Abstract
Mixtures of “template” and “adder” proteins self-assemble into large amyloid fibers of varying morphology and modulus. Fibers range from low modulus, rectangular cross-sectioned tapes to high modulus, circular cross-sectioned cylinders. Varying the proteins in the mixture can elicit “in-between” morphologies, such as elliptical cross-sectioned fibers and twisted tapes, both of which have moduli in-between rectangular tapes and cylindrical fibers. Experiments on mixtures of proteins of known amino acid sequence show that control of the large amyloid fiber morphology is dependent on the amount of glutamine repeats or “Q-blocks” relative to hydrophobic side chained amino acids such as alanine, isoleucine, leucine, and valine in the adder protein. Adder proteins with only hydrophobic groups form low modulus rectangular cross-sections and increasing the Q-block content allows excess hydrogen bonding on amide groups that results in twist and higher modulus. The experimental results show that large amyloid fibers of specific shape and modulus can be designed and controlled at the molecular level.
1. Introduction
Amyloids are self-assembled protein materials containing β-sheets. While the most common context for amyloids is in neurodegenerative diseases, there is another class of amyloid that performs beneficial functions in nature called “functional” amyloids [1,2]. Functional amyloids occur as rigid protofibrils about 2–4 nm high, 10–30 nm wide, and >100 nm long with β-sheets oriented perpendicular to the protofibril axis to yield what is traditionally called the “cross-β” protein secondary structure [1,3,4,5]. Examples include egg silks of the Chrysopidae family, barnacle cement, and bacterial and fungal hyphae [2,6,7,8,9,10]. These natural functional amyloids have been shown to display toughness as evidenced by a “sawtooth” pattern in atomic force microscopy (AFM) force-distance curves [11]. Functional amyloids have now become an inspiration for the burgeoning field of using amyloid protofibrils for high performance materials [1,12,13,14,15].
Many different proteins have been shown to form amyloid protofibrils [16]. Self-assembly begins when a protein molecule destabilizes, unfolds, and hydrogen bonds to another unfolded protein molecule to build the β-sheet. Growth of the structure occurs when more protein molecules add into the β-sheet and elongate it into the protofibril [17,18]. Protofibrils can align via liquid crystal interactions and further assemble through hydrophobic interactions into multi-stranded fibrils about 7–10 nm high and 30–200 nm wide [4]. Protofibrils and fibrils can twist based on several factors in the amino acid sequence [3,5,19]. Fibrils can further assemble into large fibrils that can be more cylindrical in cross-section and 200–700 nm wide or more rectangular in cross-section and also 200–700 nm wide but 20–500 nm high [20,21]. In certain protein mixtures, assembly can continue to yield large amyloid fibers and tapes 10–20 μm wide and the tapes are 5 μm high [22,23]. Our recent research has focused on the characteristics of these protein mixtures that allow them to self-assemble to such large scales. “Template” proteins are short and hydrophobic enough to destabilize and quickly form β-sheets. “Adder” proteins are longer, α-helical, and hydrophilic and remain this way until perturbed. Upon mixing, hydrophobic regions in the adder protein α-helices unravel on exposed hydrophobic groups in the template and “add” into the growing β-sheet structure [24]. α to β conformation change is often observed in amyloid formation [25]. Larger scale self-assembly is dictated by hydrophobic interactions on the protofibrils, fibrils, and large fibrils where the hydrophobicity is related back to the alanine (A), isoleucine (I), leucine (L), and valine (V) content [4,23,24]. Other amino acids, most notably glutamine (Q), can contribute to the structure and affect the self-assembly [26,27]. Thus, the formation of large amyloid fibrils and fibers can be affected by the type of proteins in the mixture and self-assembly conditions [26].
In this study, different template and adder proteins are mixed together to note the effect of protein amino acid sequence and template/adder interactions on large amyloid fiber morphology and properties. The experiments build on previous work performed on protein hydrolysates [26] by using pure natural proteins and engineered proteins of known amino acid sequences so that exact protein-protein interactions can be studied. Morphology is characterized by how the large fibrils bundle into the large amyloid fiber. Nanoindentation is used to measure large amyloid fiber and tape modulus. Protein secondary structure and intermolecular interactions are assessed with Fourier transform infrared (FTIR) spectroscopy.
2. Results
Large fibrils will bundle into large amyloid fibers in different ways depending on the constituent proteins in the mixture or the self-assembling solution conditions [21,23,26]. Large amyloid fibers and tapes are characterized morphologically by their overall diameter, D (fibers), or width, W (tapes), the twist angle of the large fibril with respect to the fiber or tape axis during bundling, θ and the pitch or center-to-center distance between large fibrils in the structure parallel to the fiber axis, h. In one case, large fibrils wrap around each other like yarns in a rope to bundle into a cylindrical cross-sectioned large amyloid fiber (Figure 1a). The morphological features of the cylindrical fiber can be geometrically related through:
where π/h is the helical rotation rate of the large fibrils along the large amyloid fiber axis [28]. In another case, large fibrils bundle together much more weakly than in the cylinder case, resulting in a smaller bundling angle (Figure 1b). While the large fibrils still “wrap” around one another, the structure more resembles lateral and vertical stacking of large fibrils into a rectangular cross-sectioned large amyloid tape. There are similar expressions for the wide side of the tape, W, and thin side of the tape, H, but only the wide side is considered such that the morphological features of the rectangular tape are related through:
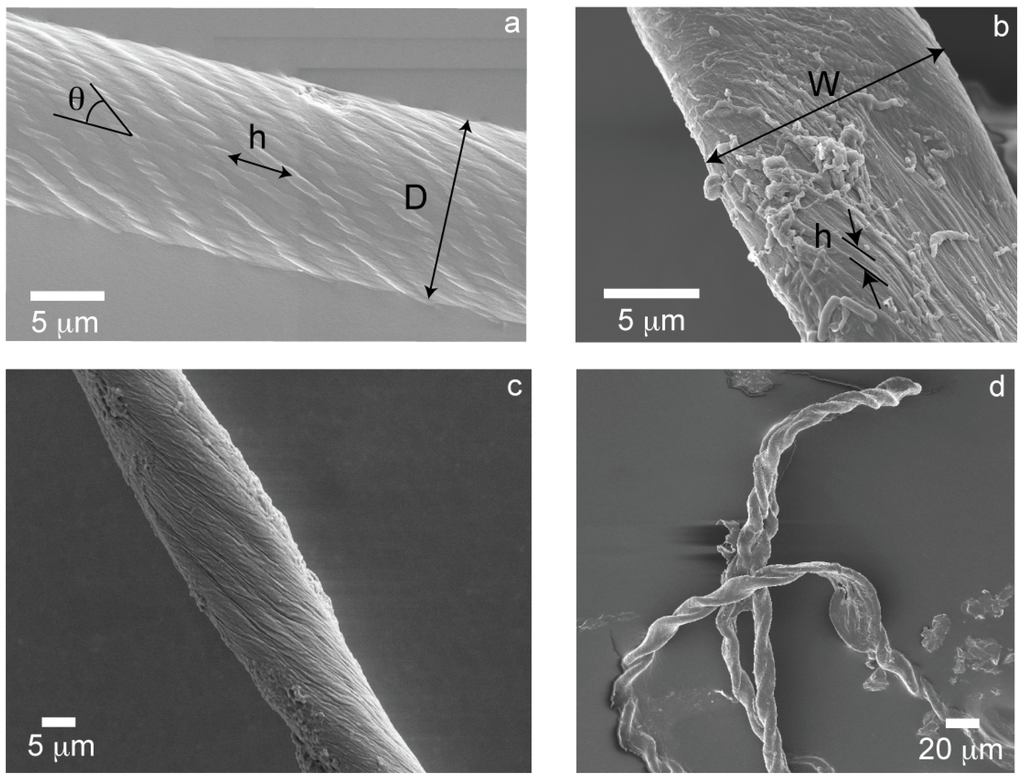
Figure 1.
