Laser-Irradiated Chlorpromazine as a Potent Anti-Biofilm Agent for Coating of Biomedical Devices
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical Solutions and Laser Irradiation
2.2. pH Measurements
2.3. Microbial Strains
2.4. Determination of Minimum Inhibitory Concentration (MIC)
2.5. Minimum Biofilm Eradication Concentration Assay (MBEC)
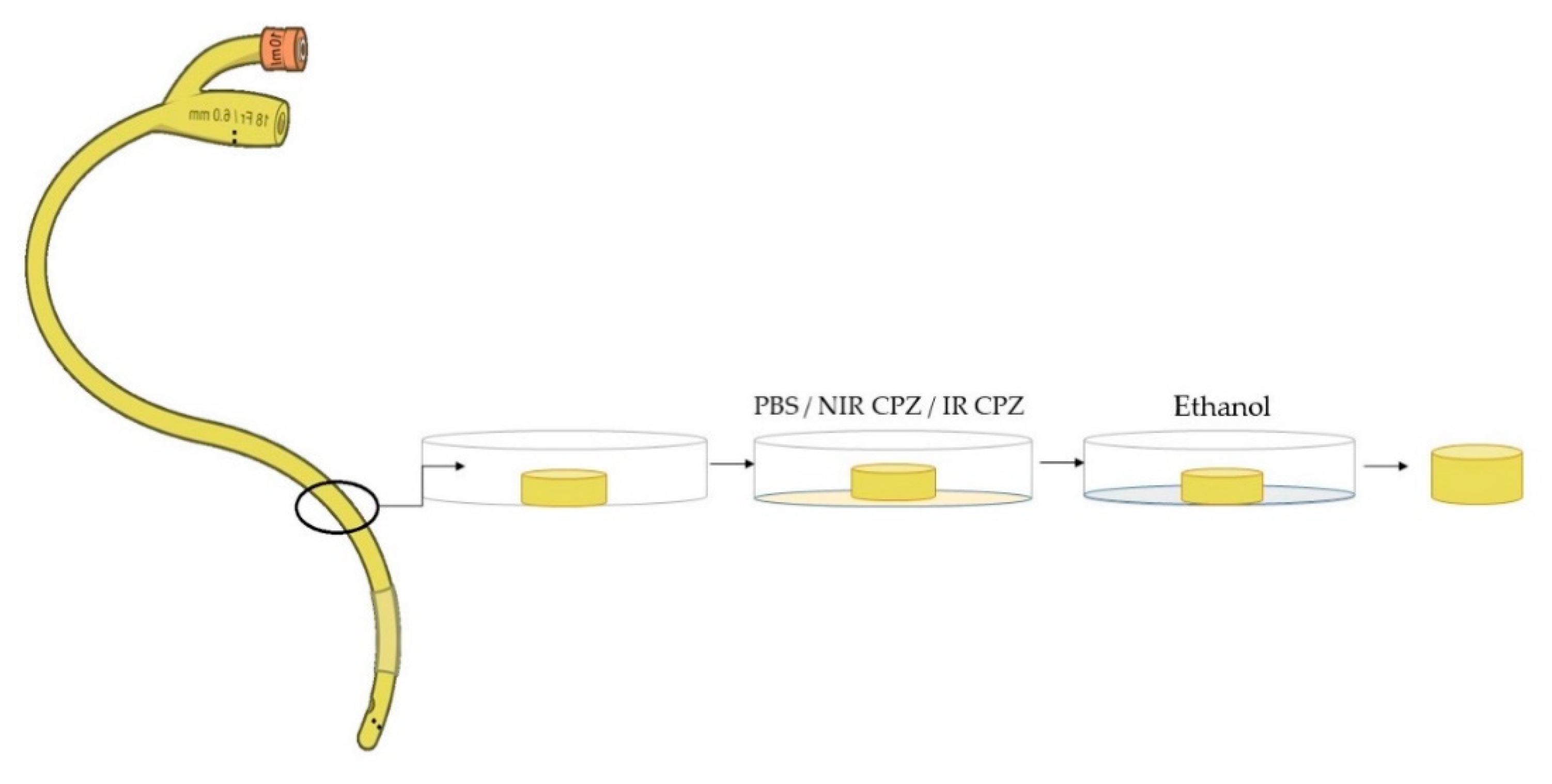
2.6. Immersion of Medical Devices
2.7. Antibiofilm Assay
2.8. Computational Approach
3. Results
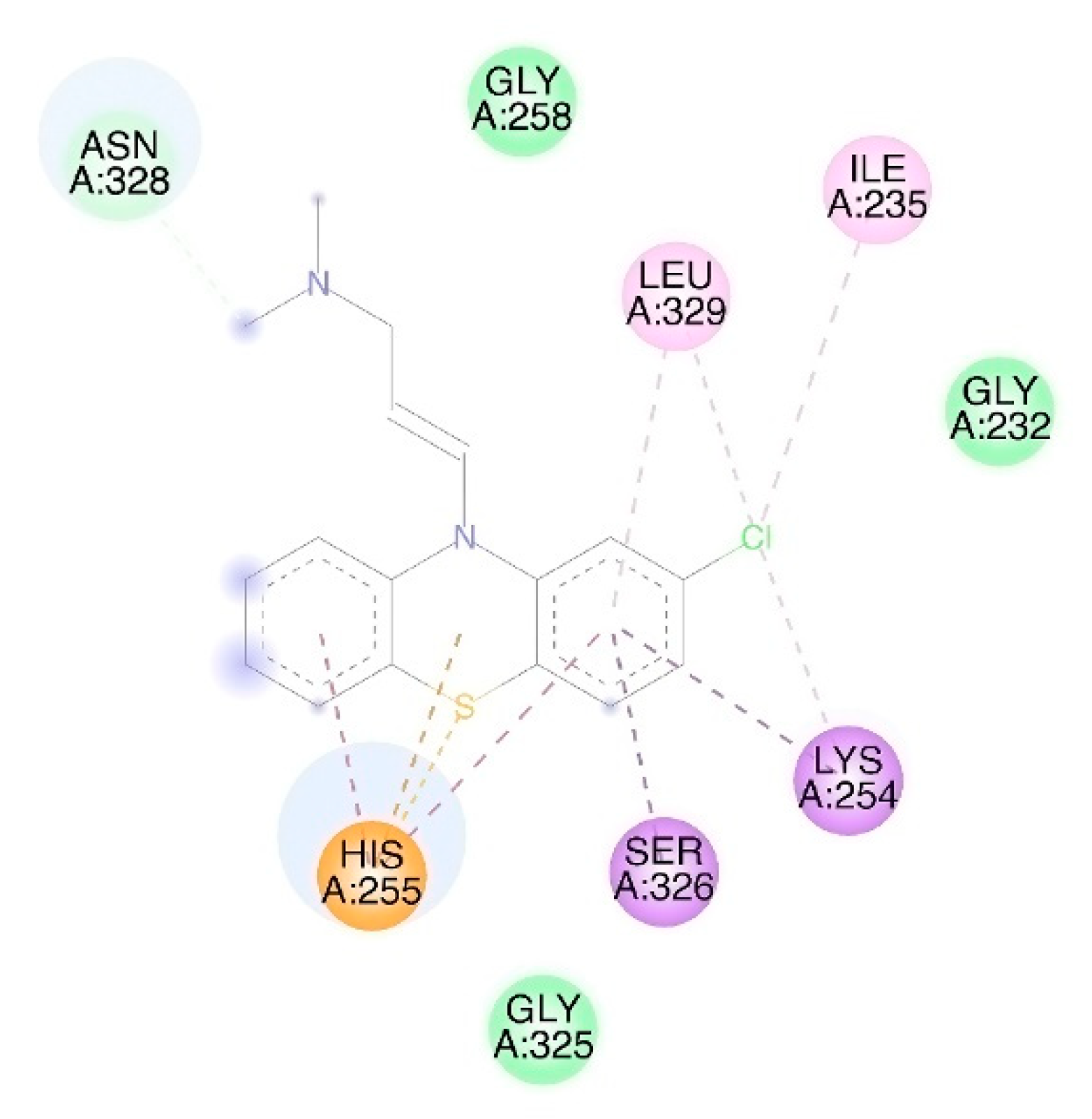
3.1. Molecular Docking
3.2. pH Values
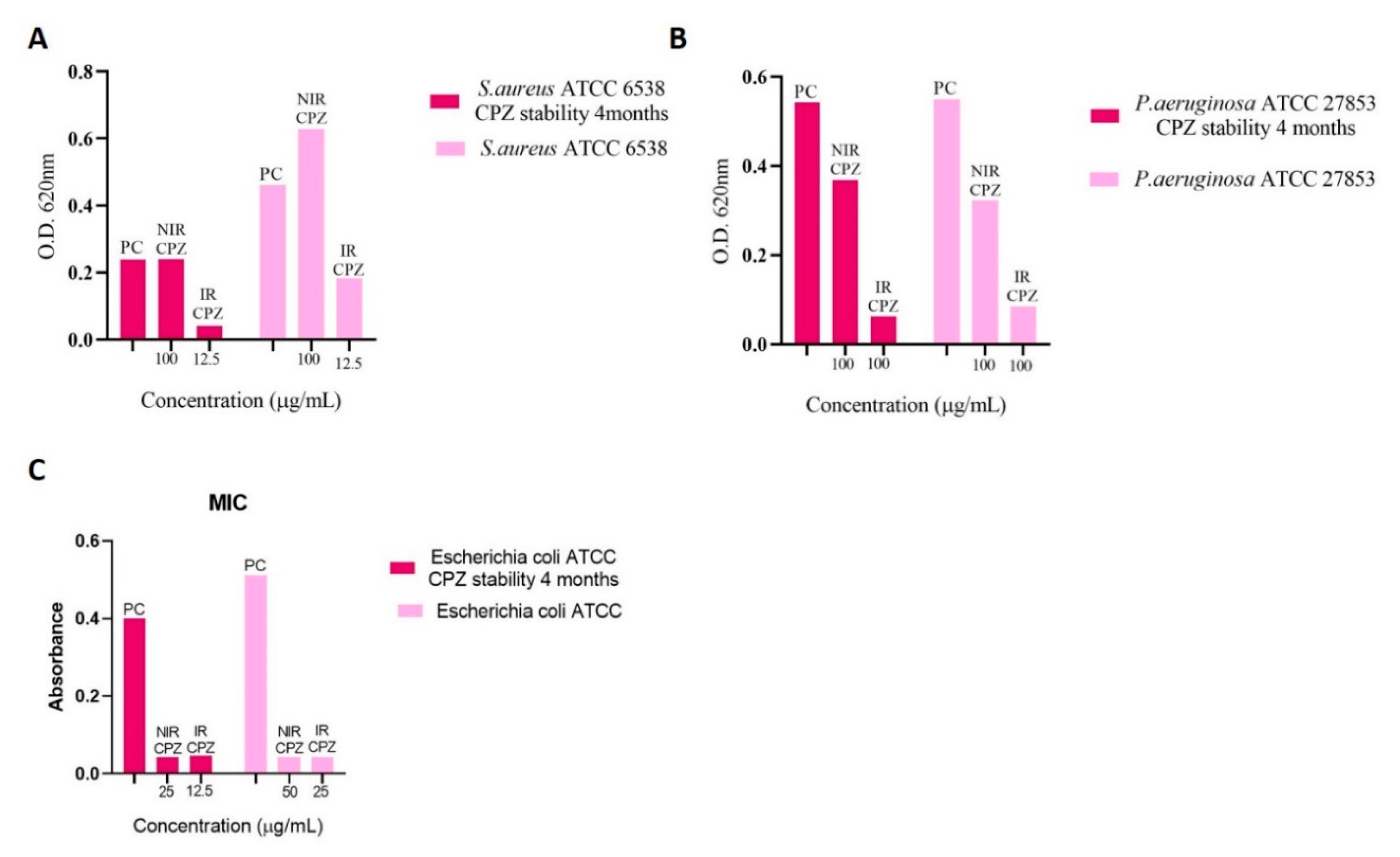
