenol/chloroform/isoamyl alcohol (P/C/I) + TRIzol method [8]; (3) a self-developed SDS-based protocol based on [8] and combined with a purification on the RNeasy spin columns; and (4) a cetyl trimethylammonium bromide (CTAB)-based procedure coupled to the RNeasy spin columns (adapted from [9,10]).
We show that the method ensuring a rapid and efficient isolation of intact, pure RNA from hemp bast fibres is the CTAB-based procedure. The RNA is suitable for both targeted gene expression profiling and RNA-Seq.
2. Experimental Section
2.1. Plant Cultivation and Sampling of Bast Fibres
The hemp plants (cv. Santhica 27, aged 1.5 months) were grown in controlled growth chambers (60% humidity) with a 16 h light 25 °C/8 h dark 20 °C cycle. At the sampling time, the plants had between six to eight internodes. The internodes of two plants were pooled per biological replicate, the cortex was quickly peeled off and put in 10 mL freshly prepared 80% (% v/v) ethanol (EtOH). Four independent biological replicates were analyzed and, for the 5′-3′ ΔCq assay hereafter shown, three technical replicates were performed. As detailed in the Results and Discussion Section, this assay enables the selective analysis of the integrity of messenger RNAs (mRNAs), which constitute only a small fraction (ca. 1%–5%) of the whole RNA population in a cell.
We separated the bast fibres in a mortar, by repeatedly pressing the cortex with a pestle (Supplementary Materials Figure S1), as described in [11,12].
The fibres were subsequently quickly blot-dried using autoclaved wipers (WypAll, Kimberley-Clark, Muller & Wegener, Luxembourg) and immediately frozen in liquid nitrogen. They were then ground to a fine powder using liquid nitrogen, a mortar and a pestle. Approximately 100–150 mg of frozen powder were aliquoted in 2 mL round-bottom Eppendorf tubes and stored at −80 °C until extraction. The ground bast fibres were then subjected to the four different RNA extraction protocols detailed below.
2.2. RNA Extraction Methods, Assessment of Purity and Integrity
For the extraction using the RNeasy mini kit (Qiagen), the manufacturer’s instructions were followed, including the on-column DNase treatment. Both RLT and RLC buffers were tested.
The SDS-P/C/I + TRIzol method is based on [8]. The extraction buffer contained: 100 mM Tris-HCl pH 8, 150 mM LiCl, 50 mM ethylenediaminetetraacetic acid (EDTA), 1.5% (% w/v) SDS. Right before use, 10 µL β-mercaptoethanol (ME)/mL buffer used were added, then 400 µL of buffer were added to 100–150 mg tissue powder and the tubes were vortexed. An equal volume of P/C/I (25:24:1) was added and the homogenate vortexed. A centrifugation at 4 °C at 10,000 g for 10 min was carried out to separate the phases, then the aqueous phase was withdrawn and 200 μL C/I (24:1) were added. The tubes were shaken and put on ice for 5 min; a centrifugation was carried out using the same parameters described above. One mL TRIzol (Thermofisher Scientific, Gent, Belgium) was added to the aqueous phase and subsequently 200 μL C/I (24:1). A short incubation of 2–3 min was performed at room temperature, then the tubes were centrifuged as previously described. To the aqueous phase 300 μL cold isopropanol plus 300 μL of a high salt buffer (1.2 M NaCl, 0.8 M sodium citrate) were added and the RNA was precipitated for 1 h at −20 °C. The tubes were spun down as previously described, the pellet was washed in EtOH 70%, air dried and resuspended in 50 μL sterile water. The RNA was cleaned following the RNeasy mini kit clean-up protocol (Qiagen).
The SDS-P/C/I + RNeasy procedure is identical to the SDS-P/C/I + TRIzol method, until the phase separation with C/I (24:1). However it omits the TRIzol step and instead precipitates the RNA directly with cold isopropanol for 1 h at −20 °C. The RNA is then bound to the silica membranes of the RNeasy columns, washed with the RW1 buffer and eluted, as detailed in the RNeasy mini kit protocol (Qiagen).
The CTAB-based protocol is based on the buffer described in [10], i.e., 2% (% w/v) CTAB, 2.5% (% w/v) polyvinylpyrrolidone (PVP-40), 2 M NaCl, 100 mM Tris-HCl pH 8, 25 mM EDTA, 20 µL β-ME/mL buffer added just prior to use. Five hundred μL of the buffer were added to 100–150 mg of frozen tissue powder and an incubation with gentle shaking was carried out at 60 °C for 10 min. Subsequently 500 μL C/I (24:1) were added, the tubes were vortexed and centrifuged as described before. The aqueous phase was withdrawn and a precipitation was done with cold isopropanol (2/3 of the volume) for 1 h at −20 °C. The RNA was then bound to the silica membranes of the RNeasy columns and washed/eluted following the manufacturer’s instructions.