(a) THWG 37 °C, pH8 cylindrical fiber; (b) THGd:Al 0.36:0.64 flat tape; (c) THGd:My 0.36:0.64 22 °C elliptical fiber; (d) P7 highly twisted tape.
Figure 1.
(a) THWG 37 °C, pH8 cylindrical fiber; (b) THGd:Al 0.36:0.64 flat tape; (c) THGd:My 0.36:0.64 22 °C elliptical fiber; (d) P7 highly twisted tape.
Given the wide variety of protein mixtures studied, other morphologies “in-between” the extremes of cylinders and tapes are also observed. Specifically, elliptical cross-sections (Figure 1c) and twisted tapes (Figure 1d), which are referred to as “mixed” morphologies. The twist angle, θ, is inversely proportional to the pitch, h, (Figure 2a) and directly proportional to the diameter or width of the large amyloid fibers or tapes (Figure 2b) as predicted by Equations (1) and (2).
The modulus of each large amyloid fiber, tape, and mixed structure is measured with nanoindentation. Fibers, tapes, and mixed structures can be differentiated by modulus, with fibers having the highest modulus, tapes the lowest modulus, and mixed morphologies in between. It is observed that θt < θf, similar to the modulus trend. Modulus, E, is plotted versus twist angle, θ (Figure 3). In general, rectangular tapes have very low twist angles and have a relatively constant modulus with θ. Mixed morphologies, which deviate from rectangular cross-section and tend toward a cylindrical morphology, generally have a higher modulus and twist angle θ. Cylindrical fibers have the highest moduli and θ values with a strong correlation (r2 = 0.98) found between E and θ.
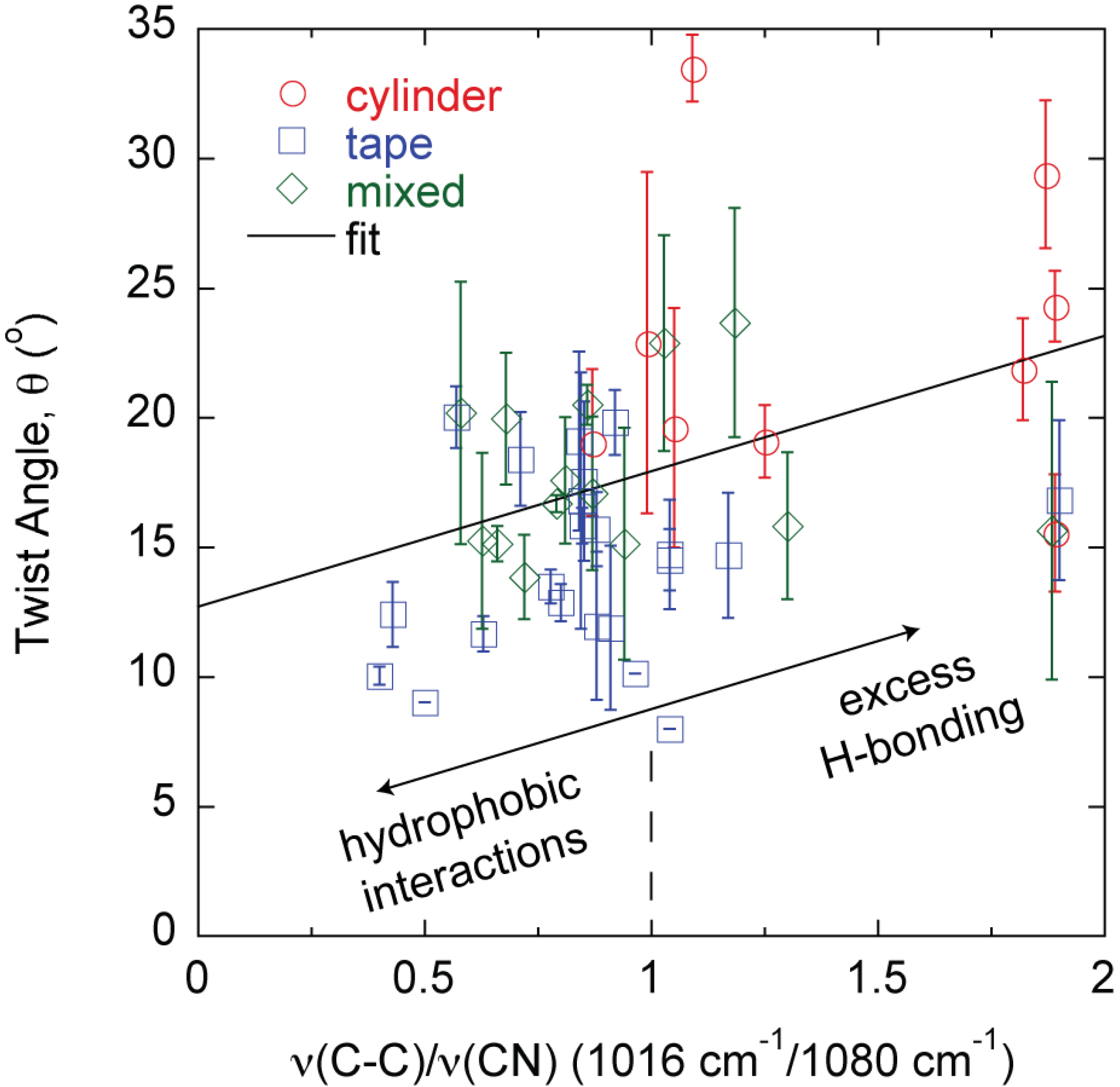
The differences in morphology and properties suggest larger scale manifestations of molecular level phenomena since each fiber, tape, and mixed structure is produced from a different protein mixture. While the signature of the amyloid is high-density β-sheet content, the amyloid structure β-sheet mole fraction cannot differentiate the different large amyloid fiber morphologies observed (Figure 4). The β-sheet mole fraction is found from deconvolution of the FTIR Amide I absorbance [27]. Changes are also observed in the FTIR spectrum between 1000 cm−1 and 1100 cm−1 [23,26,27]. The absorbance at 1080 cm−1 is assigned to the CN stretching absorbance, ν(CN), and the absorbance at 1016 cm−1 is assigned to the C–C stretching absorbance, ν(C–C) [29,30]. A correlation is found between the large amyloid fiber twist angle, θ, and the final value of the ratio ν(C–C)/ν(CN) at the end of self-assembly (Figure 5).
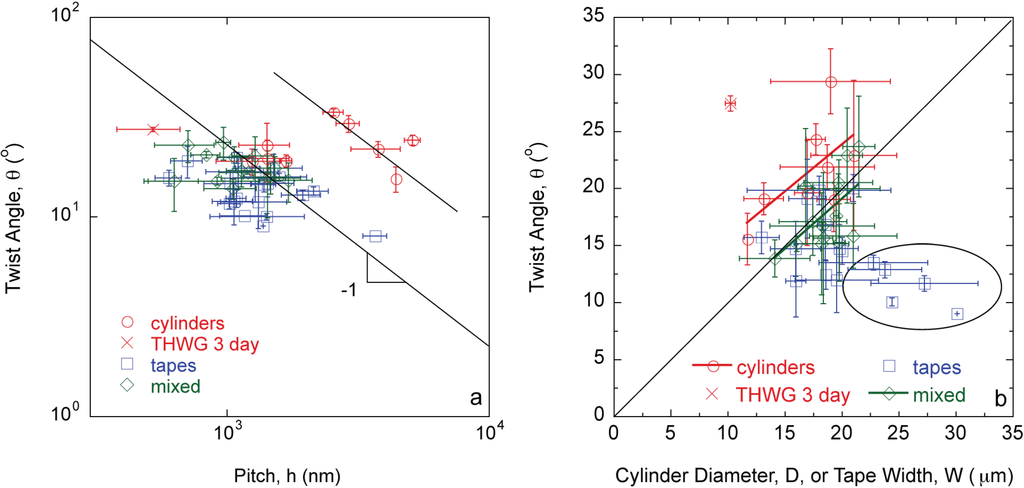
Figure 2.