3.3. Determination of Minimum Inhibitory Concentration (MIC)
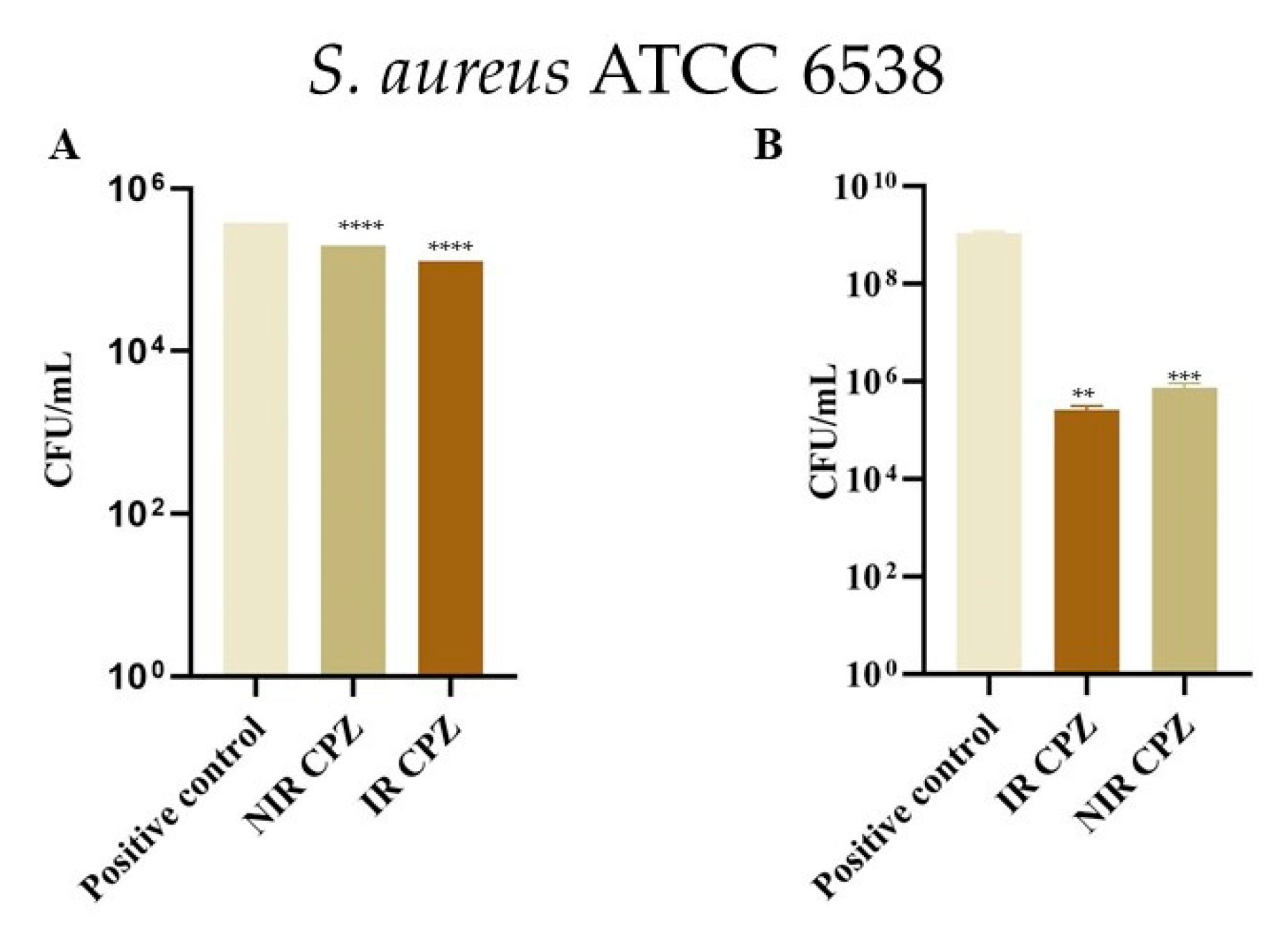
3.4. Inhibition of Bacterial Adhesion on Cotton and Metal
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Annual Report of the Chief Medical Officer Volume Two, 2011. Infections and the Rise of Antimicrobial Resistance. 2011; 154p. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/138331/CMO_Annual_Report_Volume_2_2011.pdf (accessed on 7 June 2020).
- McEwen, S.A.; Collignon, P.J. Antimicrobial Resistance: A One Health Perspective. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; Schwarz, S., Cavaco, L.M., Shen, J., Eds.; American Society of Microbiology: Washington, DC, USA, 2018; pp. 521–547. ISBN 978-1-55581-979-8. [Google Scholar]
- Sakko, M.; Tj, L. Microbiology of Root Canal Infections. Prim. Dent. J. 2016, 5, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Johani, K.; Abualsaud, D.; Costa, D.M.; Hu, H.; Whiteley, G.; Deva, A.; Vickery, K. Characterization of microbial community composition, antimicrobial resistance and biofilm on intensive care surfaces. J. Infect. Public Health 2018, 11, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.-F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef]
- Welte, T. Nosocomial Infections—A Present and Future Challenge. Dtsch. Arztebl. Int. 2013, 110, 625–626. [Google Scholar] [CrossRef]
- World Health Organization. Report on the Burden of Endemic Health Care-Associated Infection Worldwide; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Khan, H.A.; Baig, F.K.; Mehboob, R. Nosocomial infections: Epidemiology, prevention, control and surveillance. Asian Pac. J. Trop. Biomed. 2017, 7, 478–482. [Google Scholar] [CrossRef]
- Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014.
