Nanowire Aptamer-Sensitized Biosensor Chips with Gas Plasma-Treated Surface for the Detection of Hepatitis C Virus Core Antigen
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Proteins and Aptamers
2.3. Silicon-on-Insulator Nanowire (SOI-NW) Sensor Chips
2.4. Sensitization of Sensor Surface
2.5. Preparation of the Core Antigen of Hepatitis C Virus (HCVcoreAg) Solutions in Buffer
2.6. Serum Samples
2.7. Electrical Measurements
2.8. Data Representation
3. Results
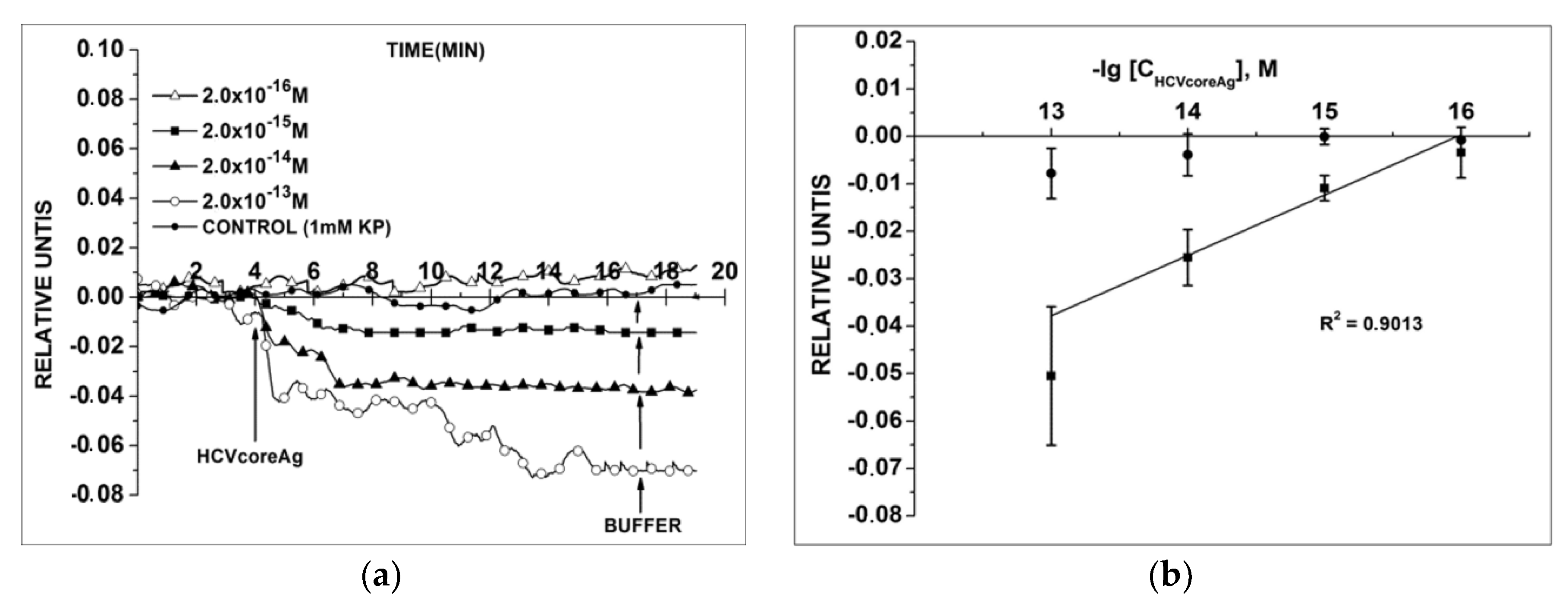
3.1. Detection of HCVcoreAg in Acidic Buffer
3.2. Detection of HCVcoreAg in Neutral Buffer