Twist angle, θ, is (a) inversely proportional to pitch, h; and (b) directly proportional to fiber diameter, D, and tape width, W, following Equations (1) and (2). Fully formed THWG fibers have higher θ and h values.
Figure 2.
Twist angle, θ, is (a) inversely proportional to pitch, h; and (b) directly proportional to fiber diameter, D, and tape width, W, following Equations (1) and (2). Fully formed THWG fibers have higher θ and h values.
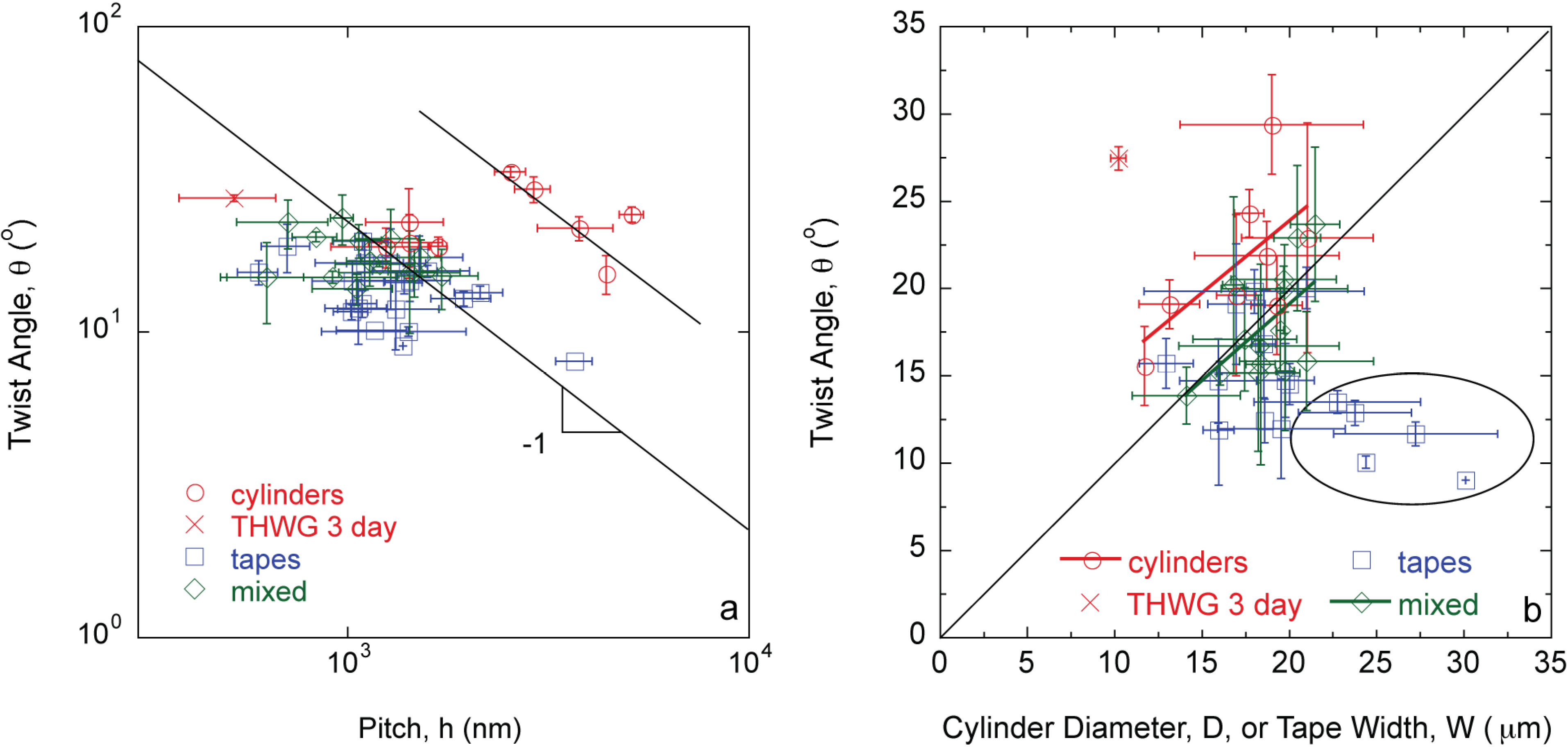
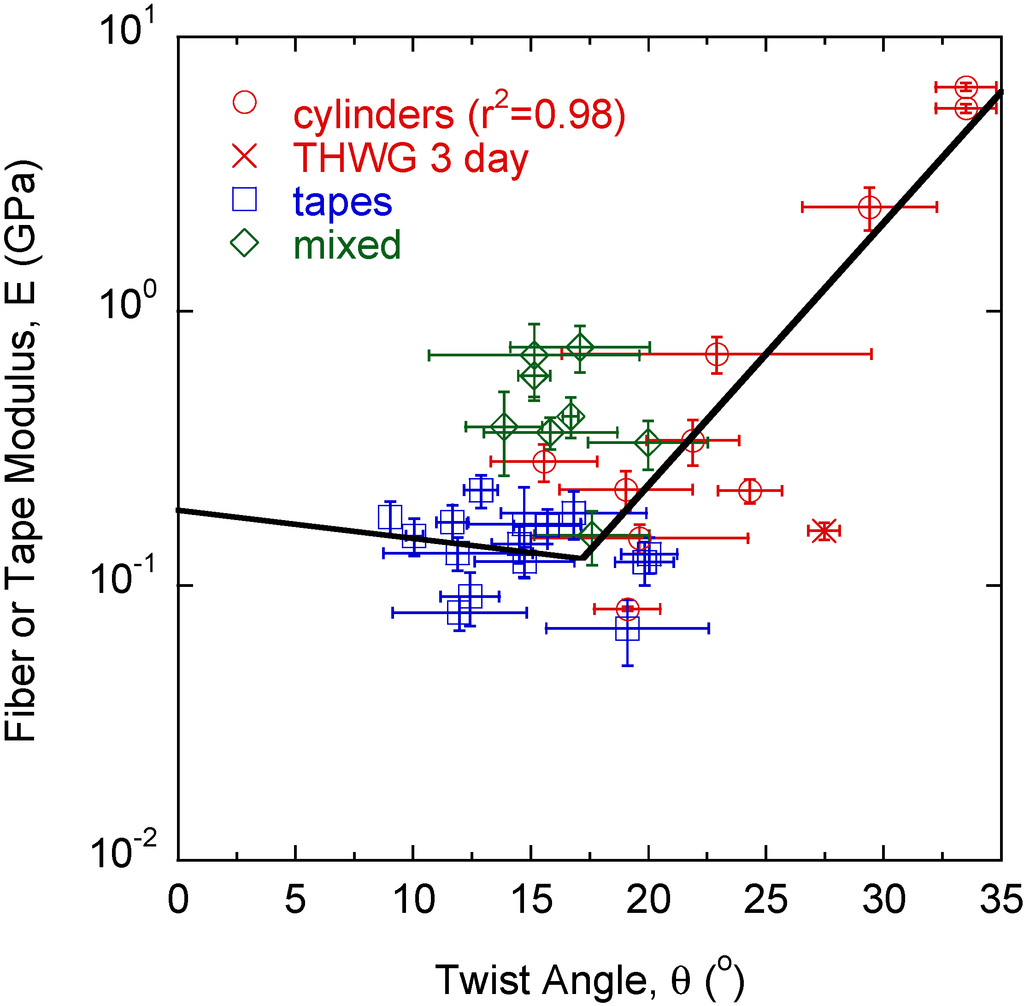
Figure 3.