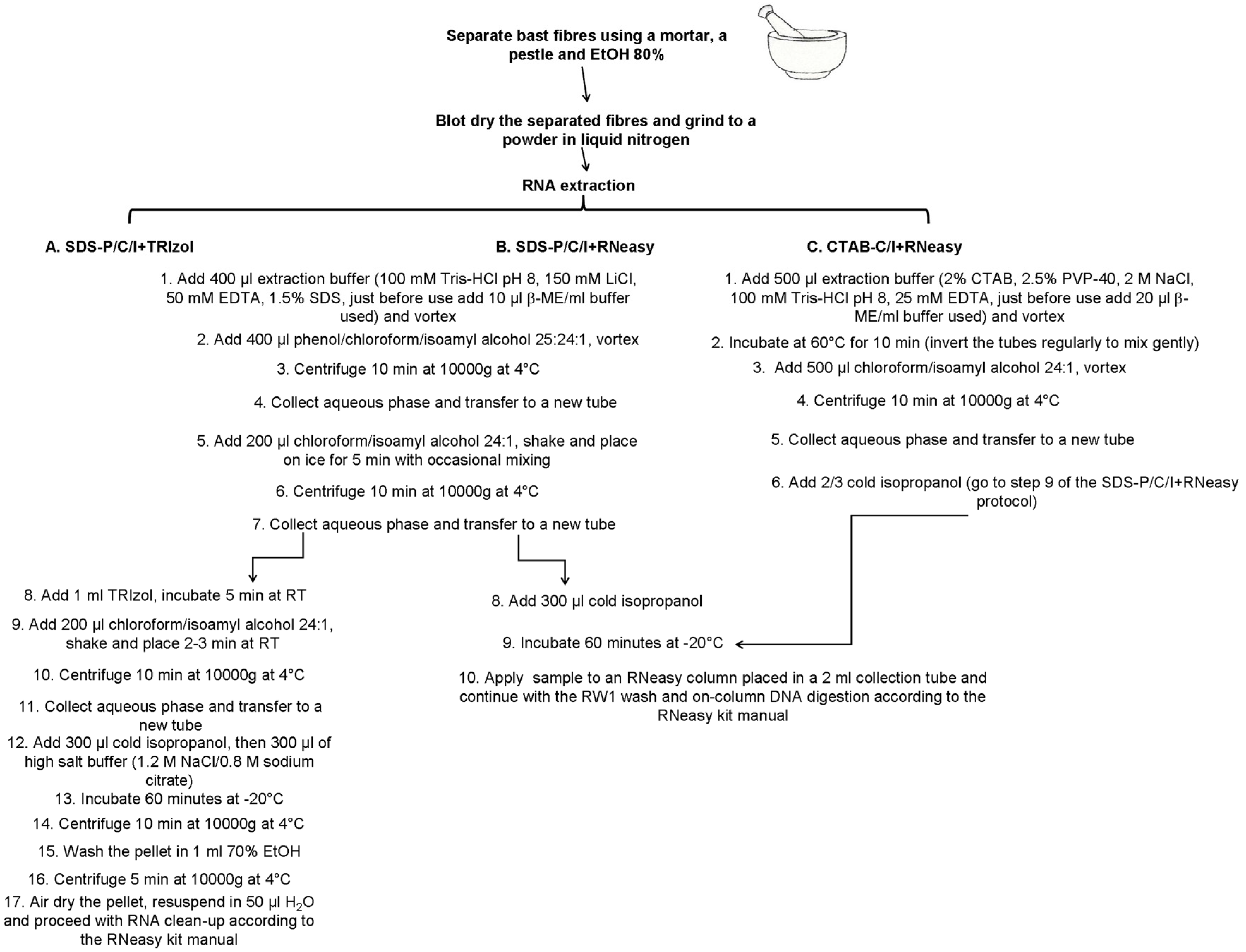
A summary of the three extraction methods described is shown in Figure 1.
