Design and Construction of Large Amyloid Fibers
Abstract
:1. Introduction
2. Results
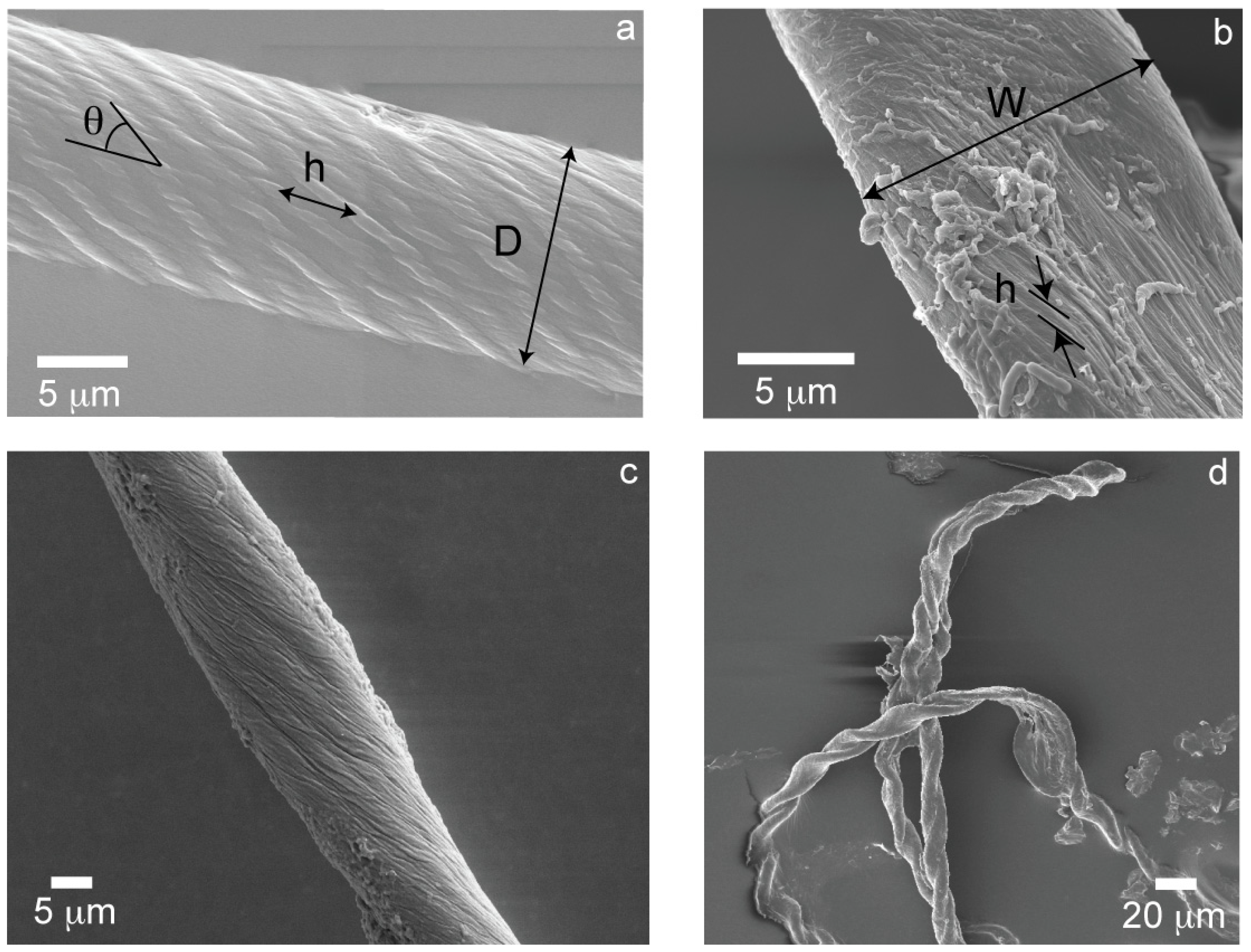

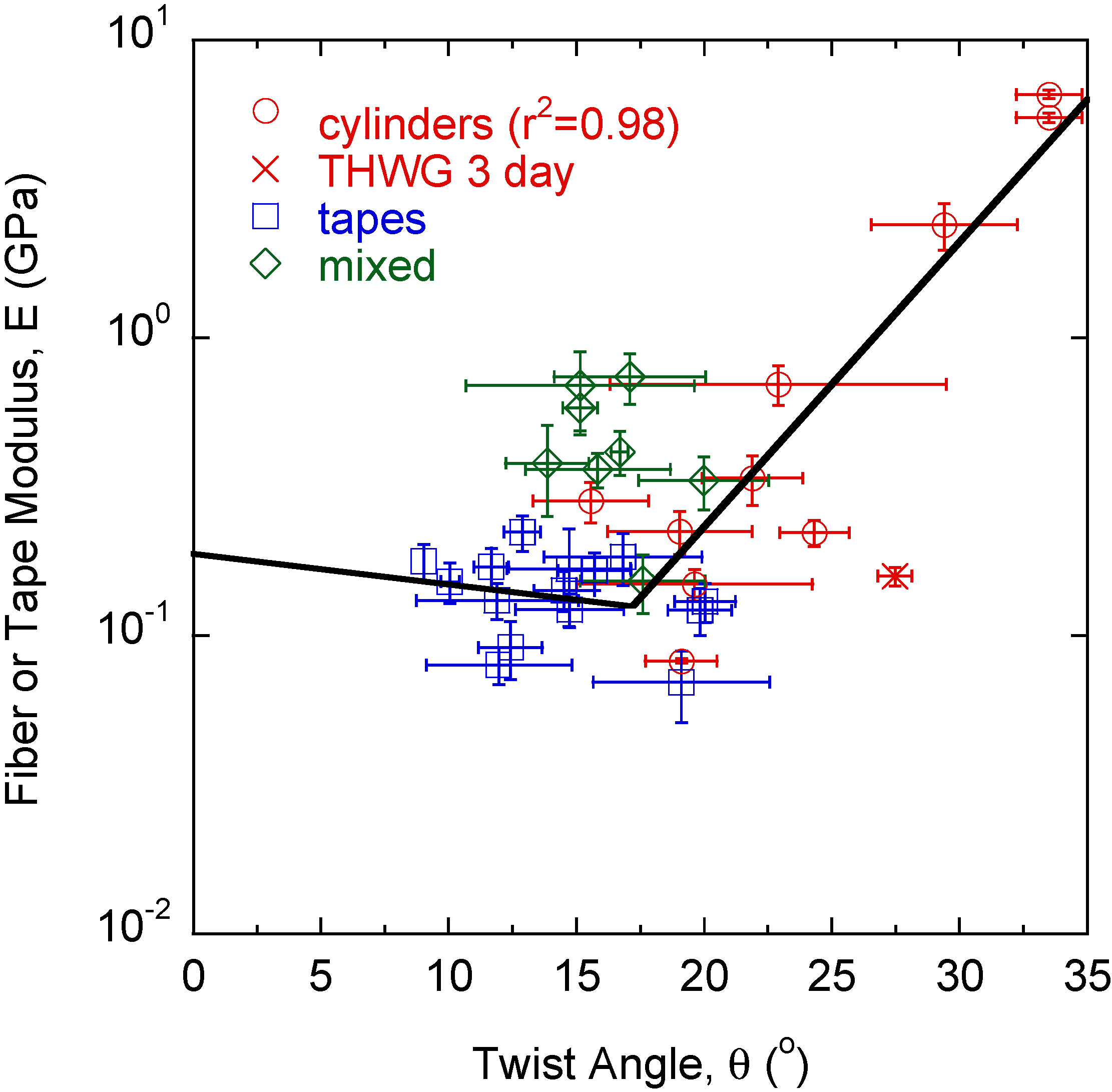
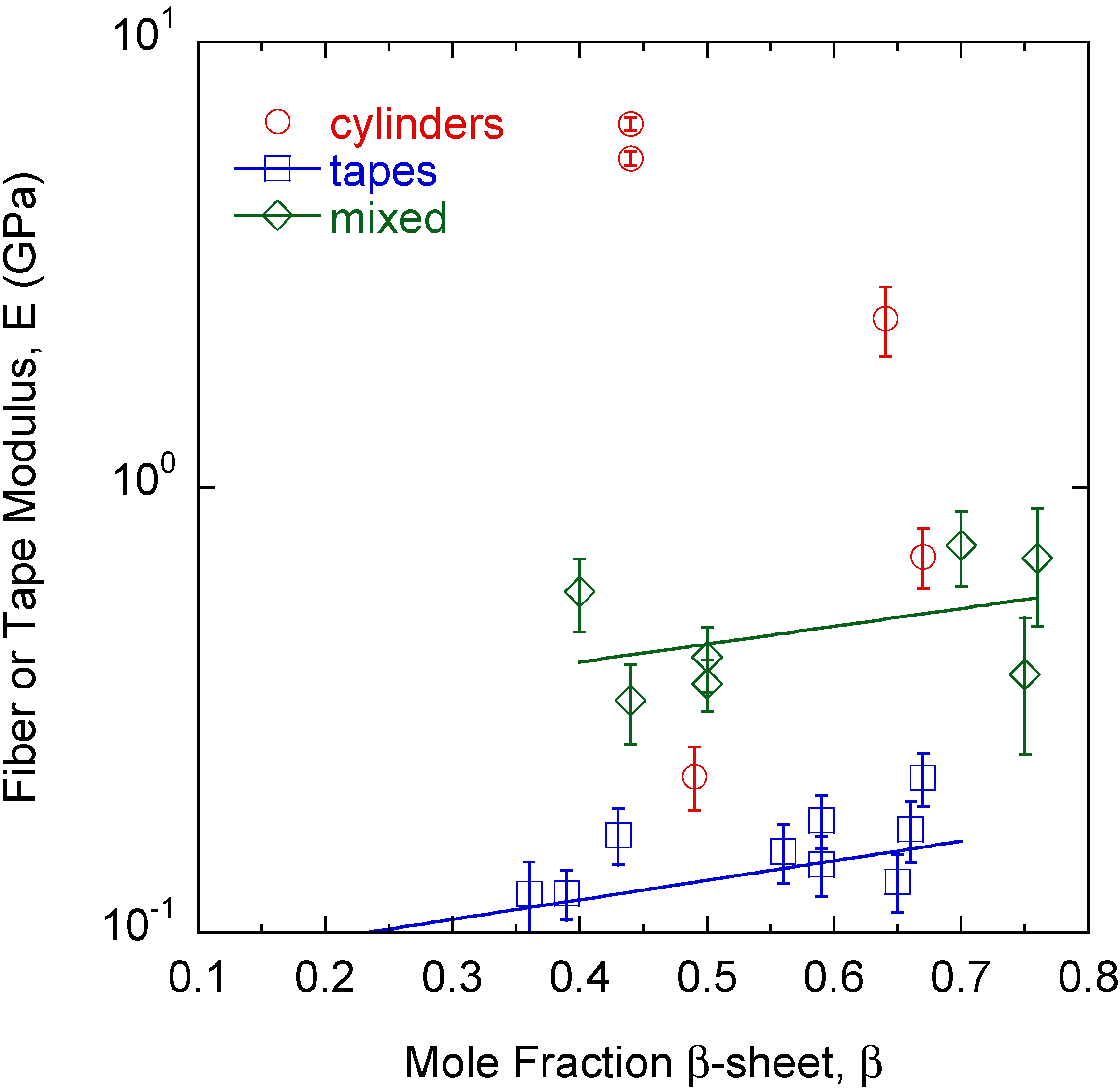
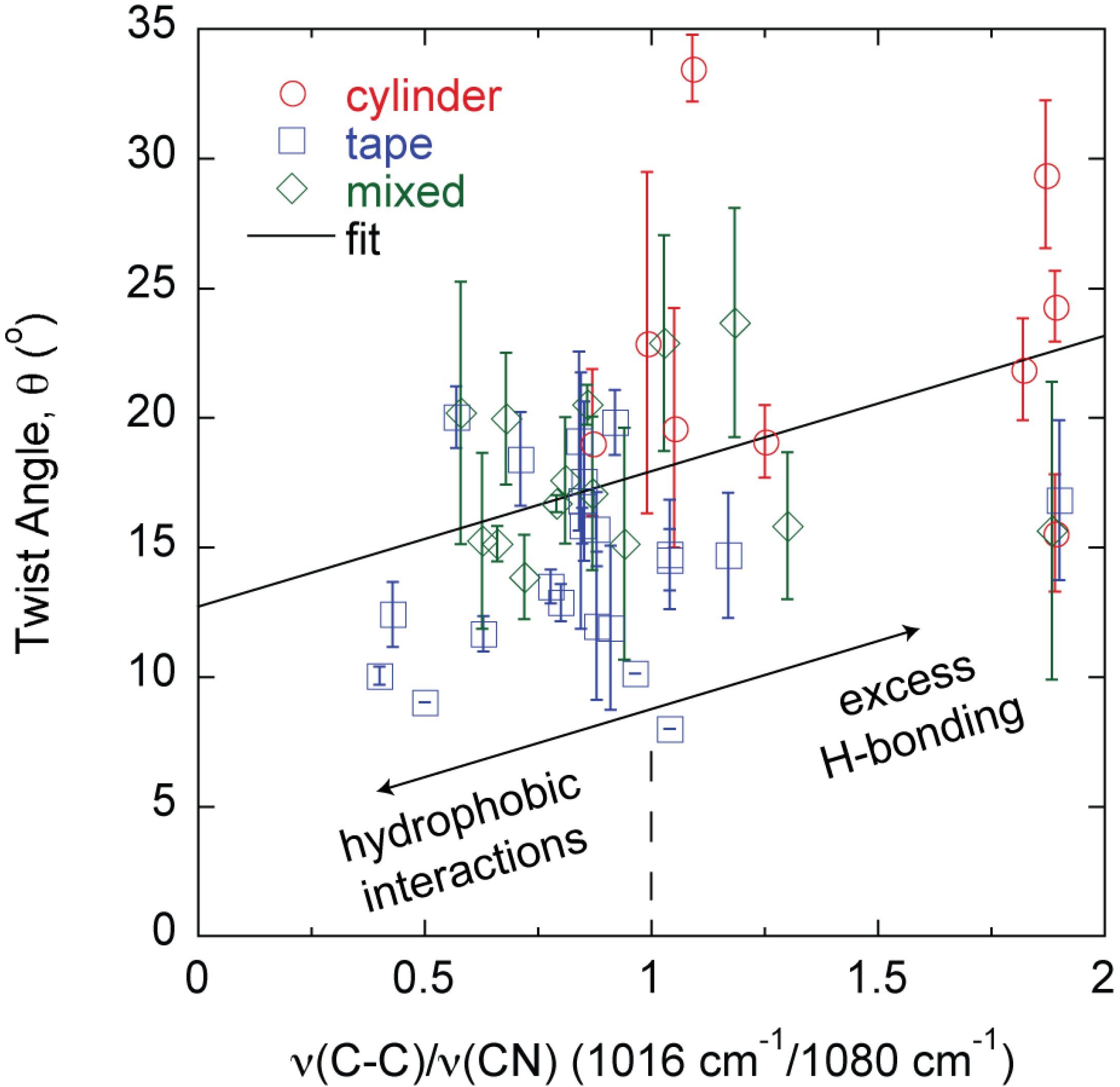
3. Discussion
4. Materials and Methods
4.1. Protein Mixtures
4.2. Fourier Transform Infrared (FTIR) Spectroscopy
4.3. Scanning Electron Microscopy (SEM)
4.4. Nanoindentation
5. Conclusions
Acknowledgments
Author Contributions
Supplementary Information
Conflicts of Interest
References
- Knowles, T.P.J.; Buehler, M.J. Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 2011, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid-from bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Adamcik, J.; Jung, J.-M.; Flakowski, J.; de Los Rios, P.; Dietler, G.; Mezzenga, R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat. Nanotechnol. 2010, 5, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Bolisetty, S.; Adamcik, J.; Mezzenga, R. Snapshots of fibrillation and aggregation kinetics in multistranded amyloid β-lactoglobulin fibrils. Soft Matter 2011, 7, 493–499. [Google Scholar] [CrossRef]
- Aggeli, A.; Nyrkova, I.A.; Bell, M.; Harding, R.; Carrick, L.; McLeish, T.C.B.; Semenov, A.N.; Boden, N. Hierarchical self-assembly of chiral rod-like molecules as a model for peptide β-sheet tapes, ribbons, fibrils, and fibers. Proc. Natl. Acad. Sci. USA 2001, 98, 11857–11862. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.E.; Dickinson, G.H.; Orihuela, B.; Kulp III, J.L.; Rittschof, D.; Wahl, K.J. Characterization of the adhesive plaque of the barnacle Balanus amphitrite: Amyloid-like nanofibrils are a major component. Langmuir 2010, 26, 6549–6556. [Google Scholar] [CrossRef] [PubMed]
- Gebbink, M.F.B.G.; Claessen, D.; Bouma, B.; Dijkhuizen, L.; Wosten, H.A.B. Amyloids—A functional coat for microorganisms. Nat. Rev. Microbiol. 2005, 3, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Schmidt, J.C.; Chapman, M.R. The curli nucleator protein, csgb, contains an amyloidogenic domain that directs csga polymerization. Proc. Natl. Acad. Sci. USA 2007, 104, 12494–12499. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Ramsook, C.B.; Lipke, P.N.; Dufrêne, Y.F. Unzipping a functional microbial amyloid. ACS Nano 2012, 6, 7703–7711. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.D.; Rudall, K.M. Structure of the silk of Chrysopa egg-stalks. Nature 1957, 179, 905–906. [Google Scholar] [CrossRef]
- Mostaert, A.; Higgins, M.; Fukuma, T.; Rindi, F.; Jarvis, S. Nanoscale mechanical characterisation of amyloid fibrils discovered in a natural adhesive. J. Biol. Phys. 2006, 32, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E. Self-assembled peptide nanostructures: The design of molecular building blocks and their technological utilization. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Oppenheim, T.W.; Buell, A.K.; Chirgadze, D.Y.; Welland, M.E. Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nat. Nanotechnol. 2010, 5, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Born, A.-K.; Schweizer, T.; Zenobi-Wong, M.; Cerruti, M.; Mezzenga, R. Amyloid-hydroxyapatite bone biomimetic composites. Adv. Mater. 2014, 26, 3207–3212. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Li, C.; Adamcik, J.; Shao, Z.; Chen, X.; Mezzenga, R. Modulating materials by orthogonally oriented β-strands: Composites of amyloid and silk fibroin fibrils. Adv. Mater. 2014, 26, 4569–4574. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Esler, W.P.; Stimson, E.R.; Jennings, J.M.; Vinters, H.V.; Ghilardi, J.R.; Lee, J.P.; Mantyh, P.W.; Maggio, J.E. Alzheimer’s disease amyloid propagation by a template-dependent dock-lock mechanism. Biochemistry 2000, 39, 6288–6295. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.R.; Douglass, A.; Vale, R.D.; Weissman, J.S. Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol. 2004, 2, 1582–1590. [Google Scholar] [CrossRef]
- Adamcik, J.; Mezzenga, R. Adjustable twisting periodic pitch of amyloid fibrils. Soft Matter 2011, 7, 5437–5443. [Google Scholar] [CrossRef]
- Lara, C.C.; Adamcik, J.; Jordens, S.; Mezzenga, R. General self-assembly mechanism converting hydrolyzed globular proteins into giant multistranded amyloid ribbons. Biomacromolecules 2011, 12, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Barone, J.R. Evolution of the amyloid fiber over multiple length scales. ACS Nano 2013, 7, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Athamneh, A.; Barone, J.R. Enzyme-mediated self-assembly of highly ordered structures from disordered proteins. Smart Mater. Struct. 