Tapes have the lowest twist angles, θ, with moduli, E, independent of θ. As more twist is induced into the structure, E and θ begin to increase. Cylinders, formed from highly twisted large fibrils, show a strong correlation between E and θ.
Figure 3.
Tapes have the lowest twist angles, θ, with moduli, E, independent of θ. As more twist is induced into the structure, E and θ begin to increase. Cylinders, formed from highly twisted large fibrils, show a strong correlation between E and θ.
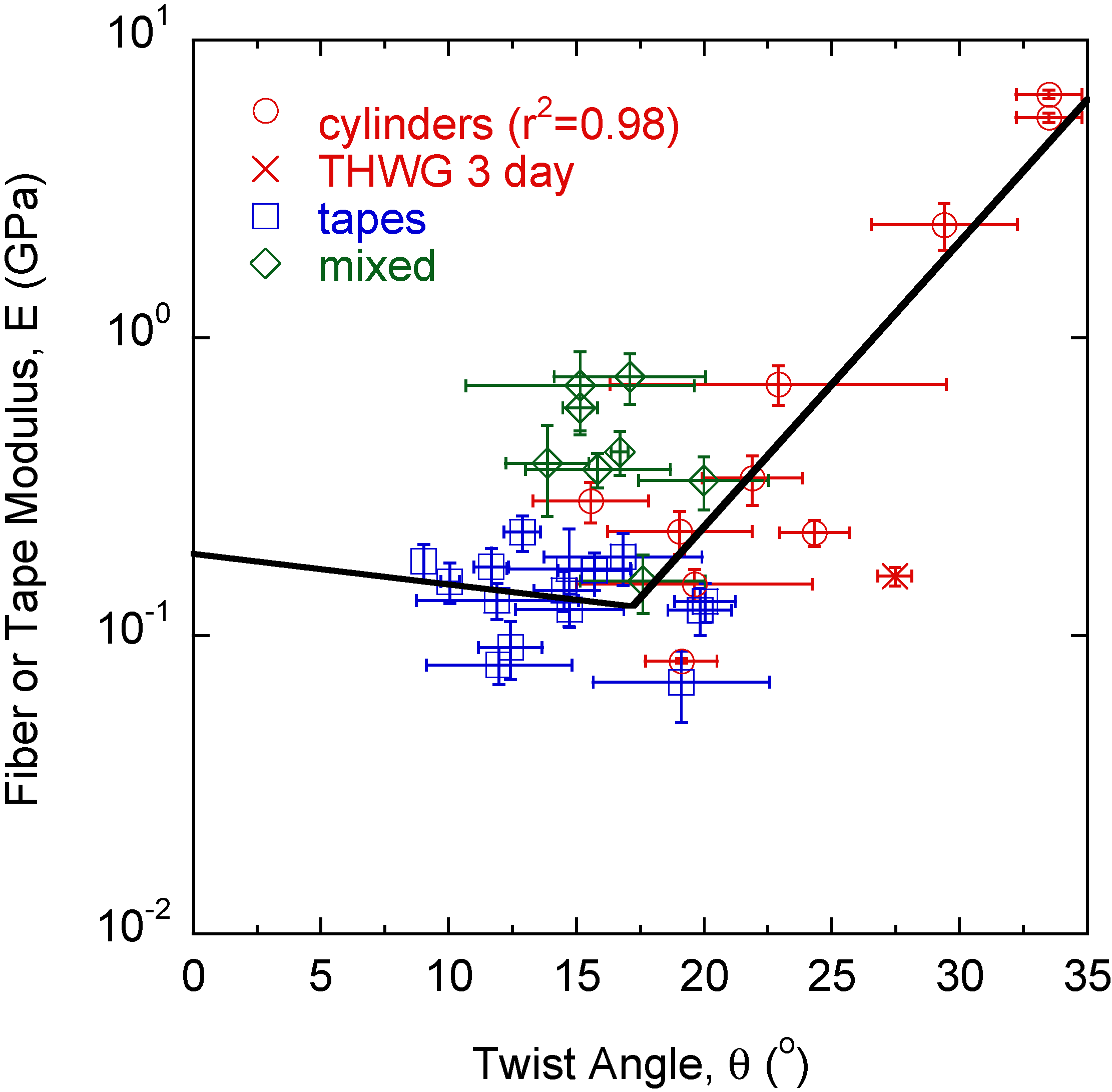
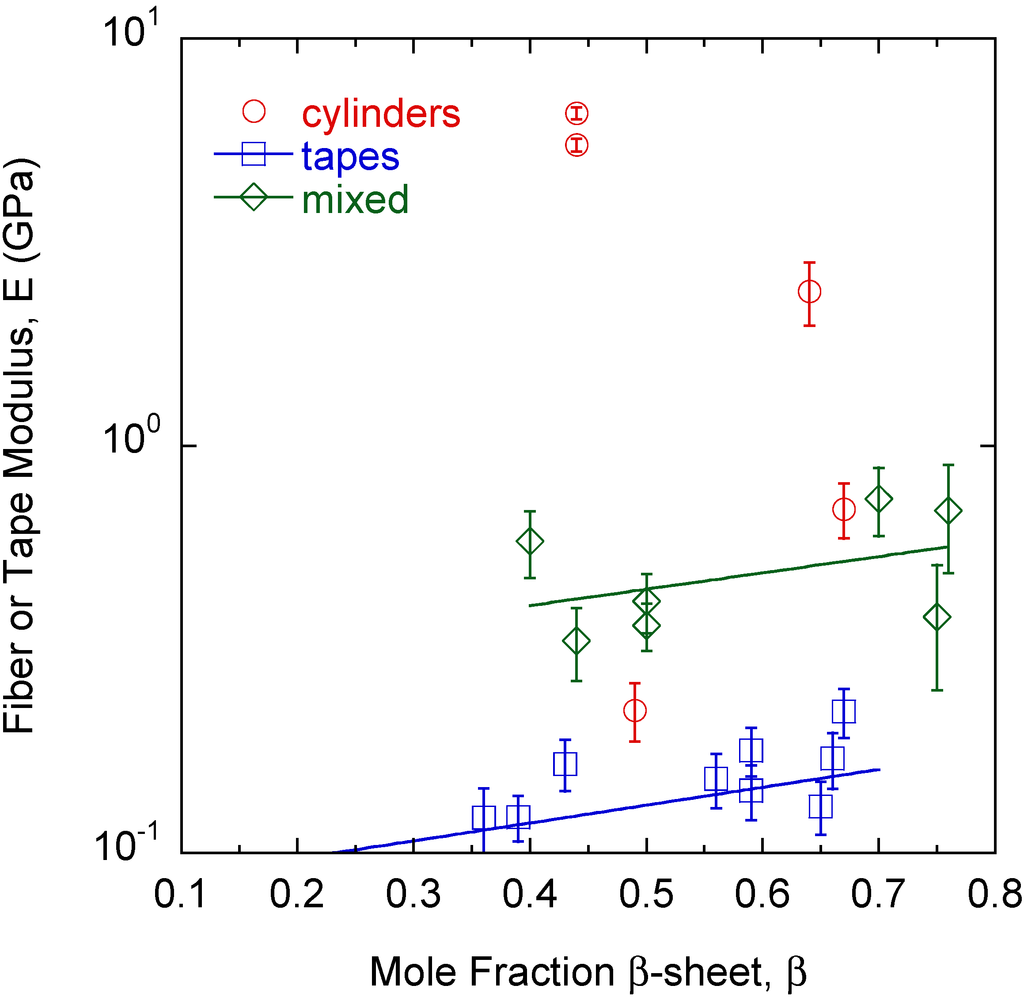
Figure 4.
