Characterization of the Bacteriophage-Derived Endolysins PlySs2 and PlySs9 with In Vitro Lytic Activity against Bovine Mastitis Streptococcus uberis
Abstract
:1. Introduction
2. Results
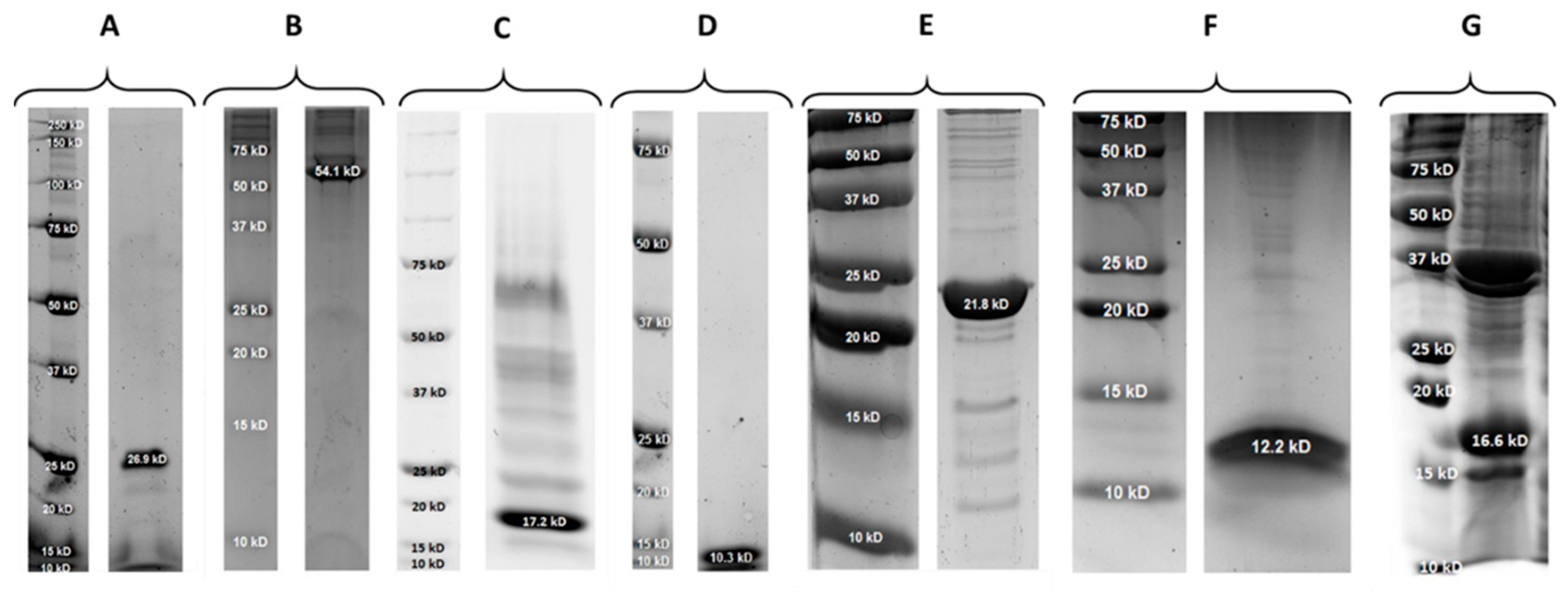
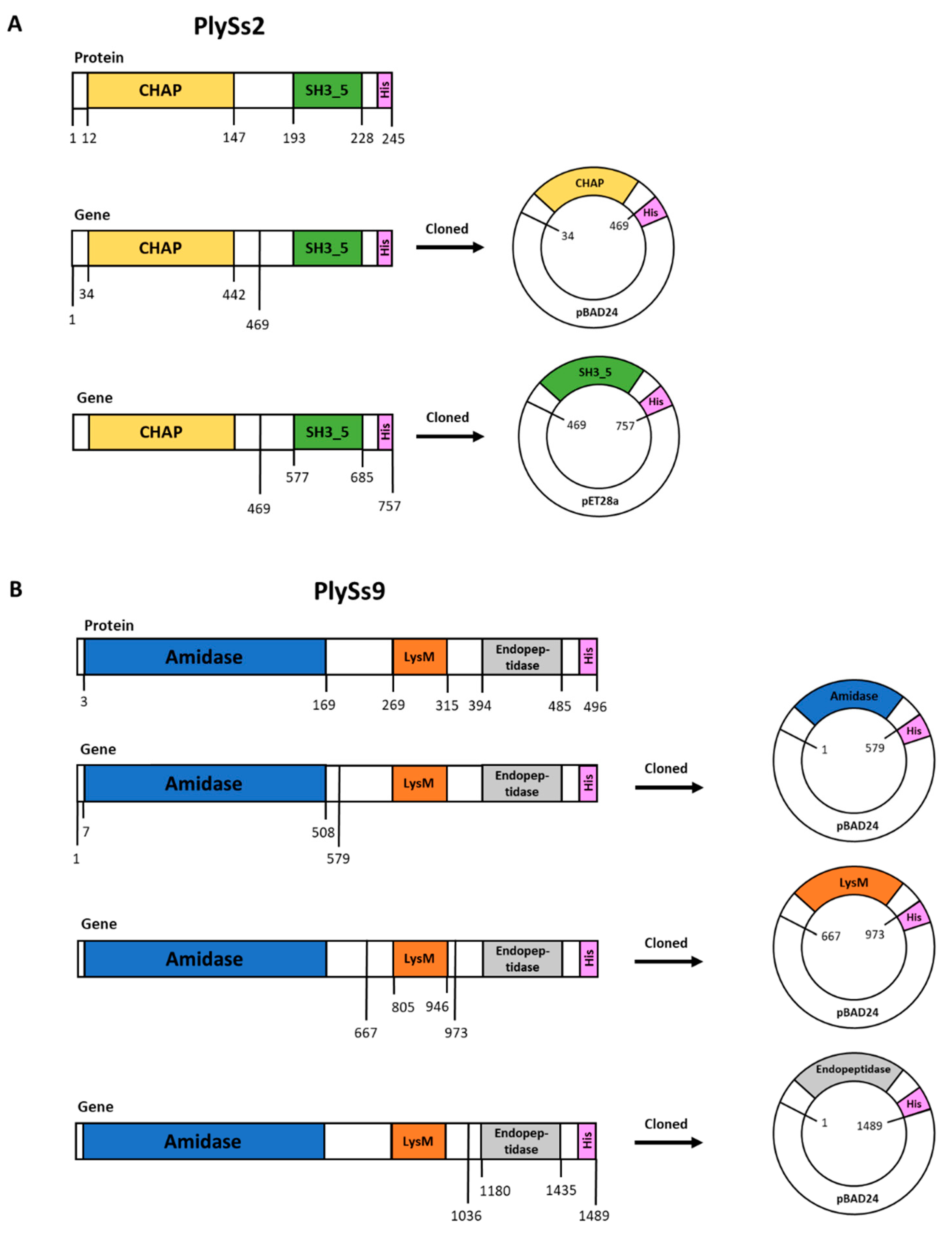
2.1. Properties of PlySs2, PlySs9 and Their Individual Subdomains
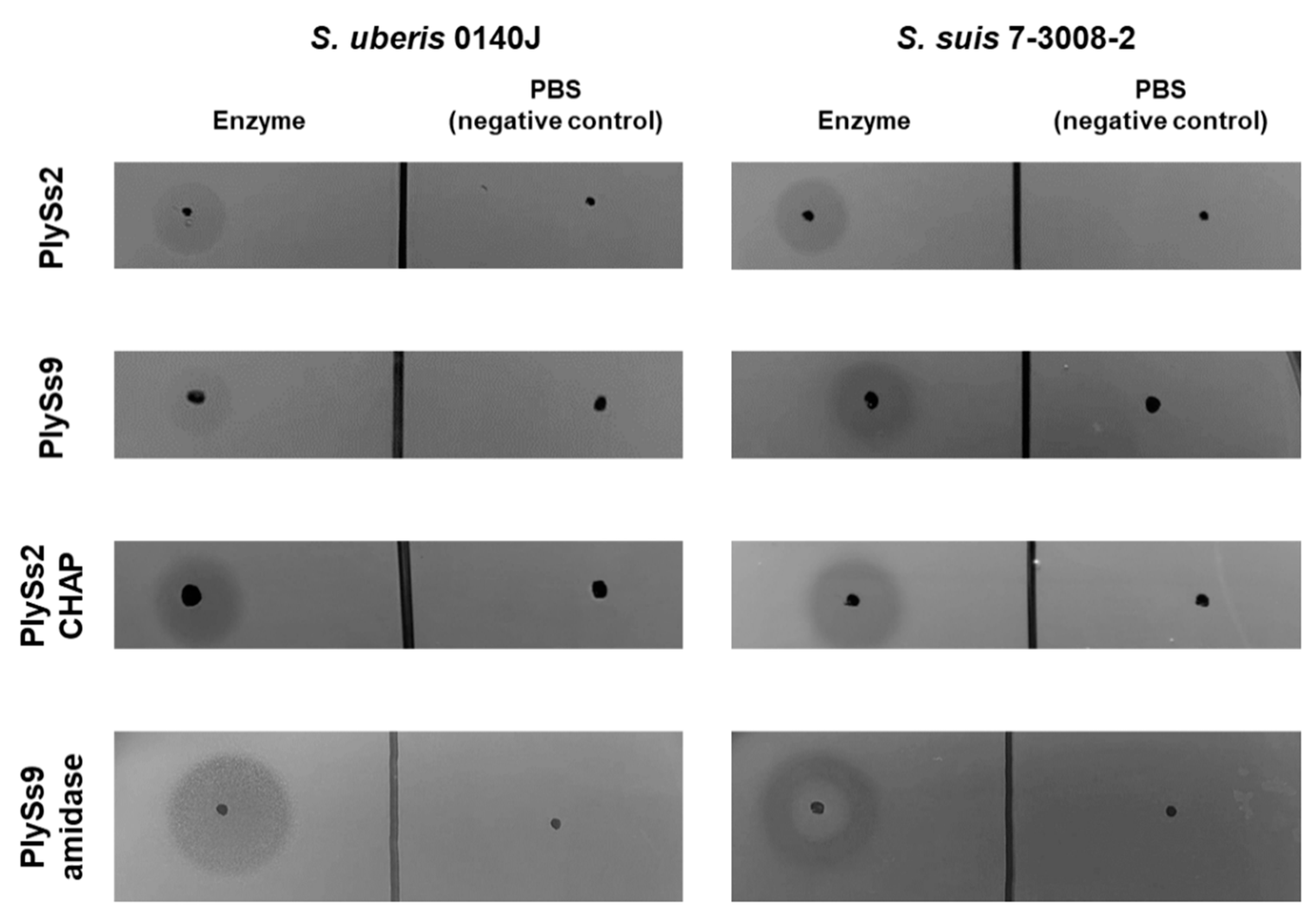
2.2. Qualitative Assessment of the Lytic Activity of PlySs2, PlySs9 and Their Catalytic Subdomains
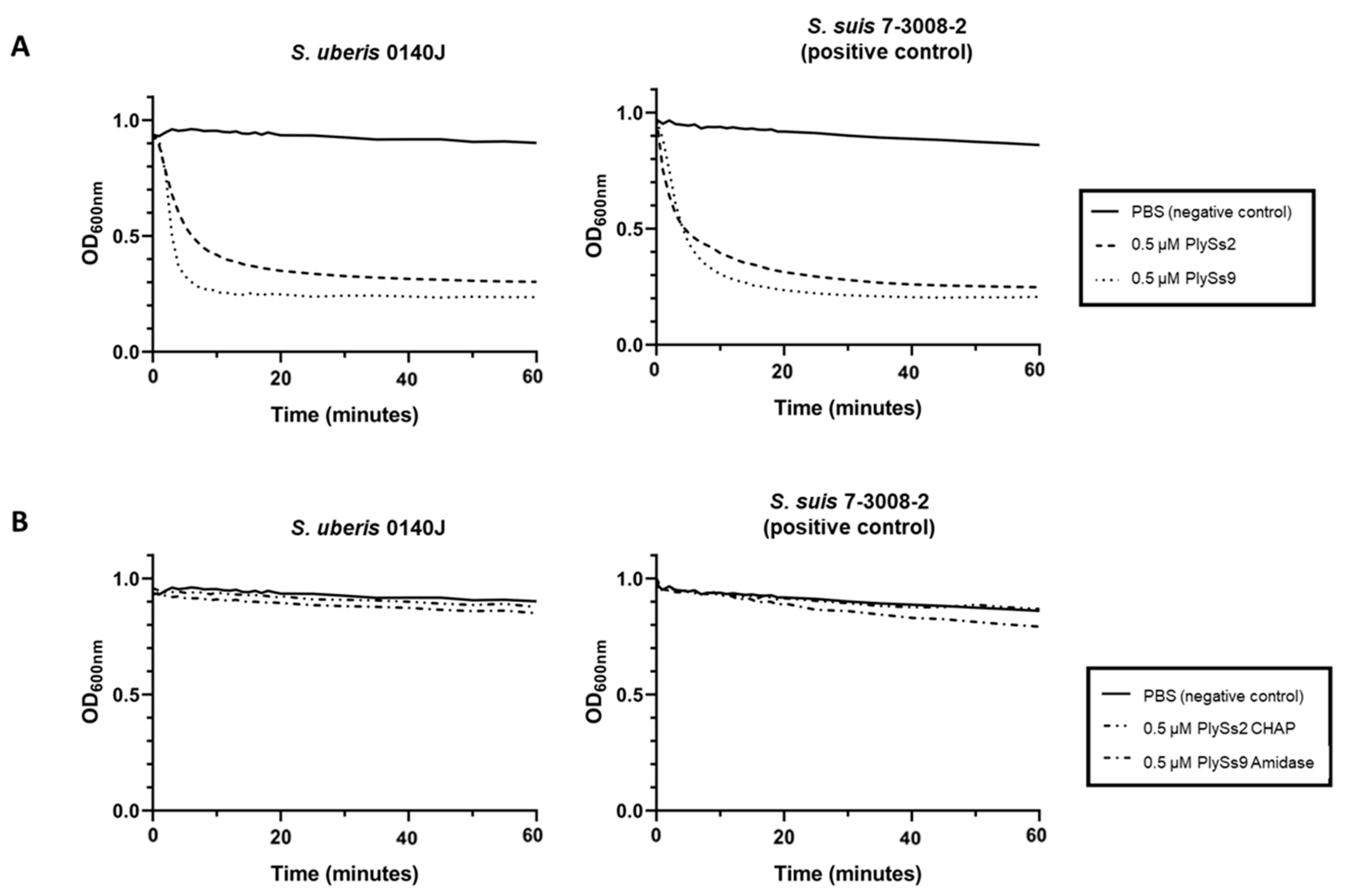
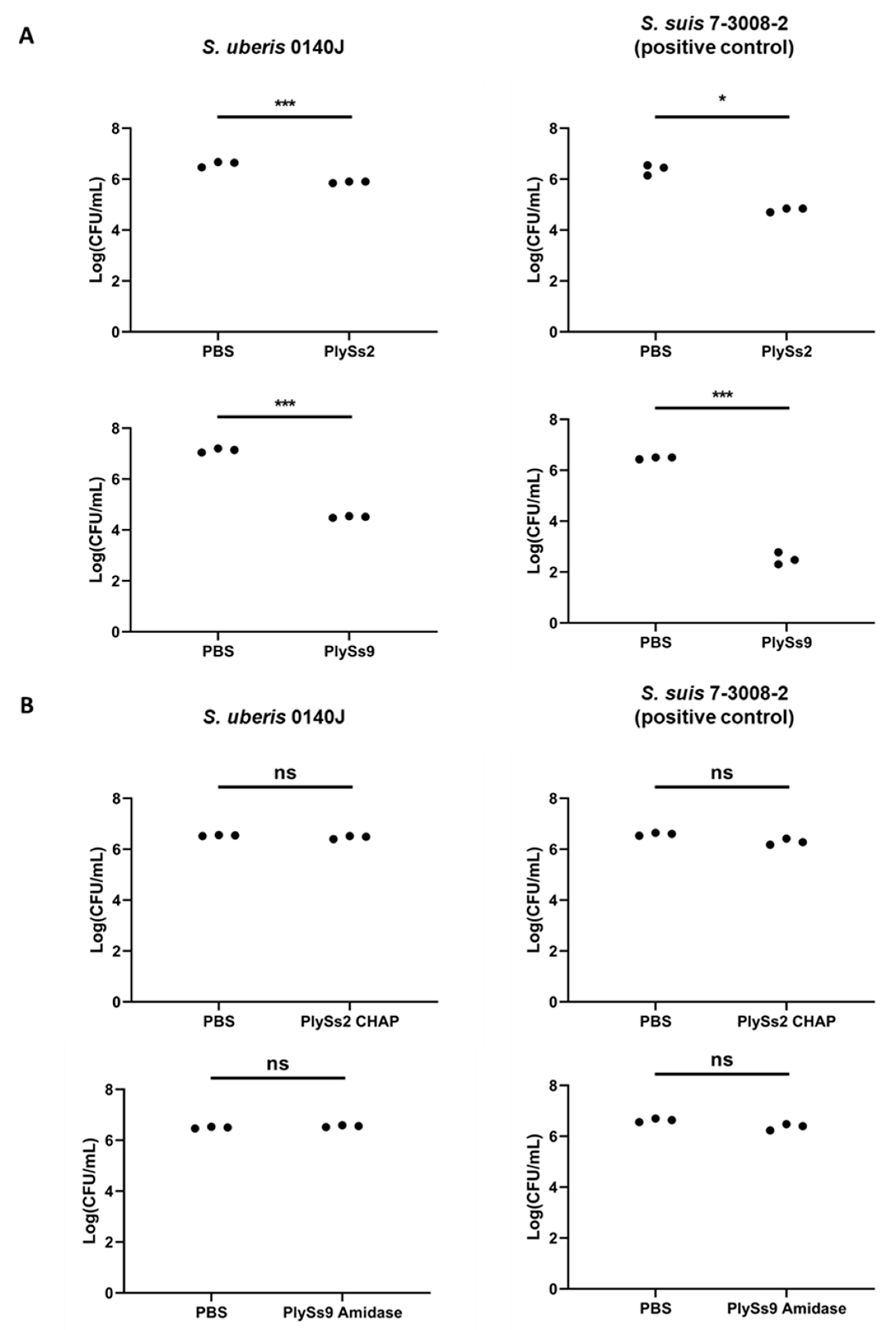
2.3. Quantitative Assessment of the Lytic Capacity of PlySs2, PlySs9 and Their Catalytic Subdomains
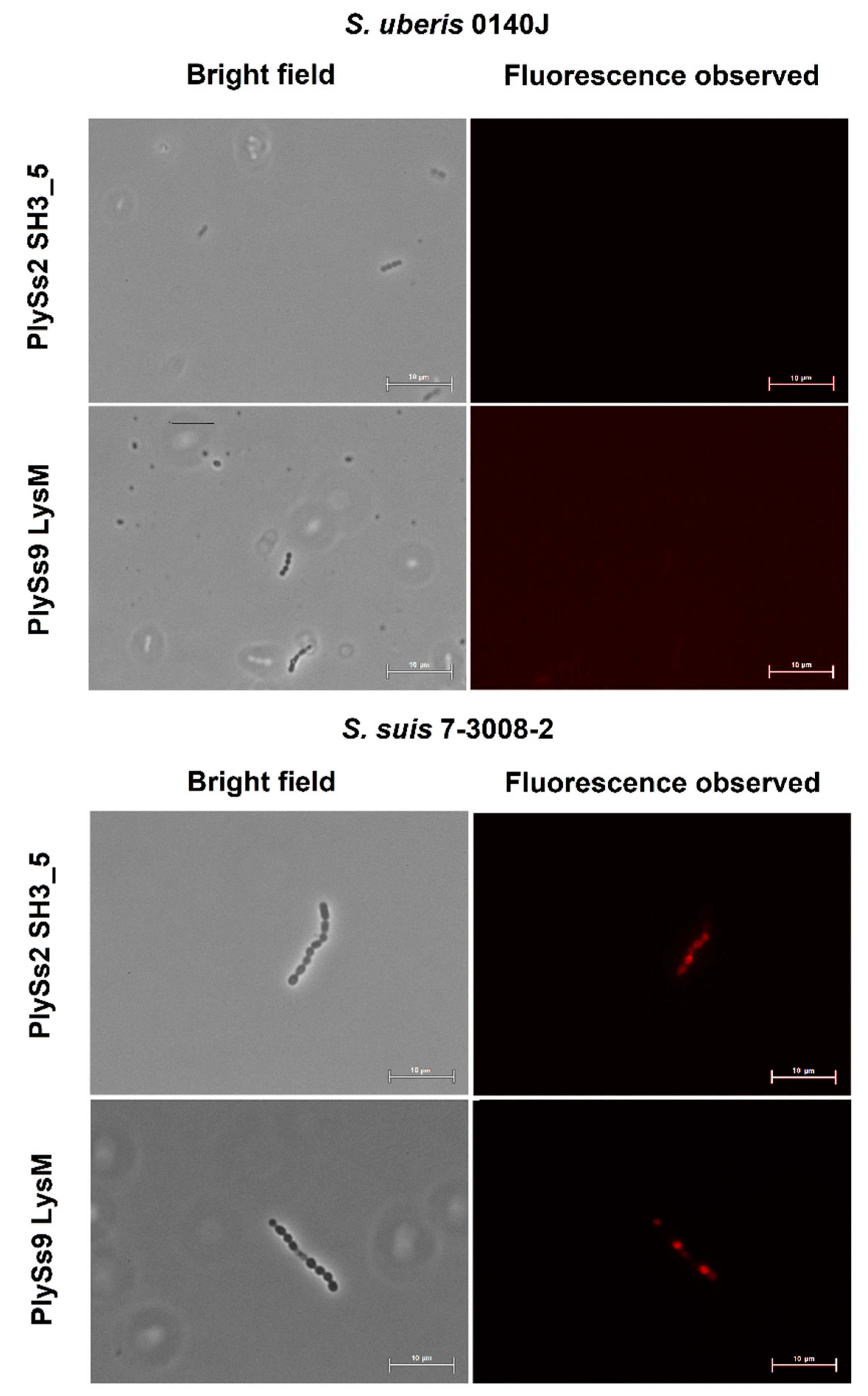