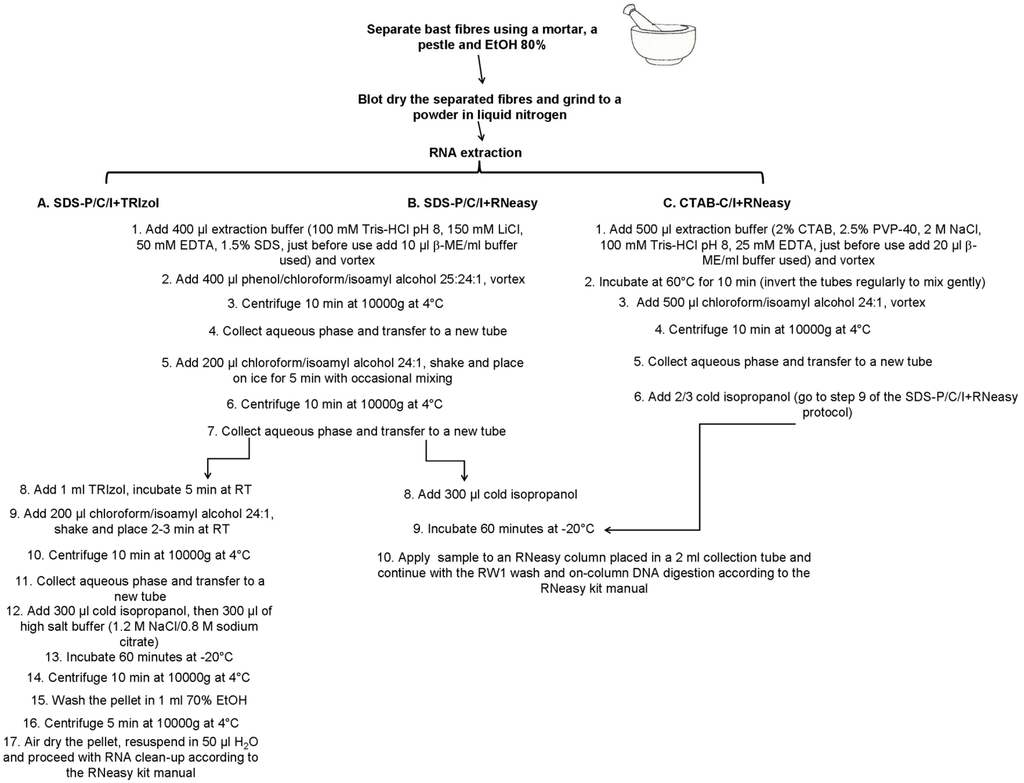
Figure 1.
Scheme showing three RNA extraction protocols used in the study. Details on the buffer composition are also provided.
The purity and quantity of the extracted RNA were measured at the NanoDrop ND-1000 (Thermo Scientific, Villebon-sur-Yvette, France). To assess the integrity of the RNA extracted with the four protocols, a quality control was carried out using a 2100 Bioanalyzer with the RNA Nano 6000 kit (Agilent Technologies, Santa Clara, CA, USA), following the manufacturer’s instructions.
For the 5′-3′ ΔCq assay, we decided to use two housekeeping genes, i.e., hemp glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin (contigs csa_locus_1460_iso_2_len_1872_ver_2 and csa_locus_2263_iso_5_len_1488_ver_2, respectively, Medicinal Plants Genomic Resource database [13]). For this test, primers amplifying the 5′ and 3′ regions of the genes were designed (Supplementary Materials Table S1).
Reverse transcription was carried out using the ProtoScript II reverse transcriptase (RTase) (NEB) with oligo d(T)23 VN (New England Biolabs, Leiden, The Netherlands), following the manufacturer’s instructions.
For the RT-qPCR, the Takyon Low ROX SYBR MasterMix dTTP Blue Kit (Eurogentec, Liège, Belgium) was used. Reactions (10 µL final volume) were set up in 384-wells plate using a liquid handling system (epMotion, Eppendorf, Hambourg, Germany) and run on a ViiA 7 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA). The PCR conditions were as described in [14]. The specificity of the amplified products was checked with a melt curve analysis.
3. Results and Discussion
3.1. Comparison of the RNA Extraction Methods
The extraction using the RNeasy mini kit (Qiagen) was not appropriate for RNA extraction from hemp bast fibres. Although a higher yield of RNA was observed with the buffer RLC, the RNA was always contaminated by a peak at 230 nm due to the guanidine isothyocyanate and guanidine HCl in the buffers used (Table 1).

Table 1.
Summary of the parameters (yield and purity measured at the NanoDrop) of the RNA extracted with the four different methods.
To avoid the contaminating the peak at 230 nm, we decided to use a solubilisation buffer devoid of guanidine salts. Therefore, we tested a procedure based on the use of SDS-P/C/I for an efficient cell lysis and RNase inactivation [15], coupled to TRIzol (Thermofisher Scientific) [8]. We used this procedure together with the subsequent RNA clean-up detailed in the RNeasy mini kit protocol (Qiagen). This procedure resulted in low 260/230 ratios, caused by TRIzol/phenol carryover (Table 1).