There is a slight dependence of tape and mixed morphology structure modulus, E, on β-sheet content, β. However, β-sheet content alone cannot differentiate the different types of structures observed.
Figure 4.
There is a slight dependence of tape and mixed morphology structure modulus, E, on β-sheet content, β. However, β-sheet content alone cannot differentiate the different types of structures observed.
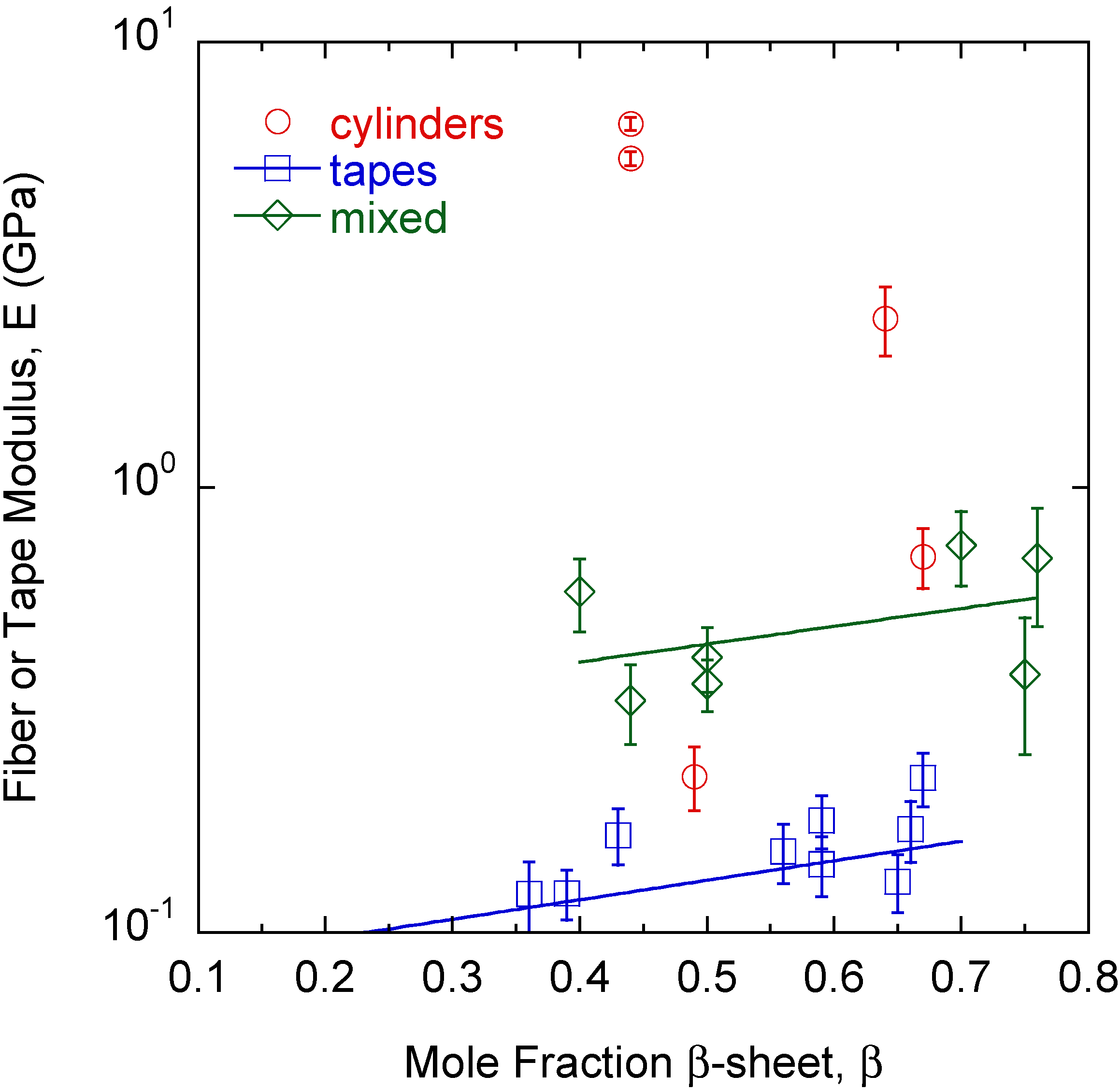
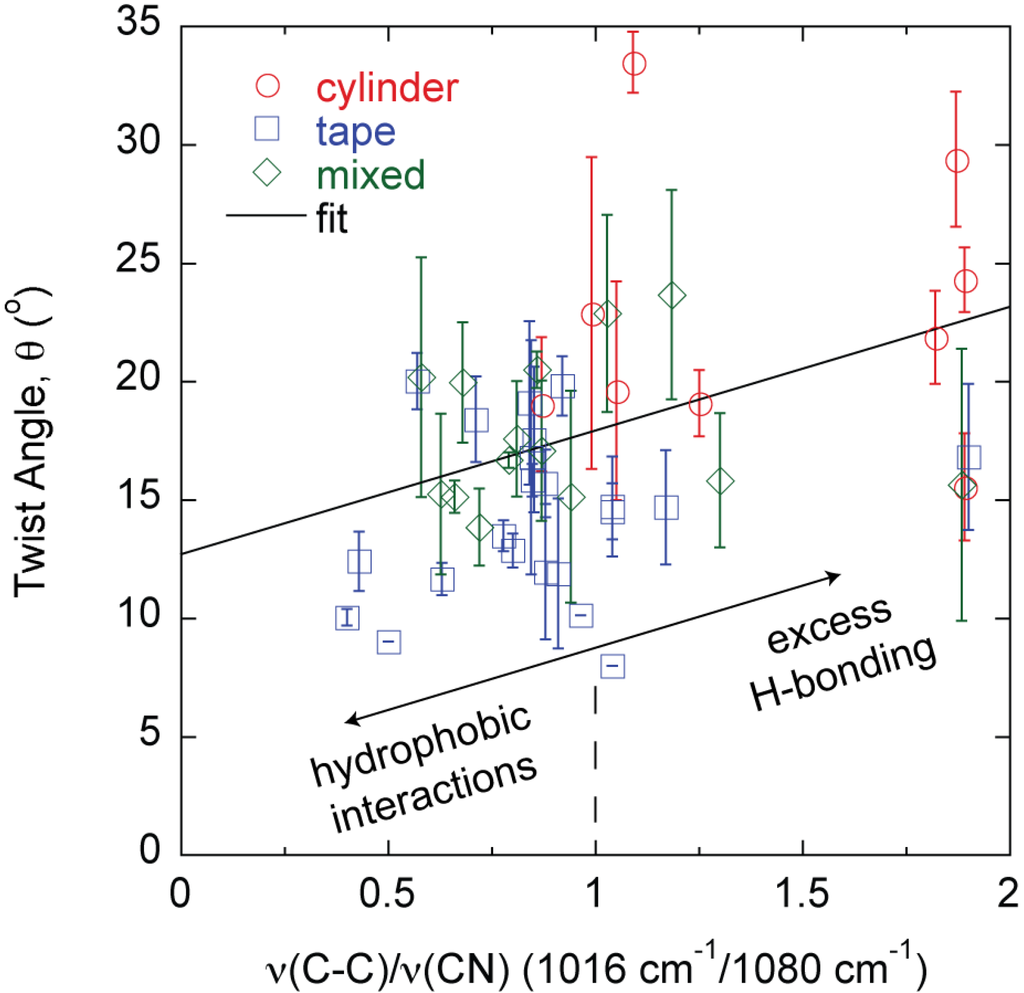
Figure 5.
Fiber morphology, θ, can be predicted by the hydrogen bonding state of constituent protein amino acid side chains as described by the FTIR absorbance ratio ν(C–C)/ν(CN).