2.4. Evaluation of the Pathogen-Binding Capacity of the PlySs2 and PlySs9 Cell-Wall Binding Domains
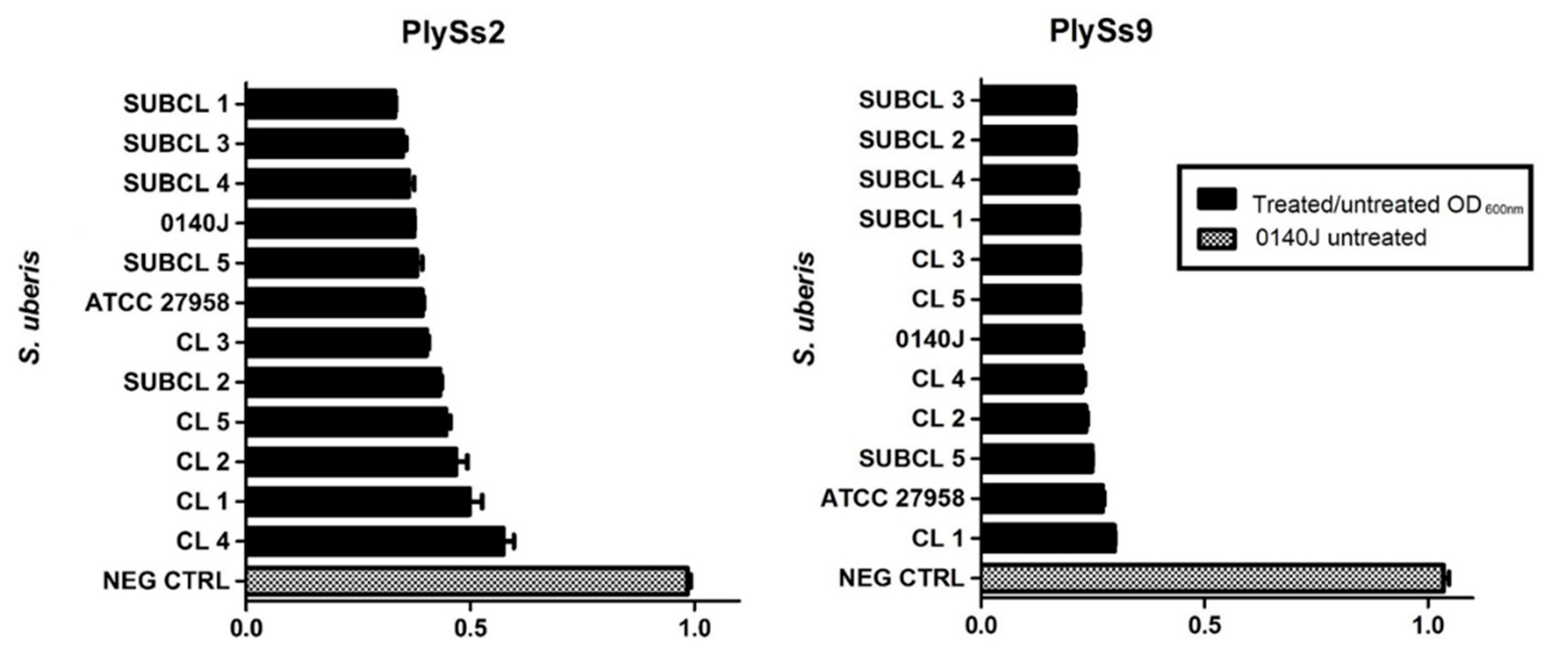
2.5. Evaluation of the Lytic Activity of PlySs2 and PlySs9 against A Panel of Isolated (Sub)Clinical S. uberis Strains
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Structural and Functional Bioinformatics of PlySs2 and PlySs9
4.3. Plasmid Construction, DNA Manipulation and Cloning
4.4. Expression Conditions and Protein Purification
4.5. Spot-on-Plate Assays, Turbidity Reduction Assays, Time Killing Assays and Minimal Inhibitory Concentrations
4.6. Alexa Fluor 555 Labeling and Fluorescence Microscopy