- Emerson, C.B.; Eyzaguirre, L.M.; Albrecht, J.S.; Comer, A.C.; Harris, A.D.; Furuno, J.P. Healthcare-Associated Infection and Hospital Readmission. Infect. Control Hosp. Epidemiol. 2012, 33, 539–544. [Google Scholar] [CrossRef]
- Safdar, N.; Maki, D.G. The Commonality of Risk Factors for Nosocomial Colonization and Infection with Antimicrobial-Resistant Staphylococcus aureus, Enterococcus, Gram-Negative Bacilli, Clostridium difficile, and Candida. Ann. Intern. Med. 2002, 136, 834–844. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Tamura, N.K.; Gasparetto, A.; Svidzinski, T.I.E. Evaluation of the Adherence of Candida Species to Urinary Catheters. Mycopathologia 2003, 156, 269–272. [Google Scholar] [CrossRef]
- Davies, J. Inactivation of antibiotics and the dissemination of resistance genes. Science 1994, 264, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Pascu, M.L. (Ed.) Laser Optofluidics in Fighting Multiple Drug Resistance; Bentham Science Publishers: Sharjah, UAE, 2017; ISBN 978-1-68108-498-5. [Google Scholar]
- van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef] [PubMed]
- USDA Agricultural Research Service; Allen, H.K. Alternatives to Antibiotics: Why and How. NAM Perspect. 2017, 7. [Google Scholar] [CrossRef]
- Sudeshna, G.; Parimal, K. Multiple non-psychiatric effects of phenothiazines: A review. Eur. J. Pharmacol. 2010, 648, 6–14. [Google Scholar] [CrossRef]
- Otręba, M.; Zdybel, M.; Pilawa, B.; Beberok, A.; Wrześniok, D.; Rok, J.; Buszman, E. EPR spectroscopy of chlorpromazine-induced free radical formation in normal human melanocytes. Eur. Biophys. J. 2015, 44, 359–365. [Google Scholar] [CrossRef]
- MacAllister, S.L.; Young, C.; Guzdek, A.; Zhidkov, N.; O’Brien, P.J. Molecular cytotoxic mechanisms of chlorpromazine in isolated rat hepatocytes. Can. J. Physiol. Pharmacol. 2013, 91, 56–63. [Google Scholar] [CrossRef]
- Pascu, M.L.; Danko, B.; Martins, A.; Jedlinszki, N.; Alexandru, T.; Nastasa, V.; Boni, M.; Militaru, A.; Andrei, I.R.; Staicu, A.; et al. Exposure of Chlorpromazine to 266 nm Laser Beam Generates New Species with Antibacterial Properties: Contributions to Development of a New Process for Drug Discovery. PLoS ONE 2013, 8, e55767. [Google Scholar] [CrossRef][Green Version]
- Alexandru, T.; Staicu, A.; Pascu, A.; Radu, E.; Stoicu, A.; Nastasa, V.; Dinache, A.; Boni, M.; Amaral, L.; Pascu, M.L. Characterization of mixtures of compounds produced in chlorpromazine aqueous solutions by ultraviolet laser irradiation: Their applications in antimicrobial assays. J. Biomed. Opt. 2014, 20, 051002. [Google Scholar] [CrossRef]
- Drucker, A.M.; Rosen, C.F. Drug-Induced Photosensitivity: Culprit Drugs, Management and Prevention. Drug Saf. 2011, 34, 821–837. [Google Scholar] [CrossRef]
- Vlad, I.M.; Nuta, D.C.; Chirita, C.; Caproiu, M.T.; Draghici, C.; Dumitrascu, F.; Bleotu, C.; Avram, S.; Udrea, A.M.; Missir, A.V.; et al. In Silico and In Vitro Experimental Studies of New Dibenz[b,e]oxepin-11(6H)one O-(arylcarbamoyl)-oximes Designed as Potential Antimicrobial Agents. Molecules 2020, 25, 321. [Google Scholar] [CrossRef]
- Limban, C.; Diţu, L.M.; Măruțescu, L.; Missir, A.V.; Chifiriuc, M.C.; Căproiu, M.T.; Morusciag, L.; Chiriţă, C.; Udrea, A.-M.; Nuţă, D.C.; et al. Design, Synthesis and Biopharmacological Profile Evaluation of New 2-((4- Chlorophenoxy)Methyl)-N-(Arylcarbamothioyl)Benzamides with Broad Spectrum Antifungal Activity. COC 2019, 23, 1365–1377. [Google Scholar] [CrossRef]
- Tozar, T.; Pascu, M.L. Time Stability of Laser Exposed Phenothiazines Aqueous Solutions in View of Antimicrobial Research. Proc. Rom. Acad. Ser. A 2018, 19, 537–544. [Google Scholar]