Figure 5.
Fiber morphology, θ, can be predicted by the hydrogen bonding state of constituent protein amino acid side chains as described by the FTIR absorbance ratio ν(C–C)/ν(CN).
3. Discussion
The results described in this study connect molecular level phenomena to macroscopic fiber behavior. Specifically, amyloid large fibrils can be self-assembled and can further bundle into large amyloid fibers and tapes in a predictable manner based on characteristics of the constituent template and adder proteins in the mixture. The majority of the fibers follow the same θ vs. h relationship except fully formed THWG fibers (Figure 2a). THWG large fibrils bundle into the final twist angle early (THWG 3 day) but it takes 20 days to develop the final THWG large amyloid fiber diameter (Figure 2b) and modulus (Figure 3) at the same conditions. THWG fibers have the highest ν(C–C)/ν(CN) values (Figure 5). High values for ν(C–C)/ν(CN) arise because there is strong hydrogen bonding on the amide at the end of the glutamine side group that results in a low value of ν(CN). The CH2–CH2 portion linking the amide to the main chain is more free to vibrate resulting in a higher value for ν(C–C) [27]. Taken together, this suggests that large fibrils are highly attracted to each other early through hydrophobic interactions on A, I, L, and V and begin twisting from excess hydrogen bonding on Q side groups but more large fibrils must add together thus adding more excess hydrogen bonding to yield fully formed fibers of D ~ 20 μm and E ~ 3–6 GPa [24]. Fully formed THWG fibers are clearly special resulting in very cylindrical cross-sections of high modulus and large fibril pitch. This is because THWG fibers have the highest Q-block content, which has been shown to be important in amyloid self-assembly [5,31,32,33,34,35]. THWG self-assembly that relies on hydrophobic interactions and excess hydrogen bonding on Q is pH independent. However, changing the temperature to lower or higher temperatures shows a deviation from the high modulus, cylindrical fiber instead favoring a mixed elliptical and tape morphology (Figure S1 and Table S1). This suggests that 37 °C is the thermodynamically and kinetically favored temperature for optimal THWG self-assembly. Fibers still self-assemble at the other temperatures showing favorable thermodynamics but kinetics must prevent the THWG large fibrils from bundling in the systematic manner they do at 37 °C.
θ is linear with D or W with cylinders having the highest θ, tapes the lowest θ, and mixed morphologies in-between (Figure 2b). There are some tape outliers where θ is not linear with W and θ is much lower than anticipated, which are circled in Figure 2b. These points are THGd:Am 0.85:0.15, THGd:Am 0.39:0.61, Am, THGd:P4An and Gd20KK:My. Am is the largest adder protein used and yields very ill-formed fibers because its high molecular weight makes it difficult to incorporate into the structure. Gd20KK:My also results in ill-formed fibers. This does not appear to be a molecular weight mismatch effect because CB4:My, P4:My, P4An:My, and P7:My have the same molecular weight mismatch but yield fully formed fibers. The biggest problem with addition of My onto Gd20KK may be the positive charge at pH 8 from the addition of K (Table S2). Gd20KK:My 0.36:0.64 has a charge mismatch and repulsive forces may precede hydrophobic interactions. Gd20KK:P4An 0.50:0.50 has a similar charge mismatch but hydrophobic interactions on these short proteins may form quickly to overcome the repulsive forces. Compared to THWG, this is the first indication that charge is a factor to consider in a molecular level model for amyloid self-assembly. ThGd:P4An yields fully formed tapes but the edges curl in and embed in one another, which slightly underestimates the tape width.
Hydrophobic interactions initiate α-helix to β-sheet conformation change in the adder protein and addition into the growing β-sheet structure and eventual protofibril [24]. Hydrophobic interactions also drive protofibril bundling into fibrils, fibril bundling into large fibrils, and large fibril bundling into large amyloid fibers [4,21]. Hydrophobic interactions alone dominate rectangular cross-section tape formation resulting in modest moduli (Figure 4) [26]. Although formed from a multitude of proteins, tape modulus remains relatively constant with θ at about E ~ 0.10–0.15 GPa. θ values are the lowest because hydrophobic interactions alone cannot induce twist into the large amyloid tape and many very weak hydrophobic interactions do not result in a high modulus. THGd:My is a mixture largely dominated by hydrophobic interactions and results in tapes over a wide range of conditions. However, self-assembly in 100 mM NaCl results in a cylinder with an increased θ and no gain in h or E. This seems to suggest some charge is screened on THGd proteins and perhaps My, which is neutral, to allow a gain in one kind of charge and like charge repulsion and twisting [19]. There are no new molecular interactions so there is no gain in modulus.
Changing THGd:My self-assembly temperature from 37 °C also changes the morphology to elliptical and twisted tape (Table S1) [26]. At some temperatures θ and h also increase. This could be a kinetic effect where certain proteins in the THGd hydrolysate are favored in the self-assembly process over others or temperature tightens large-scale interactions to cause the tape to twist or misshapen slightly. In general, mixed morphologies that begin to deviate from a tape and trend more towards a cylinder have a higher modulus with twist angle beginning to increase. All of these systems have proteins that contain Q-blocks except for P4An, which contains blocks of other amino acids with side groups capable of hydrogen bonding. Most of the cylinders are made from wheat gluten (WG), which has the highest Q content and longest Q-blocks. The only one that does not is P4An:My, which again has blocks of hydrogen bonding amino acids.
While tapes and mixed morphologies show slight increases in modulus with the β-sheet mole fraction (Figure 4), β-sheet content alone cannot distinguish one morphology from the others. This is a surprising result because traditional thinking is that β-sheet content directly correlates to fiber rigidity. The results show that other protein features can contribute to rigidity and recent theoretical work on silk indicates that structural subtleties in the amorphous regions around the β-sheets are important to properties [36]. Here, the modulus is that of the mature large amyloid fiber bundled from smaller self-assembled protofibrils. The β-sheet content is that in individual nanometer sized protofibrils and affects the modulus of the protofibril [37]. Other intermolecular interactions in the protofibril, as well as how the protofibrils bundle together and the interactions that drive the larger scale assembly appear to be important in determining the final large amyloid fiber modulus. The protein Q content seems to be an important factor when looking at how it influences modulus. However, it is simply not the Q content but the state of the amide group on glutamine that is important because the cylinders and mixed morphologies in some cases have the same Q content but different self-assembly conditions or protein mixtures have resulted in different θ, h, and E. The ratio ν(C–C/ν(CN) can differentiate the final morphology of the large amyloid fiber or tape during the self-assembly process [26]. In general, tapes, mixed morphologies, and cylinders have low, moderate, and high θ and ν(C–C)/ν(CN) values, respectively (Figure 5) [27]. In Q-containing proteins, the ν(C–C) and ν(CN) absorbances are much more pronounced than in non Q-containing proteins. As described above, as the amide on the end of the Q side group hydrogen bonds more, ν(C–C)/ν(CN) increases and for all cylinders is about 1 or greater meaning that amide hydrogen bonding increases significantly. Thus, there is excess hydrogen bonding on Q containing proteins. The value of ν(C–C)/ν(CN) in the final large amyloid fiber or tape at the end of self-assembly describes the state of θ and E. ν(C–C) and ν(CN) absorbances also appear in non Q-containing proteins but are much less pronounced and thus originate in other amino acids that still contain C–C and C–N bonds in the side group but are there in much smaller amounts [27]. The two outliers in Figure 5 are THWG 100 mM NaCl and THWG 22 °C, which have high ν(C–C)/ν(CN) but low θ. Figure 4 and Figure 5 are profound because they show that a macroscopic property like modulus can be anticipated by the large amyloid fiber morphology and molecular structure. Figure 5 in particular connects phenomena over four orders of magnitude of length scale. However, a complete model must account for other effects, particularly ones that disrupt normal self-assembly such as charge and kinetic effects. This is because THWG outliers in Figure 5 show a great deal of Q hydrogen bonding but low twist angles and tape outliers in Figure 2b show charge and molecular weight effects.