4.7. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Halasa, T.; Huijps, K.; Østerås, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Huijps, K.; Lam, T.J.; Hogeveen, H. Costs of mastitis: Facts and perception. J. Dairy Res. 2008, 75, 113–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Käppeli, N.; Morach, M.; Zurfluh, K.; Corti, S.; Nüesch-Inderbinen, M.; Stephan, R. Sequence types and antimicrobial resistance profiles of Streptococcus uberis isolated from bovine mastitis. Front. Vet. Sci. 2019, 6, 234. [Google Scholar] [CrossRef] [Green Version]
- Davies, P.L.; Leigh, J.A.; Bradley, A.J.; Archer, S.C.; Emes, R.D.; Green, M.J. Molecular epidemiology of Streptococcus uberis clinical mastitis in dairy herds: Strain heterogeneity and transmission. J. Clin. Microbiol. 2015, 54, 68–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taponen, S.; Liski, E.; Heikkilä, A.-M.; Pyörälä, S. Factors associated with intramammary infection in dairy cows caused by coagulase-negative staphylococci, Staphylococcus aureus, Streptococcus uberis, Streptococcus dysgalactiae, Corynebacterium bovis, or Escherichia coli. J. Dairy Sci. 2017, 100, 493–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zigo, F.; Elečko, J.; Farkašová, Z.; Zigová, M.; Vasiľ, M.; Ondrašovičová, S.; Lenka, K. Preventive methods in reduction of mastitis pathogens in dairy cows. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 121–126. [Google Scholar] [CrossRef]
- Al-Qumber, M.; Tagg, J. Commensal bacilli inhibitory to mastitis pathogens isolated from the udder microbiota of healthy cows. J. Appl. Microbiol. 2006, 101, 1152–1160. [Google Scholar] [CrossRef]