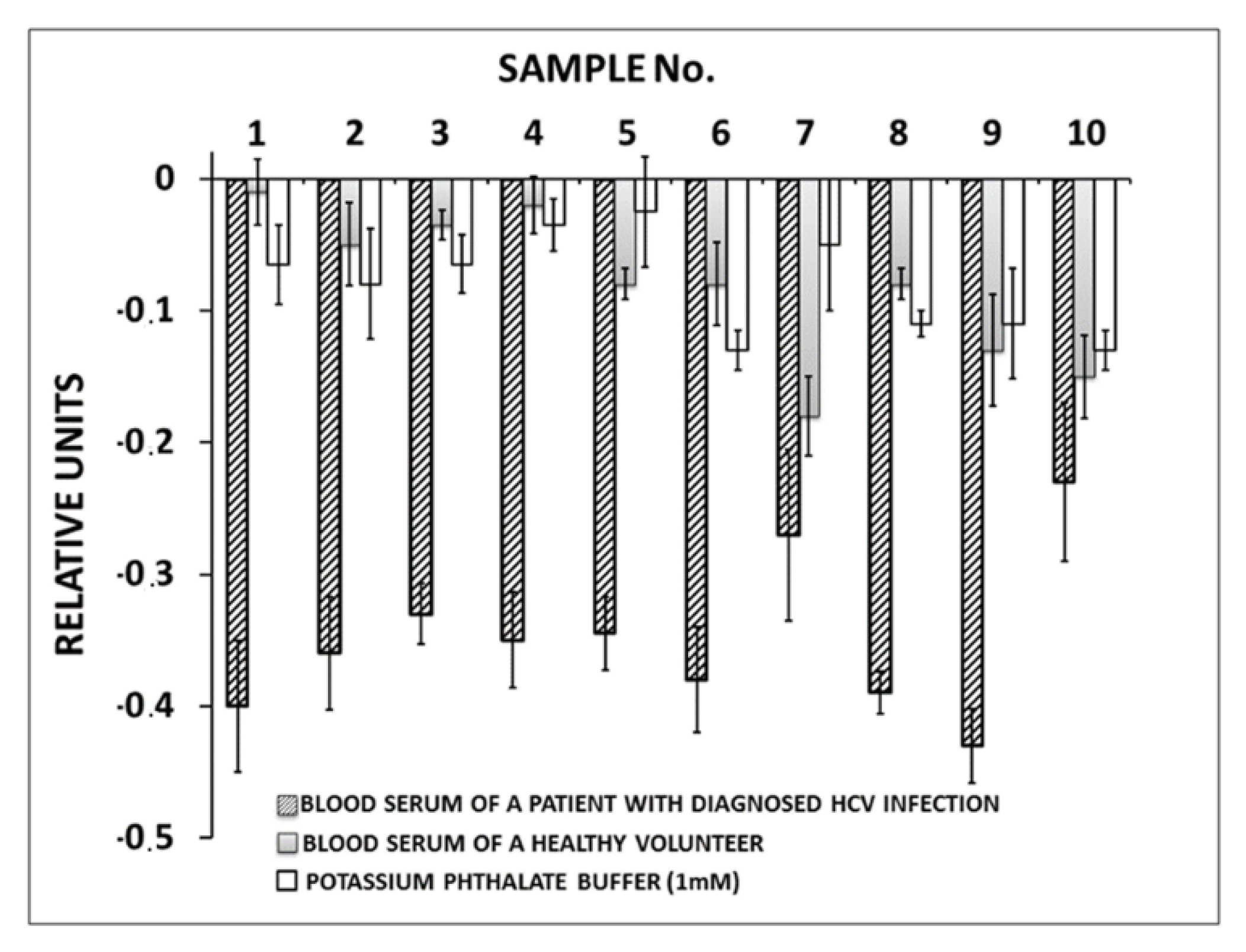
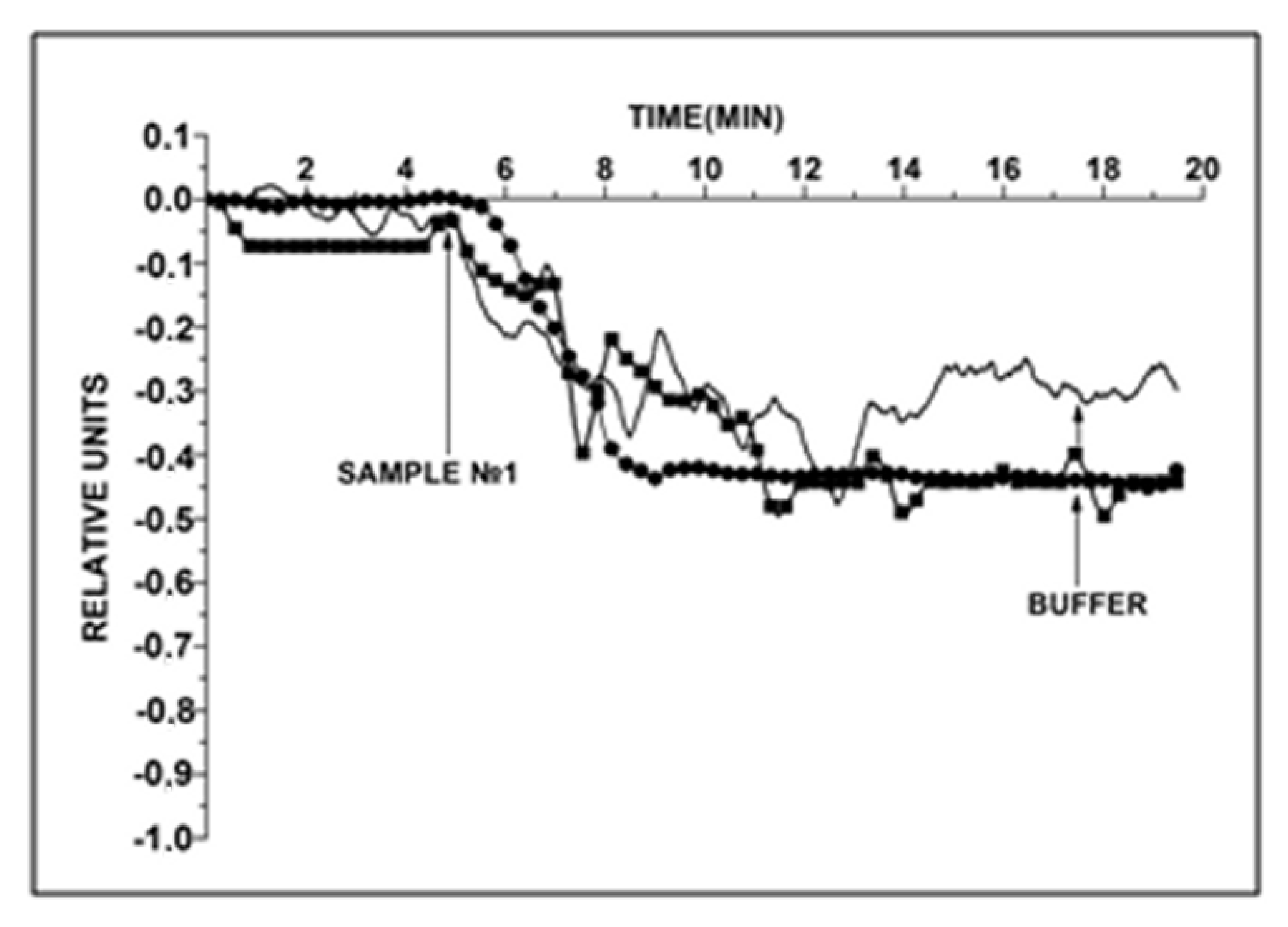
3.3. HCVcoreAg Detection in Human Serum Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alavi, M.; Law, M.G.; Grebely, J.; Amin, J.; Hajarizadeh, B.; George, J.; Dore, G.J. Time to decompensated cirrhosis and hepatocellular carcinoma after an HBV or HCV notification: A population-based study. J. Hepatol. 2016, 65, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Santantonio, T.; Wiegand, J.; Gerlach, J.T. Acute hepatitis C: Current status and remaining challenges. J. Hepatol. 2008, 49, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Maasouny, B.; Wedemeyer, H. Natural history of acute and chronic hepatitis. Best. Pract. Res. Clin. Gastroenterol. 2012, 26, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Global Hepatitis Report 2017. Available online: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/ (accessed on 12 May 2020).
- World Health Organization. Progress Report on Access to Hepatitis C Treatment, Focus on Overcoming Barriers in Low and Middle Income Countries. Available online: http://apps.who.int/iris/bitstream/10665/260445/1/WHO-CDS-HIV-18.4-eng.pdf?ua=1 (accessed on 12 May 2020).