4. Materials and Methods
4.1. Protein Mixtures
Detailed procedures for the preparation of trypsin hydrolyzed wheat gluten (THWG), tryspin hydrolyzed gliadin (THGd), and its mixtures with the following natural proteins: gliadin:α-lactalbumin (THGd:Al), gliadin:amylase (THGd:Am), gliadin:hemoglobin (THGd:Hm), gliadin:insulin (THGd:In), and gliadin:myoglobin (THGd:My) can be found elsewhere [23,24,26]. THWG-95 °C was prepared the same as described previously except the solution was heated to 95 °C for 30 min at the end of the reaction [22]. α-chymotrypsin hydrolyzed wheat gluten (AHWG) was prepared the same as THWG but at 25 °C and pH 7.5, the optimal conditions for the enzyme. Individual protein properties are given in Table S2. Protein properties can be found using the ProtParam tool in ExPASy [38]. The tendency for β-sheet aggregation is found using the AGG parameter in TANGO [39]. TANGO only works for 500 amino acid sequences. For Am, cutting the 12 amino acids at the N-terminus gives AGG = 2325 and cutting the 12 amino acids at the C-terminus gives AGG = 2219. The value given is an average of the two. The sequences of the natural proteins can be found from the UniProt number, which is also given in Table S2. Wheat gluten is a mixture of gliadin (Gd, UniProt P04721, 0.49 mol fraction), high molecular weight glutenin (GtH, UniProt P08488, 0.06 mol fraction), and low molecular weight glutenin (GtL, UniProt P10386, 0.45 mol fraction). Hydrolyzing WG with trypsin or α-chymotrypsin yields two different hydrolysis products and two different self-assembled large amyloid fibers.
A series of engineered template and adder proteins was synthesized by Peptide 2.0 (Chantilly, VA, USA) with amino acid sequences confirmed by mass spectrometry. Pure proteins and 0.5:0.5 mol:mol mixtures of the synthetic proteins P4, P4An, and P7 with myoglobin (My) were dissolved in water, adjusted to pH 8 with small volumes of NaOH, and incubated at 37 °C in centrifuge tubes and monitored for 35 days (P4 and P7) and 20 days (P4An). Mixture volumes were 10 mL. P4 and P7 were monitored for longer than 20 days to see if they changed over long times but no further changes occurred so all changes happen very quickly. THGd was also mixed with P4An and P7 at 0.07:0.93 mol:mol THGd:P4An or P7 and incubated at the same conditions. Previous work comparing THWG to Gd:My has shown the importance of glutamine repeats or Q-blocks in large amyloid fiber self-assembly. P4 and P7 are designed to vary Q-block size. P4An replaces Q in P4 with other hydrophilic amino acids capable of hydrogen bonding.
Pure Gd20KK, CB4, and a 0.36:0.64 mol:mol mixture of each with My as well as 0.5:0.5 mol:mol Gd20KK:P4An and Gd20KK:P7 were prepared in a similar way and monitored for 20 days. Gd20KK is a synthetic protein based on Gd20, the gliadin 1–20 trypsin hydrolysis product with a high predicted tendency for β-sheet formation. Two lysines (K) are added at the end to increase solubility. CB4 is a synthetic protein modeled on a protein obtained from the hydrolysis of barnacle cement proteins, which are known amyloid formers [6,40]. At the end of incubation, liquid mixtures were dried at room temperature on Teflon®-coated aluminum foil in a fume hood. The sequences of the engineered proteins are given in Table S3. All protein mixtures considered in this study are listed in Table S1 as are the E, θ, and h values for cross-reference with the figures.
4.2. Fourier Transform Infrared (FTIR) Spectroscopy
Attenuated total reflectance (ATR) FTIR spectra of the incubating solutions were recorded daily on a Thermo Nicolet 6700 FTIR Spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA) with a 45° ZnSe crystal trough. The spectra were collected using 256 scans at 4 cm−1 resolution from 4000–525 cm−1. The spectrum of each protein solution was ratioed against the aqueous solvent background to reveal the protein absorbance spectrum. Three spectra were acquired at each point in time and averages ± standard error reported for each condition. Procedures for detailed quantitative analysis of the FTIR spectrum can be found elsewhere [27]. All reported FTIR values were obtained at 20 days of incubation.
4.3. Scanning Electron Microscopy (SEM)
Large amyloid fibers formed from dried solution were mounted onto aluminum SEM stubs with double-sided tape. Scanning electron micrographs were obtained using a LEO 1550 field-emission SEM (Zeiss, Peabody, MA, USA) with a 4–6 mm working distance, 5 kV accelerating voltage, and an In-lens SE-detector. Fiber dimensions were obtained on 5 fibers for samples with template and adder proteins in the mixture. Fibers formed from single proteins were not prolific fiber formers in the absence of a complementary template or adder protein. In the case of fibers formed from single proteins, measurements are based on 1 or 2 fibers. Dimensions are reported as averages ± standard error.
4.4. Nanoindentation
Fibers were mounted on stainless steel stubs and secured to the surface by a small amount of epoxy such that the fiber laid flat on the surface and was not submerged in the epoxy. Nanoindentation experiments were performed at room temperature using a Hysitron Triboindenter (Minneapolis, MN, USA) with a 0.738 μm radius, 90° conical diamond tip except for THWG-95 °C, which was tested with a Berkovich diamond 142.3 degree, 3-sided pyramidal tip of similar width. The appropriate indentation depth was found using the epoxy as a blank prior to sample indentation. Experiments were in displacement-controlled (DC) mode with a maximum displacement of 1000 nm at a rate of 100 nm/s. Indentations were performed 20–30 times on each of 3–5 fibers for each protein mixture and on 1–2 fibers for single proteins. The notable exceptions are THWG pH 8 and THWG-95 °C, which have been measured many different times from many different preparations and consistently show the highest moduli. Fiber or tape elastic (Young’s) modulus, E, was determined as previously reported and is presented as averages ± standard error [23,41].
5. Conclusions
The results of this study show that macroscopic features of self-assembled protein fibers can be designed and controlled at the molecular level. Large amyloid fibers are self-assembled protein fibers that can have rectangular or cylindrical cross-sections and morphologies in-between. Rectangular large amyloid tapes of low modulus originate in hydrophobic interactions. Cylindrical large amyloid fibers of high modulus have hydrophobic interactions and excess hydrogen bonding on Q amino acid side groups. Varying the ratio of hydrophobic groups to Q-blocks in the proteins can control morphology and modulus.
Acknowledgments
Some sample preparation and characterization was performed by Simone Gelinas, Emily Harker, David Roth, and Sydney Shaouy as part of the NSF REU program NSF-EEC-1156645. Generous funding through NSF-CMMI-0856262, USDA-NIFA #2010-65504-20429, the Virginia Tech Institute for Critical Technology and Applied Science, and the Virginia Tech Biodesign and Bioprocessing Research Center is greatly appreciated.
Author Contributions
Justin R. Barone conceived and designed the experiments, analyzed data, and wrote the manuscript. Devin M. Ridgley performed sample preparation, nanoindentation, FTIR spectroscopy, and SEM experiments, analyzed data, supervised the REU students, and wrote portions of the manuscript. Caitlin M. W. Rippner performed sample preparation, FTIR spectroscopy, data analysis, and wrote portions of the manuscript.