- Prado, M.; Almeida, R.; Özen, C.; Luther, D.; Lewis, M.; Headrick, S.; Oliver, S. Vaccination of dairy cows with recombinant Streptococcus uberis adhesion molecule induces antibodies that reduce adherence to and internalization of S. uberis into bovine mammary epithelial cells. Vet. Immunol. Immunopathol. 2011, 141, 201–208. [Google Scholar] [CrossRef]
- Tiwari, H.; Babra, C.; Williams, V.; De Wet, S.; Gibson, J.; Paxman, A.; Morgan, E.; Sunagar, R.; Isloor, S.; Hegde, N.R.; et al. Trends intherapeutic and prevention strategies for management of bovine mastitis: An overview. J. Vaccines Vaccin. 2013, 4, 1000176. [Google Scholar] [CrossRef] [Green Version]
- Garcia, S.; Osburn, B.I.; Cullor, J.S. A one health perspective on dairy production and dairy food safety. One Health 2019, 7, 100086. [Google Scholar] [CrossRef]
- Bradley, A.; De Vliegher, S.; Green, M.J.; Larrosa, P.; Payne, B.; Van De Leemput, E.S.; Samson, O.; Valckenier, D.; Van Werven, T.; Waldeck, H.; et al. An investigation of the dynamics of intramammary infections acquired during the dry period on European dairy farms. J. Dairy Sci. 2015, 98, 6029–6047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organisation. Critically Important Antimicrobials for Human Medicine. 2018. Available online: https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf (accessed on 3 August 2020).