We then decided to combine the SDS-P/C/I solubilisation/extraction step with the purification using the silica membranes of the RNeasy spin columns (Qiagen), thereby omitting the TRIzol step (Figure 1). This combined protocol resulted in the recovery of pure RNA with better 260/230 ratios (Table 1).
We finally tested a CTAB-C/I + RNeasy spin column–based procedure previously described for the efficient and straight-forward extraction of RNA from basal angiosperms [9]. The CTAB buffer was prepared according to [10], since it is optimal for tissues containing lignin, as is the case for older hemp internodes.
This method is faster than the two previously described and it resulted in the extraction of pure RNA with better 260/230 ratios (Table 1).
The integrities of the RNAs extracted with the four methods were subsequently assessed. As can be seen in Supplementary Materials Figures S2 and S3, the RNAs were all intact. The RNA integrity number (RIN) was, however, higher for the CTAB-C/I + RNeasy (average RIN: 9.225, n = 4), as compared to the SDS-P/C/I + RNeasy (average RIN: 8.875, n = 4), the SDS-P/C/I + TRIzol (average RIN: 8.6, n = 4) and the RNeasy kit (average RIN: 8.45, n = 4).
3.2. Integrity of Representative Hemp Bast Fibre mRNAs
Since the Bioanalyzer estimates the total RNA integrity by measuring the integrity of ribosomal RNAs, we decided to assess the integrity by using the 5′-3′ ratio mRNA integrity test (5′-3′ ΔCq assay) [16]. This test is based on a reverse transcription (RT) reaction carried out using an oligod(T) primer (mature mRNAs possess a polyA tail) and on the assumption that the RTase will not always extend to the 5′ of the gene when the target mRNA is degraded [17].
The 5′ and 3′ amplicons targeted were 1155 bp and 1067 bp away from each other for GAPDH and actin, respectively. The amplification efficiencies of the primers were 89.6% and 102.6% for the 5′ and 3′ GAPDH primer pairs, and 102.9% and 100.2% for the 5′ and 3′ actin primer pairs.
We obtained a 5′-3′ ΔCq of 2.98 and 2.71 for GAPDH and a 5′-3′ ΔCq of 1.41 and 1.12 for actin with the SDS-P/C/I + RNeasy and CTAB-C/I + RNeasy protocols, respectively (Table 2). The difference in values between GAPDH and actin are most likely due to the different amplification efficiencies of the 5′ and 3′ GAPDH primer pairs. The primers of the actin 5’ and 3’ amplicons show, however, comparable amplification efficiencies, and in this case a 5′-3′ ΔCq slightly >1 was obtained (with 1 denoting an intact mRNA).

Table 2.
The 5′-3′ ratio mRNA integrity test. The test was carried out using primers designed on the 5′ and 3′ regions of GAPDH and actin genes from hemp (n = 4). The mean Cq values for each gene correspond, in the order shown, to the 5′ and 3′ amplicons.
The CTAB-C/I + RNeasy protocol shows the smallest differences in 5′-3′ ΔCq, thereby supporting the data obtained with the Bioanalyzer (i.e., higher RIN values; Supplementary Materials Figure S2).
4. Conclusions
This study has validated four different RNA extraction protocols from C. sativa bast fibres and has shown the suitability and rapidity of the CTAB-C/I + RNeasy protocol to extract pure, intact RNA. We therefore propose that the CTAB-C/I + RNeasy protocol is the most suitable for an efficient extraction of pure RNA from hemp bast fibres to carry out molecular investigations.
In our experience, the critical points needing special care by the researcher to achieve a good purity of bast fibre RNA using the CTAB-C/I + RNeasy protocol are: (1) grinding of the separated bast fibres to a powder (as fine as possible, although the material is fibrous in nature); (2) using the indicated extraction buffer/frozen powder ratios after phase separation (increasing the frozen tissue powder amount does not result in increased RNA yields); (3) performing the withdrawal of the aqueous phase without delay and taking care to avoid aspiration of the debris at the interface.
The RNA extracted from hemp bast fibres with the CTAB-C/I + RNeasy procedure is pure and intact and therefore suitable for both downstream targeted gene expression analysis via RT-qPCR and high-throughput transcriptomics.
Supplementary Materials
The following are available online at http://www.mdpi.com/2079-6439/4/3/23/s1. Figure S1: Isolation of hemp bast fibres, Figure S2: Integrity of the purified RNAs, Figure S3: Gel electrophoresis of the RNAs extracted with the four different methods, Table S1: Primers used for the 5′-3′ ratio mRNA integrity test.