2009, 18, 104024. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Ebanks, K.C.; Barone, J.R. Peptide mixtures can self-assemble into large amyloid fibers of varying size and morphology. Biomacromolecules 2011, 12, 3770–3779. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Claunch, E.C.; Lee, P.W.; Barone, J.R. The role of protein hydrophobicity in conformation change and self-assembly into large amyloid fibers. Biomacromolecules 2014, 15, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Kirkitadze, M.D.; Condron, M.M.; Teplow, D.B. Identification and characterization of key kinetic intermediates in amyloid β-protein fibrillogenesis. J. Mol. Biol. 2001, 312, 1103–1119. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Claunch, E.C.; Barone, J.R. The effect of processing on large, self-assembled amyloid fibers. Soft Matter 2012, 8, 10298–10306. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Claunch, E.C.; Barone, J.R. Characterization of amyloid structures by FT-IR and raman spectroscopy. Appl. Spectrosc. 2013, 67, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Grason, G.M. Defects in crystalline packings of twisted filament bundles. I. Continuum theory of disclinations. Phys. Rev. E 2012, 85, 031603. [Google Scholar] [CrossRef]
- Gunzler, H.; Gremlich, H.-U. IR Spectroscopy; Wiley-VCH: Weinheim, Germany, 2002. [Google Scholar]
- Barth, A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 2000, 74, 141–173. [Google Scholar] [CrossRef] [PubMed]
- Perutz, M.F.; Johnson, T.; Suzuki, M.; Finch, J.T. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. USA 1994, 91, 5355–5358. [Google Scholar] [CrossRef] [PubMed]
- DePace, A.H.; Santoso, A.; Hillner, P.; Weissman, J.S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 1998, 93, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Scherzinger, E.; Sittler, A.; Schweiger, K.; Heiser, V.; Lurz, R.; Hasenbank, R.; Bates, G.P.; Lehrach, H.; Wanker, E.E. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: Implications for huntington’s disease pathology. Proc. Natl. Acad. Sci. USA 1999, 96, 4604–4609. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Berthelier, V.; Bradley Hamilton, J.; O’Nuallain, B.; Wetzel, R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry 2002, 41, 7391–7399. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, P.; Atkins, E. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules 2005, 6, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement controls stiffness, strength and mechanical toughness of β-sheet crystals in silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.S.; Adamcik, J.; Jeong, J.S.; Lashuel, H.A.; Mezzenga, R.; Dietler, G. Influence of the β-sheet content on the mechanical properties of aggregates during amyloid fibrillization. Angew. Chem. Int. Ed. 2015, 54, 2462–2466. [Google Scholar] [CrossRef]