- Andrei, I.R.; Tozar, T.; Dinache, A.; Boni, M.; Nastasa, V.; Pascu, M.L. Chlorpromazine transformation by exposure to ultraviolet laser beams in droplet and bulk. Eur. J. Pharm. Sci. 2016, 81, 27–35. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48 (Suppl. 1), 5–16. [Google Scholar] [CrossRef] [PubMed]
- Fisher, L.E.; Hook, A.L.; Ashraf, W.; Yousef, A.; Barrett, D.A.; Scurr, D.J.; Chen, X.; Smith, E.F.; Fay, M.; Parmenter, C.D.J.; et al. Biomaterial modification of urinary catheters with antimicrobials to give long-term broadspectrum antibiofilm activity. J. Control. Release 2015, 202, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.; Alexandru, T.; Boni, M.; Damian, V.; Stoicu, A.; Dutschk, V.; Pascu, M.L. Interaction of solutions containing phenothiazines exposed to laser radiation with materials surfaces, in view of biomedical applications. Int. J. Pharm. 2014, 475, 270–281. [Google Scholar] [CrossRef] [PubMed]
- Morán, M.C.; Tozar, T.; Simon, A.; Dinache, A.; Smarandache, A.; Andrei, I.R.; Boni, M.; Pascu, M.L.; Cirisano, F.; Ferrari, M. Toxicity study in blood and tumor cells of laser produced medicines for application in fabrics. Colloids Surf. B Biointerfaces 2016, 137, 91–103. [Google Scholar] [CrossRef]
- Ananthakrishnan, N.; Rao, R.S.; Shivam, S. Bacterial adherence to cotton and silk sutures. Natl. Med. J. India. 1992, 5, 217–218. [Google Scholar]
- Fage, C.D.; Lathouwers, T.; Vanmeert, M.; Gao, L.; Vrancken, K.; Lammens, E.; Weir, A.N.M.; Degroote, R.; Cuppens, H.; Kosol, S.; et al. The Kalimantacin Polyketide Antibiotics Inhibit Fatty Acid Biosynthesis in Staphylococcus aureus by Targeting the Enoyl-Acyl Carrier Protein Binding Site of FabI. Angew. Chem. Int. Ed. 2020, 59, 10549–10556. [Google Scholar] [CrossRef]
- Fujita, J.; Maeda, Y.; Nagao, C.; Tsuchiya, Y.; Miyazaki, Y.; Hirose, M.; Mizohata, E.; Matsumoto, Y.; Inoue, T.; Mizuguchi, K.; et al. Crystal structure of FtsA from Staphylococcus aureus. FEBS Lett. 2014, 588, 1879–1885. [Google Scholar] [CrossRef]
- Mahasenan, K.V.; Molina, R.; Bouley, R.; Batuecas, M.T.; Fisher, J.F.; Hermoso, J.A.; Chang, M.; Mobashery, S. Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis. J. Am. Chem. Soc. 2017, 139, 2102–2110. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Kawai, F.; Obayashi, E.; Akashi, S.; Roper, D.I.; Tame, J.R.H.; Park, S.-Y. Crystal Structures of Penicillin-Binding Protein 3 (PBP3) from Methicillin-Resistant Staphylococcus aureus in the Apo and Cefotaxime-Bound Forms. J. Mol. Biol. 2012, 423, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.A.N.; Chatterjee, S.S.; Hamilton, S.M.; Eltis, L.D.; Chambers, H.F.; Strynadka, N.C.J. Structural and kinetic analyses of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance in Staphylococcus aureus. J. Biol. Chem. 2018, 293, 19854–19865. [Google Scholar] [CrossRef] [PubMed]
- Sainsbury, S.; Bird, L.; Rao, V.; Shepherd, S.M.; Stuart, D.I.; Hunter, W.N.; Owens, R.J.; Ren, J. Crystal Structures of Penicillin-Binding Protein 3 from Pseudomonas aeruginosa: Comparison of Native and Antibiotic-Bound Forms. J. Mol. Biol. 2011, 405, 173–184. [Google Scholar] [CrossRef]
- Smith, J.D.; Kumarasiri, M.; Zhang, W.; Hesek, D.; Lee, M.; Toth, M.; Vakulenko, S.; Fisher, J.F.; Mobashery, S.; Chen, Y. Structural Analysis of the Role of Pseudomonas aeruginosa Penicillin-Binding Protein 5 in β-Lactam Resistance. Antimicrob. Agents Chemother. 2013, 57, 3137–3146. [Google Scholar] [CrossRef]
- Sauvage, E.; Derouaux, A.; Fraipont, C.; Joris, M.; Herman, R.; Rocaboy, M.; Schloesser, M.; Dumas, J.; Kerff, F.; Nguyen-Distèche, M.; et al. Crystal Structure of Penicillin-Binding Protein 3 (PBP3) from Escherichia coli. PLoS ONE 2014, 9, e98042. [Google Scholar] [CrossRef]