Acknowledgments
The authors wish to thank Aude Corvisy and Laurent Solinhac for the excellent technical support. The Fonds National de la Recherche, Luxembourg, (Project CANCAN C13/SR/5774202), is gratefully acknowledged for financial support.
Author Contributions
Gea Guerriero conceived and designed the experiments; Gea Guerriero and Lauralie Mangeot-Peter performed the experiments; Gea Guerriero, Lauralie Mangeot-Peter and Sylvain Legay analyzed the data; Jean-Francois Hausman leads the project CANCAN; Gea Guerriero, Lauralie Mangeot-Peter, Jean-Francois Hausman and Sylvain Legay wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| RT-qPCR | real-time PCR |
| SDS | sodium dodecyl sulfate |
| P/C/I | phenol/chloroform/isoamyl alcohol |
| C/I | chloroform/isoamyl alcohol |
| CTAB | cetyl trimethylammonium bromide |
| EDTA | ethylenediaminetetraacetic acid |
| ME | β-mercaptoethanol |
| PVP-40 | polyvinylpyrrolidone |
| EtOH | ethanol |
| RNA-Seq | RNA sequencing |
| RIN | RNA integrity number |
| RT | reverse transcription |
| RTase | reverse transcriptase |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| Cq | quantitation cycle |
References
- Nolan, T.; Bustin, S. Procedures for quality control of RNA samples for use in quantitative reverse transcription PCR. In Essentials of Nucleic Acid Analysis: A Robust Approach; Keer, J.T., Birch, L., Eds.; The Royal Society of Chemistry: London, UK, 2008; pp. 189–207. [Google Scholar]
- Die, J.V.; Román, B. RNA quality assessment: A view from plant qPCR studies. J. Exp. Bot. 2012, 63, 6069–6077. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Sergeant, K.; Hausman, J.F. Integrated-omics: A powerful approach to understanding the heterogeneous lignification of fibre crops. Int. J. Mol. Sci. 2013, 14, 10958–10978. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Hausman, J.F.; Cai, G. No stress! Relax! Mechanisms governing growth and shape in plant cells. Int. J. Mol. Sci. 2014, 15, 5094–5114. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Sergeant, K.; Hausman, J.F. Wood biosynthesis and typologies: A molecular rhapsody. Tree Physiol. 2014, 34, 839–855. [Google Scholar] [CrossRef] [PubMed]
- Andre, C.M.; Hausman, J.F.; Guerriero, G. Cannabis sativa: The plant of the thousand and one molecules. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Hausman, J.F.; Strauss, J.; Ertan, H.; Siddiqui, K.S. Lignocellulosic biomass: Biosynthesis, degradation and industrial utilization. Eng. Life Sci. 2015, 16, 1–16. [Google Scholar] [CrossRef]
- Li, Z.; Trick, H.N. Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. Biotechniques 2005, 38, 872, 874 and 876. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Koh, J.; Yoo, M.J.; Kong, H.; Hu, Y.; Ma, H.; Soltis, P.S.; Soltis, D.E. Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 2005, 43, 724–744. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A Rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Mokshina, N.; Gorshkova, T.; Deyholos, M.K. Chitinase-like (CTL) and cellulose synthase (CESA) gene expression in gelatinous-type cellulosic walls of flax (Linum usitatissimum L.) bast fibers. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Ibragimova, N.N.; Mokshina, N.E.; Gorshkova, T.A. Cell wall proteins of flax phloem fibers. Russ. J. Bioorg. Chem. 2012, 38, 117–125. [Google Scholar] [CrossRef]
- Medicinal Plants Genomic Resource. Available online: http://medicinalplantgenomics.msu.edu/index.shtml (accessed on 12 July 2016).
- Behr, M.; Legay, S.; Hausman, J.F.; Guerriero, G. Analysis of cell wall-related genes in organs of Medicago sativa L. under different abiotic stresses. Int. J. Mol. Sci. 2015, 16, 16104–16124. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Yiap, B.C. Erratum to “DNA, RNA, and Protein Extraction: The Past and The Present”. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Vermeulen, J.; de Preter, K.; Lefever, S.; Nuytens, J.; de Vloed, F.; Derveaux, S.; Hellemans, J.; Speleman, F.; Vandesompele, J. Measurable impact of RNA quality on gene expression results from quantitative PCR. Nucleic Acids Res. 2011, 39. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K.; Czechowski, T.; Scheible, W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell. 2008, 20, 1736–1737. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).