- De Jong, A.; El Garch, F.; Simjee, S.; Moyaert, H.; Rose, M.; Youala, M.; Siegwart, E. Monitoring of antimicrobial susceptibility of udder pathogens recovered from cases of clinical mastitis in dairy cows across Europe: VetPath results. Vet. Microbiol. 2018, 213, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Elst, N.V.; Meyer, E. Potential therapeutic application of bacteriophages and phage-derived endolysins as alternative treatment of bovine mastitis. Vlaams Diergeneeskd. Tijdschr. 2018, 87, 181–187. [Google Scholar] [CrossRef]
- Dams, D.; Briers, Y. Enzybiotics: Enzyme-based antibacterials as therapeutics. Adv. Exp. Med. Biol. 2019, 1148, 233–253. [Google Scholar] [CrossRef] [PubMed]
- Radzikowski, D.; Kalińska, A.; Ostaszewska, U.; Gołębiewski, M. Alternative solutions to antibiotics in mastitis. Anim. Sci. Pap. Rep. 2020, 38, 117–133. [Google Scholar]
- Zduńczyk, S.; Janowski, T. Bacteriophages and associated endolysins in therapy and prevention of mastitis and metritis in cows: Current knowledge. Anim. Reprod. Sci. 2020, 218, 106504. [Google Scholar] [CrossRef]
- Angelopoulou, A.; Warda, A.K.; Hill, C.; Ross, R.P. Non-antibiotic microbial solutions for bovine mastitis—Live biotherapeutics, bacteriophage, and phage lysins. Crit. Rev. Microbiol. 2019, 45, 564–580. [Google Scholar] [CrossRef]
- Nilsson, A.S. Pharmacological limitations of phage therapy. Upsala J. Med Sci. 2019, 124, 218–227. [Google Scholar] [CrossRef]
- Gill, J.J.; Pacan, J.C.; Carson, M.E.; Leslie, K.E.; Griffiths, M.W.; Sabour, P.M. Efficacy and pharmacokinetics of bacteriophage therapy in treatment of subclinical Staphylococcus aureus mastitis in lactating dairy cattle. Antimicrob. Agents Chemother. 2006, 50, 2912–2918. [Google Scholar] [CrossRef] [Green Version]
- Gill, J.J.; Sabour, P.; Leslie, K.; Griffiths, M. Bovine whey proteins inhibit the interaction of Staphylococcus aureus and bacteriophage K. J. Appl. Microbiol. 2006, 101, 377–386. [Google Scholar] [CrossRef]
- Nelson, D.C.; Schmelcher, M.; Rodríguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Appl. Microbiol. 2012, 83, 299–365. [Google Scholar] [CrossRef] [Green Version]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashani, H.H.; Schmelcher, M.; Sabzalipoor, H.; Hosseini, E.S.; Moniri, R. Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: Current status of research and novel delivery strategies. Clin. Microbiol. Rev. 2017, 31, 00071-17. [Google Scholar] [CrossRef] [Green Version]
- Totté, J.; Van Doorn, M.B.; Pasmans, S. Successful treatment of chronic Staphylococcus aureus-related dermatoses with the topical Endolysin Staphefekt SA.100: A report of 3 cases. Case Rep. Dermatol. 2017, 9, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Oh, J.T.; Sauve, K.; Bradford, P.A.; Cassino, C.; Schuch, R. Antimicrobial activity of Exebacase (Lysin CF-301) against the most common causes of infective endocarditis. Antimicrob. Agents Chemother. 2019, 63, 01078-19. [Google Scholar] [CrossRef]
- Donovan, D.M.; Lardeo, M.; Foster-Frey, J. Lysis of staphylococcal mastitis pathogens by bacteriophage phi11 endolysin. FEMS Microbiol. Lett. 2006, 265, 133–139. [Google Scholar] [CrossRef] [Green Version]
- Schmelcher, M.; Powell, A.M.; Camp, M.J.; Pohl, C.S.; Donovan, D.M. Synergistic streptococcal phage λSA2 and B30 endolysins kill streptococci in cow milk and in a mouse model of mastitis. Appl. Microbiol. Biotechnol. 2015, 99, 8475–8486. [Google Scholar] [CrossRef]
- Fan, J.; Zeng, Z.; Mai, K.; Yang, Y.; Feng, J.; Bai, Y.; Sun, B.; Xie, Q.; Tong, Y.; Ma, J. Preliminary treatment of bovine mastitis caused by Staphylococcus aureus, with Trx-SA1, recombinant endolysin of S. aureus bacteriophage IME-SA1. Vet. Microbiol. 2016, 191, 65–71. [Google Scholar] [CrossRef]
- Obeso, J.M.; Martínez, B.; Rodríguez, A.; García, P. Lytic activity of the recombinant staphylococcal bacteriophage ΦH5 endolysin active against Staphylococcus aureus in milk. Int. J. Food Microbiol. 2008, 128, 212–218. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, D.; Garrido, V.; Fernández, L.; Portilla, S.; Rodríguez, A.; Grilló, M.J.; García, P. Phage lytic protein LysRODI prevents staphylococcal mastitis in mice. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Gilmer, D.B.; Schmitz, J.E.; Euler, C.W.; Fischetti, V.A. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2013, 57, 2743–2750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Yang, H.; Yu, J.; Wei, H. Molecular dissection of phage lysin PlySs2: Integrity of the catalytic and cell wall binding domains is essential for its broad lytic activity. Virol. Sin. 2015, 30, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Nilebäck, L.; Widhe, M.; Seijsing, J.; Bysell, H.; Sharma, P.K.; Hedhammar, M. Bioactive silk coatings reduce the adhesion of staphylococcus aureus while supporting growth of osteoblast-like cells. ACS Appl. Mater. Interfaces 2019, 11, 24999–25007. [Google Scholar] [CrossRef] [PubMed]