- ExPASy Bioinformatics Resource Portal. Available online: http://web.expasy.org/protparam/ (accessed on 1 July 2014).
- Tango-A computer algorithm for prediction of aggregating regions in unfolded polypeptide chains. Available online: tango.crg.es (accessed on 1 July 2014).
- Kamino, K.; Odo, S.; Maruyama, T. Cement proteins of the acorn-barnacle, Megabalanus rosa. Biol. Bull. 1996, 190, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Oliver, W.C.; Pharr, G.M. An improved technique for determining hardness and elastic modulus using load and displacement sensing indentation experiments. J. Mater. Res. 1992, 7, 1564–1583. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ridgley, D.M.; Rippner, C.M.W.; Barone, J.R. Design and Construction of Large Amyloid Fibers. Fibers 2015, 3, 90-102. https://doi.org/10.3390/fib3020090
Ridgley DM, Rippner CMW, Barone JR. Design and Construction of Large Amyloid Fibers. Fibers. 2015; 3(2):90-102. https://doi.org/10.3390/fib3020090
Chicago/Turabian StyleRidgley, Devin M., Caitlin M. W. Rippner, and Justin R. Barone. 2015. "Design and Construction of Large Amyloid Fibers" Fibers 3, no. 2: 90-102. https://doi.org/10.3390/fib3020090
APA StyleRidgley, D. M., Rippner, C. M. W., & Barone, J. R. (2015). Design and Construction of Large Amyloid Fibers. Fibers, 3(2), 90-102. https://doi.org/10.3390/fib3020090