Supplementary Information
Supplementary materials can be accessed at: http://www.mdpi.com/2079-6439/3/2/0090/s1.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Knowles, T.P.J.; Buehler, M.J. Nanomechanics of functional and pathological amyloid materials. Nat. Nanotechnol. 2011, 6, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional amyloid-from bacteria to humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Adamcik, J.; Jung, J.-M.; Flakowski, J.; de Los Rios, P.; Dietler, G.; Mezzenga, R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat. Nanotechnol. 2010, 5, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Bolisetty, S.; Adamcik, J.; Mezzenga, R. Snapshots of fibrillation and aggregation kinetics in multistranded amyloid β-lactoglobulin fibrils. Soft Matter 2011, 7, 493–499. [Google Scholar] [CrossRef]
- Aggeli, A.; Nyrkova, I.A.; Bell, M.; Harding, R.; Carrick, L.; McLeish, T.C.B.; Semenov, A.N.; Boden, N. Hierarchical self-assembly of chiral rod-like molecules as a model for peptide β-sheet tapes, ribbons, fibrils, and fibers. Proc. Natl. Acad. Sci. USA 2001, 98, 11857–11862. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.E.; Dickinson, G.H.; Orihuela, B.; Kulp III, J.L.; Rittschof, D.; Wahl, K.J. Characterization of the adhesive plaque of the barnacle Balanus amphitrite: Amyloid-like nanofibrils are a major component. Langmuir 2010, 26, 6549–6556. [Google Scholar] [CrossRef] [PubMed]
- Gebbink, M.F.B.G.; Claessen, D.; Bouma, B.; Dijkhuizen, L.; Wosten, H.A.B. Amyloids—A functional coat for microorganisms. Nat. Rev. Microbiol. 2005, 3, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Schmidt, J.C.; Chapman, M.R. The curli nucleator protein, csgb, contains an amyloidogenic domain that directs csga polymerization. Proc. Natl. Acad. Sci. USA 2007, 104, 12494–12499. [Google Scholar] [CrossRef] [PubMed]
- Alsteens, D.; Ramsook, C.B.; Lipke, P.N.; Dufrêne, Y.F. Unzipping a functional microbial amyloid. ACS Nano 2012, 6, 7703–7711. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.D.; Rudall, K.M. Structure of the silk of Chrysopa egg-stalks. Nature 1957, 179, 905–906. [Google Scholar] [CrossRef]
- Mostaert, A.; Higgins, M.; Fukuma, T.; Rindi, F.; Jarvis, S. Nanoscale mechanical characterisation of amyloid fibrils discovered in a natural adhesive. J. Biol. Phys. 2006, 32, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Gazit, E. Self-assembled peptide nanostructures: The design of molecular building blocks and their technological utilization. Chem. Soc. Rev. 2007, 36, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Oppenheim, T.W.; Buell, A.K.; Chirgadze, D.Y.; Welland, M.E. Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nat. Nanotechnol. 2010, 5, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Born, A.-K.; Schweizer, T.; Zenobi-Wong, M.; Cerruti, M.; Mezzenga, R. Amyloid-hydroxyapatite bone biomimetic composites. Adv. Mater. 2014, 26, 3207–3212. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Li, C.; Adamcik, J.; Shao, Z.; Chen, X.; Mezzenga, R. Modulating materials by orthogonally oriented β-strands: Composites of amyloid and silk fibroin fibrils. Adv. Mater. 2014, 26, 4569–4574. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Esler, W.P.; Stimson, E.R.; Jennings, J.M.; Vinters, H.V.; Ghilardi, J.R.; Lee, J.P.; Mantyh, P.W.; Maggio, J.E. Alzheimer’s disease amyloid propagation by a template-dependent dock-lock mechanism. Biochemistry 2000, 39, 6288–6295. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.R.; Douglass, A.; Vale, R.D.; Weissman, J.S. Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol. 2004, 2, 1582–1590. [Google Scholar] [CrossRef]
- Adamcik, J.; Mezzenga, R. Adjustable twisting periodic pitch of amyloid fibrils. Soft Matter 2011, 7, 5437–5443. [Google Scholar] [CrossRef]
- Lara, C.C.; Adamcik, J.; Jordens, S.; Mezzenga, R. General self-assembly mechanism converting hydrolyzed globular proteins into giant multistranded amyloid ribbons. Biomacromolecules 2011, 12, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Barone, J.R. Evolution of the amyloid fiber over multiple length scales. ACS Nano 2013, 7, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Athamneh, A.; Barone, J.R. Enzyme-mediated self-assembly of highly ordered structures from disordered proteins. Smart Mater. Struct. 2009, 18, 104024. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Ebanks, K.C.; Barone, J.R. Peptide mixtures can self-assemble into large amyloid fibers of varying size and morphology. Biomacromolecules 2011, 12, 3770–3779. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Claunch, E.C.; Lee, P.W.; Barone, J.R. The role of protein hydrophobicity in conformation change and self-assembly into large amyloid fibers. Biomacromolecules 2014, 15, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Kirkitadze, M.D.; Condron, M.M.; Teplow, D.B. Identification and characterization of key kinetic intermediates in amyloid β-protein fibrillogenesis. J. Mol. Biol. 2001, 312, 1103–1119. [Google Scholar] [CrossRef] [PubMed]
- Ridgley, D.M.; Claunch, E.C.; Barone, J.R. The effect of processing on large, self-assembled amyloid fibers. Soft Matter 2012, 8, 10298–10306. [Google Scholar] [CrossRef]
- Ridgley, D.M.; Claunch, E.C.; Barone, J.R. Characterization of amyloid structures by FT-IR and raman spectroscopy. Appl. Spectrosc. 2013, 67, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Grason, G.M. Defects in crystalline packings of twisted filament bundles. I. Continuum theory of disclinations. Phys. Rev. E 2012, 85, 031603. [Google Scholar] [CrossRef]
- Gunzler, H.; Gremlich, H.-U. IR Spectroscopy; Wiley-VCH: Weinheim, Germany, 2002. [Google Scholar]
- Barth, A. The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 2000, 74, 141–173. [Google Scholar] [CrossRef] [PubMed]
- Perutz, M.F.; Johnson, T.; Suzuki, M.; Finch, J.T. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. USA 1994, 91, 5355–5358. [Google Scholar] [CrossRef] [PubMed]
- DePace, A.H.; Santoso, A.; Hillner, P.; Weissman, J.S. A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell 1998, 93, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Scherzinger, E.; Sittler, A.; Schweiger, K.; Heiser, V.; Lurz, R.; Hasenbank, R.; Bates, G.P.; Lehrach, H.; Wanker, E.E. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: Implications for huntington’s disease pathology. Proc. Natl. Acad. Sci. USA 1999, 96, 4604–4609. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Berthelier, V.; Bradley Hamilton, J.; O’Nuallain, B.; Wetzel, R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry 2002, 41, 7391–7399. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, P.; Atkins, E. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules 2005, 6, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement controls stiffness, strength and mechanical toughness of β-sheet crystals in silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.S.; Adamcik, J.; Jeong, J.S.; Lashuel, H.A.; Mezzenga, R.; Dietler, G. Influence of the β-sheet content on the mechanical properties of aggregates during amyloid fibrillization. Angew. Chem. Int. Ed. 2015, 54, 2462–2466. [Google Scholar] [CrossRef]
- ExPASy Bioinformatics Resource Portal. Available online: http://web.expasy.org/protparam/ (accessed on 1 July 2014).
- Tango-A computer algorithm for prediction of aggregating regions in unfolded polypeptide chains. Available online: tango.crg.es (accessed on 1 July 2014).
- Kamino, K.; Odo, S.; Maruyama, T. Cement proteins of the acorn-barnacle, Megabalanus rosa. Biol. Bull. 1996, 190, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Oliver, W.C.; Pharr, G.M. An improved technique for determining hardness and elastic modulus using load and displacement sensing indentation experiments. J. Mater. Res. 1992, 7, 1564–1583. [Google Scholar] [CrossRef]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