- Waheed, Y.; Siddiq, M.; Jamil, Z.; Najmi, M.H. Hepatitis elimination by 2030: Progress and challenges. World J. Gastroenterol. 2018, 24, 4959–4961. [Google Scholar] [CrossRef]
- de Lemos, A.S.; Chung, R.T. Hepatitis C treatment: An incipient therapeutic revolution. Trends Mol. Med. 2014, 20, 315–321. [Google Scholar] [CrossRef]
- Ross, R.S.; Viazov, S.; Salloum, S.; Hilgard, P.; Gerken, G.; Roggendorf, M. Analytical performance characteristics and clinical utility of a novel assay for total hepatitis c virus core antigen quantification. J. Clin. Microbiol. 2010, 48, 1161–1168. [Google Scholar] [CrossRef]
- Hadziyannis, E.; Minopetrou, M.; Georgiou, A.; Spanou, F.; Koskinas, J. Is HCV core antigen a reliable marker of viral load? An evaluation of HCV core antigen automated immunoassay. Ann. Gastroenterol. 2013, 26, 146–149. [Google Scholar] [PubMed]
- Wasitthankasem, R.; Vichaiwattana, P.; Auphimai, C.; Siripon, N.; Klinfueng, S.; Tangkijvanich, P.; Vongpunsawad, S.; Poovorawan, Y. HCV core antigen is an alternative marker to HCV RNA for evaluating active HCV infection: Implications for improved diagnostic option in an era of affordable DAAs. Peer J. 2017, 5, e4008. [Google Scholar] [CrossRef]
- Soliman, H.; Hozayen, W.; Mahmoud, A.; Abo-Seif, M.; Fayede, N.A. Significance of the hepatitis C virus core antigen testing as an alternative marker for hepatitis diagnosis in Egyptian patients. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 2240–2245. [Google Scholar] [PubMed]
- Buket, C.; Ayşe, A.; Selçuk, K.; Süleyman, Ö.; Emel, S. Comparison of HCV core antigen and anti-HCV with HCV RNA results. Afr. Health Sci. 2014, 14, 816–820. [Google Scholar] [CrossRef]
- Murayama, A.; Sugiyama, N.; Watashi, K.; eMasaki, T.; Suzuki, R.; Alizaki, H.; Mizuochi, T.; Wakita, T.; Kato, T. Japanese reference panel of blood specimens for evaluation of hepatitis C virus RNA and core antigen quantitative assays. J. Clin. Microbiol. 2012, 50, 1943–1949. [Google Scholar] [CrossRef] [PubMed]
- Cresswell, F.V.; Fisher, M.; Hughes, D.J.; Shaw, S.G.; Hometr, G.; Halssan-Ibrahim, M.O. Hepatitis C core antigen testing: A reliable, quick, and potentially cost-effective alternative to hepatitis C polymerase chain reaction in diagnosing acute hepatitis C virus infection. Clin. Infect. Dis. 2014, 60, 263–266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C 2016. J. Hepatol. 2017, 66, 153–194. [Google Scholar]
- World Health Organization. WHO Guidelines on Hepatitis B and C Testing; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Miédougé, M.; Sauné, K.; Kamar, N.; Rieu, M.; Rostaing, L.; Izopet, J. Analytical evaluation of HCV core antigen and interest for HCV screening in haemodialysis patients. J. Clin. Virol. 2010, 48, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Garcia-D’Angeli, A.; Brennan, J.P.; Huo, Q. Predicting detection limits of enzyme-linked immunosorbent assay (ELISA) and bioanalytical techniques in general. Analyst 2014, 139, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Kaysheva, A.L.; Ivanov, Y.D.; Zgoda, V.G.; Frantsuzov, P.; Pleshakova, T.O.; Krokhin, N.V.; Ziborov, V.S.; Archakov, A.I. Visualization and identification of hepatitis C viral particles by atomic force microscopy combined with MS/MS analysis. Biochem. (Moscow) Suppl. Ser. B Biomed. Chem. 2010, 4, 15–24. [Google Scholar] [CrossRef]
- Xu, J.; Hu, Y.; Wang, S.; Ma, X.; Guo, J. Nanomaterials in electrochemical cytosensors. Analyst 2020, 145, 2058–2069. [Google Scholar] [CrossRef]