- Harris, J.R.; Marles-Wright, J. (Eds.) Macromolecular Protein Complexes II: Structure and Function; Subcellular Biochemistry; Springer International Publishing: Cham, Switzerland, 2019; Volume 93, ISBN 978-3-030-28150-2. [Google Scholar]
- Haenni, M.; Majcherczyk, P.A.; Barblan, J.-L.; Moreillon, P. Mutational Analysis of Class A and Class B Penicillin-Binding Proteins in Streptococcus gordonii. Antimicrob. Agents Chemother. 2006, 50, 4062–4069. [Google Scholar] [CrossRef][Green Version]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Y.-M.; Davies, C. Penicillin-Binding Protein 3 Is Essential for Growth of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 61. [Google Scholar] [CrossRef]
- Ropy, A.; Cabot, G.; Sánchez-Diener, I.; Aguilera, C.; Moya, B.; Ayala, J.A.; Oliver, A. Role of Pseudomonas aeruginosa Low-Molecular-Mass Penicillin-Binding Proteins in AmpC Expression, β-Lactam Resistance, and Peptidoglycan Structure. Antimicrob. Agents Chemother. 2015, 59, 3925–3934. [Google Scholar] [CrossRef]
- BIOVIA. Dassault Systèmes, [Discovery Studio Visualizer], [V20.1.0.19295]; Dassault Systèmes: San Diego, CA, USA, 2019. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Udrea, A.-M.; Avram, S.; Nistorescu, S.; Pascu, M.-L.; Romanitan, M.O. Laser irradiated phenothiazines: New potential treatment for COVID-19 explored by molecular docking. J. Photochem. Photobiol. B Biol. 2020, 211, 111997. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M. An Introduction to Genetic Algorithms; Complex Adaptive Systems; MIT Press: Cambridge, MA, USA, 1996; ISBN 978-0-262-13316-6. [Google Scholar]
- Kabir-ud-Din; Al-Ahmadi, M.D.A.; Naqvi, A.Z.; Akram, M. Micellar Properties of a Phenothiazine Drug in Presence of Additives. Colloid J. 2009, 71, 498–502. [Google Scholar] [CrossRef]
- Tozar, T.; Nastasa, V.; Stoicu, A.; Chifiriuc, M.C.; Popa, M.; Kamerzan, C.; Pascu, M.L. In vitro antimicrobial efficacy of laser exposed chlorpromazine against Gram-positive bacteria in planktonic and biofilm growth state. Microb. Pathog. 2019, 129, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Grimsey, E.M.; Piddock, L.J.V. Do phenothiazines possess antimicrobial and efflux inhibitory properties? FEMS Microbiol. Rev. 2019, fuz017. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, M.V.; Bosne-David, S.; Amaral, L. Comparative in vitro activity of phenothiazines against multidrug-resistant Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 2000, 16, 69–71. [Google Scholar] [CrossRef]
- Nehme, H.; Saulnier, P.; Ramadan, A.A.; Cassisa, V.; Guillet, C.; Eveillard, M.; Umerska, A. Antibacterial activity of antipsychotic agents, their association with lipid nanocapsules and its impact on the properties of the nanocarriers and on antibacterial activity. PLoS ONE 2018, 13, e0189950. [Google Scholar] [CrossRef]
- Sidrim, J.J.; Amando, B.R.; Gomes, F.I.; do Amaral, M.S.; de Sousa, P.C.; Ocadaque, C.J.; Brilhante, R.S.; A Cordeiro, R.D.; Rocha, M.F.; SCM Castelo-Branco, D.D. Chlorpromazine-impregnated catheters as a potential strategy to control biofilm-associated urinary tract infections. Future Microbiol. 2019, 14, 1023–1034. [Google Scholar] [CrossRef]

| Protein | PDB Code | AA Residues from Situs | Grid Points in Dimension | Coordinates of Central Grid Point of Map |
|---|---|---|---|---|
| S. aureus | ||||
| PBP3 MRSA | 3VSL [37] | Active site: Ser392, Asn450, Ser448, Gln524, Thr603, Lys618, Gly620, Thr621, Glu623, Pro660 [37] | x 96 | x 19.57 |
| y 96 | y −48.21 | |||
| z 92 | z 25.08 | |||
| FtsA | 3WQU [35] | Active site: Glu251, Lys254, His255 [35] | x 80 | x 4.46 |
| y 78 | y 34.68 | |||
| z 80 | z −15.30 | |||
| PBP2a MRSA | 5M18 [36] | Alosteric site: Ile314, Lys 316, Lys318, Asp320 | x 86 | x 12.81 |
| y 80 | y −24.85 | |||
| z 92 | z −63.08 | |||
| Active site: Lys28, Lys86, Lys90, Lys148, Lys180, Lys181, Lys219, Met220, Tyr 223, Glu222, Glu268, Glu378, Lys382, Lys436, Tyr446, Gln502, Ser504, Asn505, Lys506, Asn507, Glu566, Glu602, Leu603, Lys604, Met605 Lys606, Gln607, Gly608, Glu609, Thr610, Gly611, Arg612 [36] | x 84 | x 24.59 | ||
| y 84 | y −27.21 | |||
| z 80 | z −5.68 | |||
| PBP4 | 5TW8 [38] | Active site: Ser75, Lys78, Ser139, Asn141, Lys259, Thr260 [38] | x 66 | x 33.58 |
| y 66 | y −66.46 | |||
| z 66 | z 37.31 | |||
| FabL | 6TBB [34] | Active site: Ala96, Phe96, Ala97, Met99, Leu102, Tyr147, Gln155 Asn156, Tyr157, Met160, Pro192, Leu196, Ser197, Ala198, Phe204, Ile207 [34] | x 70 | x −14.45 |
| y 66 | y −17.81 | |||
| z 70 | z −76.36 | |||
| E. coli | ||||
| PBP3 | 4BJP [41] | Active site: Ser307, Lys310, Ser359, Asn361, Phe417, Gly418, Tyr419, Lys494, Thr495, Thr497, Lys499 Tyr514 [41] | x 68 | x −6.55 |
| y 68 | y 22.76 | |||
| z 68 | z 13.37 | |||
| P. aeruginosa | ||||
| PBP3 | 3OC2 [39] | Active site: Glu291; Ser294, Lys297, Ser349, Asn351, Tyr407, Tyr409, Lys484, Ser485, Thr487, Arg489, Tyr501, Tyr530, Phe531 [39] | x 66 | x −0.93 |
| y 66 | y 1.95 | |||
| z 66 | z −17.89 | |||
| PBP5 | 4K91 [40] | Active site: Ser41, Lys44, Ser101, Ser203 [40] | x 56 | x 41.05 |
| y 56 | y −5.76 | |||
| z 56 | z 12.71 | |||
| Lowest EFEB kcal/mol | CPZ | CPZ-SO | PZ | PZ-SO | HOPO | 2HOPZSO | P1 | P2 |
|---|---|---|---|---|---|---|---|---|
| S. aureus | ||||||||
| PBP3 MRSA | −7.6 | −6.0 | −6.5 | −5.6 | −7.3 | −6.7 | −6.9 | −7.2 |
| FtsA | −8.7 | −8.6 | −7.1 | −8.3 | −7.5 | −6.9 | −7.7 | −7.2 |
| PBP2a MRSA (alosteric situs) | −7.6 | −6.7 | −6.3 | −6.3 | −6.8 | −6.4 | −7.2 | −7.0 |
| PBP2a MRSA (active situs) | −6.5 | −6.0 | −6.0 | −5.7 | −6.7 | −6.8 | −7.1 | −5.9 |
| PBP4 | −6.70 | −6.19 | −5.67 | −5.45 | −6.23 | −6.10 | −6.25 | −6.50 |
| FabL | −8.30 | −6.87 | −7.16 | −6.34 | −7.74 | −7.24 | −7.74 | −7.38 |
| P. aeruginosa | ||||||||
| PBP3 | −6.48 | −4.55 | −5.60 | −3.70 | −6.04 | −5.77 | −6.59 | −6.63 |
| PBP5 | −6.79 | −5.58 | −5.45 | - | −5.89 | −5.61 | −5.91 | −5.80 |
| E. coli | ||||||||
| PBP3 | −6.77 | −5.37 | −5.88 | −4.88 | −5.96 | −5.81 | −6.06 | −5.96 |
| Items | S. aureus ATCC 6538 | MRSA1 | MRSA2 | E. faecalis ATCC 29212 | P. aeruginosa ATCC 27853 | P. aeruginosa clinic1 | P. aeruginosa clinic2 | E. coli ATCC 8739 | C. parapsilosis ATCC 22019 |
|---|---|---|---|---|---|---|---|---|---|
| NIR CPZ | 100 | 100 | 100 | 50 | >100 | >100 | >100 | 50 | 25 |
| IR CPZ | 12.5 | 12.5 | 6.25 | 6.26 | 100 | 100 | 100 | 25 | 6.25 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nistorescu, S.; Gradisteanu Pircalabioru, G.; Udrea, A.-M.; Simon, Á.; Pascu, M.L.; Chifiriuc, M.-C. Laser-Irradiated Chlorpromazine as a Potent Anti-Biofilm Agent for Coating of Biomedical Devices. Coatings 2020, 10, 1230. https://doi.org/10.3390/coatings10121230
Nistorescu S, Gradisteanu Pircalabioru G, Udrea A-M, Simon Á, Pascu ML, Chifiriuc M-C. Laser-Irradiated Chlorpromazine as a Potent Anti-Biofilm Agent for Coating of Biomedical Devices. Coatings. 2020; 10(12):1230. https://doi.org/10.3390/coatings10121230
Chicago/Turabian StyleNistorescu, Simona, Gratiela Gradisteanu Pircalabioru, Ana-Maria Udrea, Ágota Simon, Mihail Lucian Pascu, and Mariana-Carmen Chifiriuc. 2020. "Laser-Irradiated Chlorpromazine as a Potent Anti-Biofilm Agent for Coating of Biomedical Devices" Coatings 10, no. 12: 1230. https://doi.org/10.3390/coatings10121230
APA StyleNistorescu, S., Gradisteanu Pircalabioru, G., Udrea, A.-M., Simon, Á., Pascu, M. L., & Chifiriuc, M.-C. (2020). Laser-Irradiated Chlorpromazine as a Potent Anti-Biofilm Agent for Coating of Biomedical Devices. Coatings, 10(12), 1230. https://doi.org/10.3390/coatings10121230