- Gao, X.P.A.; Zheng, G.; Lieber, C.M. Subthreshold regime has the optimal sensitivity for nanowire FET biosensors. Nano Lett. 2010, 10, 547–552. [Google Scholar] [CrossRef]
- Stern, E.; Klemic, J.; Routenberg, D.; Wyrembak, P.; Turner-Evans, D.; Hamilton, A.; LaVan, D.; Fahmy, T.; Reed, M. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature 2007, 445, 519–522. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.O.; Kozlov, A.F.; Malsagova, K.A.; Krohin, N.V.; Shumyantseva, V.; Shumov, I.D.; Popov, V.; Naumova, O.V.; Fomin, B.; et al. SOI nanowire for the high-sensitive detection of HBsAg and α-fetoprotein. Lab Chip 2012, 12, 5104–5111. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.O.; Kozlov, A.F.; Malsagova, K.A.; Krokhin, N.V.; Kaisheva, A.L.; Shumov, I.D.; Archakov, A.; Popov, V.P.; Naumova, O.V.; et al. SOI nanowire transistor for detection of D-NFATc1 molecules. Optoelectron. Instrum. Data Process. 2013, 49, 520–525. [Google Scholar] [CrossRef]
- Malsagova, K.A.; Ivanov, Y.D.; Pleshakova, T.O.; Kaysheva, A.L.; Shumov, I.D.; Kozlov, A.; Archakov, A.I.; Popov, V.; Fomin, B.I.; Latyshev, A.V. A SOI-nanowire biosensor for the multiple detection of D-NFATc1 protein in the serum. Anal. Methods. 2015, 7, 8078–8085. [Google Scholar] [CrossRef]
- Malsagova, K.A.; Pleshakova, T.O.; Galiullin, R.A.; Kaysheva, A.L.; Shumov, I.D.; Ilnitskii, M.A.; Popov, V.P.; Glukhov, A.V.; Archakov, A.I.; Ivanov, Y.D. Ultrasensitive nanowire-based detection of HCVcoreAg in the serum using a microwave generator. Anal. Meth. 2018, 2740–2749. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.; Malsagova, K.; Kozlov, A.L.; Kaysheva, A.; Shumov, I.; Galiullin, R.; Kurbatov, L.; Popov, V.; Naumova, O.; et al. Detection of marker miRNAs in plasma using SOI-NW biosensor. Sens. Actuators B Chem. 2018, 261, 566–571. [Google Scholar] [CrossRef]
- Laborde, C.; Pittino, F.; Verhoeven, H.A.; Lemay, S.G.; Selmi, L.; Jongsma, M.A.; Widdershoven, F.P. Real-time imaging of microparticles and living cells with CMOS nanocapacitor arrays. Nat. Nanotechnol. 2015, 10, 791–795. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lee, H.; Yang, J.; Jo, M.; Hahn, S. The fabrication, characterization and application of aptamer-functionalized Si-nanowire FET biosensors. J. Nanotechnol. 2009, 20, 235501. [Google Scholar] [CrossRef]
- Savory, N.; Abe, K.; Sode, K.; Ikebukuro, K. Selection of DNA aptamer against prostate specific antigen using a genetic algorithm and application to sensing. Biosens. Bioelectron. 2010, 26, 1386–1391. [Google Scholar] [CrossRef]
- Kaysheva, A.L.; Ivanov, Y.D.; Bukharina, N.S.; Pleshakova, T.; Frantsuzov, P.; Andreeva, E.; Kaysheva, A.; Zgoda, V.; Izotov, A.; Pavlova, T.; et al. Atomic force microscopy fishing and mass spectrometry identification of gp120 on immobilized aptamers. Int. J. Nanomed. 2014, 9, 4659–4670. [Google Scholar] [CrossRef][Green Version]
- Shi, S.; Yu, X.; Gao, Y.; Xue, B.; Wu, X.; Wang, X.; Yang, D.; Zhu, H. Inhibition of hepatitis C virus production by aptamers against the core protein. J. Virol. 2014, 88, 1990–1999. [Google Scholar] [CrossRef]
- Tyschenko, I.E.; Popov, V.P. Silicon-on-insulator structures produced by ion-beam synthesis and hydrogen transfer. In Advances in Semiconductor Nanostructures; Alexander, V., Latyshev, A.V., Dvurechenskii, A.L.A., Eds.; Elsevier BV: Amsterdam, The Netherlands, 2017; Volume 552, pp. 409–433. ISBN 978-0-12-810512-2. [Google Scholar]
- Available online: http://www.chem.agilent.com/Library/applications/5991-3207EN.pdf (accessed on 5 March 2020).
- Ivanov, Y.D.; Kozlov, A.; Galiullin, R.; Kanashenko, S.; Usanov, S.; Ivanova, N.; Ziborov, V.; Pleshakova, T. Spontaneous generation of charge in the flow-based AFM fishing system. J. Electrost. 2018, 91, 16–20. [Google Scholar] [CrossRef]
- Morota, K.; Fujinami, R.; Kinukawa, H.; Machida, T.; Ohno, K.; Saegusa, H.; Takeda, K. A new sensitive and automated chemiluminescent microparticle immunoassay for quantitative determination of hepatitis C virus core antigen. J. Virol. Meth. 2009, 157, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Liang, M.; Wang, L.; Xiang, H.; Jiang, Y.; Li, Y.; Xie, G. MultisHRP-DNA-coated CMWNTs as signal labels for an ultrasensitive hepatitis C virus core antigen electrochemical immunosensor. Biosens. Bioelectron. 2013, 47, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Wang, L.; Ma, C.; Zhang, M.; Xie, G. Sandwich immunoassay for hepatitis C virus non-structural 5A protein using a glassy carbon electrode modified with an Au-MoO3/chitosan nanocomposite. Anal. Lett. 2013, 46, 1241–1254. [Google Scholar] [CrossRef]
- Moraes, M.; Lima, L.R.; Da Silva, R.R.; Cavicchioli, M.; Ribeiro, S.J. Immunosensor based on immobilization of antigenic peptide NS5A-1 from HCV and silk fibroin in nanostructured films. Langmuir 2013, 29, 3829–3834. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yuan, L.; Liu, Q.; Tong, C.; Wang, W.; Xiao, F.; Liu, B.; Liu, X. An ultrasensitive and simple assay for the Hepatitis C virus using a reduced graphene oxide-assisted hybridization chain reaction. Analyst 2019, 144, 3972–3979. [Google Scholar] [CrossRef] [PubMed]



| Applied Method | Target Protein | Detection Limit | Detection Time |
|---|---|---|---|
| Chemiluminescence immunoassay [36] | HCVcoreAg | 0.06 pg/mL | 1 h |
| Glassy carbon electrode modified with a nanocomposite (gold/zirconia/chitosan) [37] | HCVcoreAg | 0.01 pg/mL | 25 min |
| Glassy carbon electrode was modified with an Au-MoO3/Chitosan nanocomposite [38] | HCV non-structural 5A protein | 1.0 ng/mL | 30 min |
| Silk fibroin nanostructured films on screen-printed carbon electrode [39] | HCV non-structural 5A protein | 28 μA·μg−1·mL | 10 min |
| Reduced graphene oxide nanosheets and hybridization chain reaction amplification technique [40] | HCV RNA | 0.3 pg/mL | 8–10 h |
| SOI-NW-based aptasensor (this study) | HCVcoreAg | 0.3 pg/mL | 17 min |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malsagova, K.A.; Pleshakova, T.O.; Galiullin, R.A.; Shumov, I.D.; Kozlov, A.F.; Romanova, T.S.; Popov, V.P.; Glukhov, A.V.; Konev, V.A.; Archakov, A.I.; et al. Nanowire Aptamer-Sensitized Biosensor Chips with Gas Plasma-Treated Surface for the Detection of Hepatitis C Virus Core Antigen. Coatings 2020, 10, 753. https://doi.org/10.3390/coatings10080753
Malsagova KA, Pleshakova TO, Galiullin RA, Shumov ID, Kozlov AF, Romanova TS, Popov VP, Glukhov AV, Konev VA, Archakov AI, et al. Nanowire Aptamer-Sensitized Biosensor Chips with Gas Plasma-Treated Surface for the Detection of Hepatitis C Virus Core Antigen. Coatings. 2020; 10(8):753. https://doi.org/10.3390/coatings10080753
Chicago/Turabian StyleMalsagova, Kristina A., Tatyana O. Pleshakova, Rafael A. Galiullin, Ivan D. Shumov, Andrey F. Kozlov, Tatyana S. Romanova, Vladimir P. Popov, Alexander V. Glukhov, Vladimir A. Konev, Alexander I. Archakov, and et al. 2020. "Nanowire Aptamer-Sensitized Biosensor Chips with Gas Plasma-Treated Surface for the Detection of Hepatitis C Virus Core Antigen" Coatings 10, no. 8: 753. https://doi.org/10.3390/coatings10080753
APA StyleMalsagova, K. A., Pleshakova, T. O., Galiullin, R. A., Shumov, I. D., Kozlov, A. F., Romanova, T. S., Popov, V. P., Glukhov, A. V., Konev, V. A., Archakov, A. I., & Ivanov, Y. D. (2020). Nanowire Aptamer-Sensitized Biosensor Chips with Gas Plasma-Treated Surface for the Detection of Hepatitis C Virus Core Antigen. Coatings, 10(8), 753. https://doi.org/10.3390/coatings10080753