- Gilmer, D.B.; Schmitz, J.E.; Thandar, M.; Euler, C.W.; Fischetti, V.A. The phage lysin PlySs2 decolonizes Streptococcus suis from murine intranasal mucosa. PLoS ONE 2017, 12, e0169180. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F.; Daraspe, J.; Giddey, M.; Moreillon, P.; Resch, G. In Vitro characterization of PlySK1249, a novel phage lysin, and assessment of its antibacterial activity in a mouse model of Streptococcus agalactiae bacteremia. Antimicrob. Agents Chemother. 2013, 57, 6276–6283. [Google Scholar] [CrossRef] [Green Version]
- Briers, Y.; Lavigne, R.; Volckaert, G.; Hertveldt, K. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J. Biochem. Biophys. Methods 2007, 70, 531–533. [Google Scholar] [CrossRef]
- Alnakip, M.E.; Rhouma, N.R.; Abd-Elfatah, E.N.; Quintela-Baluja, M.; Böhme, K.; Fernández-No, I.; Bayoumi, M.A.; Abdelhafez, M.M.; Taboada-Rodríguez, A.; Calo-Mata, P.; et al. Discrimination of major and minor streptococci incriminated in bovine mastitis by MALDI-TOF MS fingerprinting and 16S rRNA gene sequencing. Res. Vet. Sci. 2020, 132, 426–438. [Google Scholar] [CrossRef]
- Schmelcher, M.; Shen, Y.; Nelson, D.C.; Eugster, M.R.; Eichenseher, F.; Hanke, D.C.; Loessner, M.J.; Dong, S.; Pritchard, D.G.; Lee, J.C.; et al. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J. Antimicrob. Chemother. 2015, 70, 1453–1465. [Google Scholar] [CrossRef] [Green Version]
- Horgan, M.; O’Flynn, G.; Garry, J.; Cooney, J.; Coffey, A.; Fitzgerald, G.F.; Ross, R.P.; McAuliffe, O. Phage lysin LysK can be truncated to its CHAP domain and retain lytic activity against live antibiotic-resistant staphylococci. Appl. Environ. Microbiol. 2008, 75, 872–874. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Q.; Fischetti, V.A. Mutagenesis of a bacteriophage lytic enzyme PlyGBS significantly increases its antibacterial activity against group B streptococci. Appl. Microbiol. Biotechnol. 2007, 74, 1284–1291. [Google Scholar] [CrossRef]
- Gerstmans, H.; Grimon, D.; Gutiérrez, D.; Lood, C.; Rodríguez, A.; Van Noort, V.; Lammertyn, J.; Lavigne, R.; Briers, Y. A versaTile-driven platform for rapid hit-to-lead development of engineered lysins. Sci. Adv. 2020, 6, eaaz1136. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Bossers, A.; Harders, F.; Lu, C.; Smith, H. Comparative genomic analysis of twelve Streptococcus suis (pro) phages. Genomics 2013, 101, 336–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Protein | Molecular Size (kDa) | pI | Expression Yield (mg/L) |
|---|---|---|---|
| PlySs2 | 26.9 | 9.06 | 0.25 |
| PlySs2 CHAP | 17.2 | 7.10 | 2.60 |
| PlySs2 SH3_5 | 10.3 | 9.57 | 1.10 |
| PlySs9 | 54.1 | 9.08 | 0.12 |
| PlySs9 Amidase | 21.8 | 6.75 | 20.0 |
| PlySs9 LysM | 12.2 | 9.84 | 1.10 |
| PlySs9 Endopeptidase | 16.6 | 6.42 | Not determined |
| Subdomain Amplified by PCR | Forward Primer | Reverse Primer | Tm (°C) |
|---|---|---|---|
| PlySs2 CHAP | GAATTCATTATGGCA CAGGTTGGTAGCGGT | TCTAGATTAATGATGATGATGAT GATGCTGACGATAGCTTGCTGC | 72 |
| PlySs2 SH3_5 | GAATTCATTATGAGC CGTAGCTATCGTGAA | TCTAGATTAATGATGATGATGAT GATGTTTAAAGGTGCCCCATGC | 69 |
| PlySs9 Amidase | GAATTCATTATGGGC AAACATCTGGTGATT | TCTAGATTAATGATGATGAT GATGATGCTCGGGGTGCCGCTG | 68 |
| PlySs9 LysM | GAATTCATTATGAAA GGTCGCACCTACAAA | TCTAGATTAATGATGATGATGAT GATGGGTCGGAATTGCGGTGGT | 68 |
| PlySs9 Endopeptidase | GAATTCATTATGAGC AATCCGTATGGTGGT | GATTTAATCTGTATCAGG | 51 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vander Elst, N.; Linden, S.B.; Lavigne, R.; Meyer, E.; Briers, Y.; Nelson, D.C. Characterization of the Bacteriophage-Derived Endolysins PlySs2 and PlySs9 with In Vitro Lytic Activity against Bovine Mastitis Streptococcus uberis. Antibiotics 2020, 9, 621. https://doi.org/10.3390/antibiotics9090621
Vander Elst N, Linden SB, Lavigne R, Meyer E, Briers Y, Nelson DC. Characterization of the Bacteriophage-Derived Endolysins PlySs2 and PlySs9 with In Vitro Lytic Activity against Bovine Mastitis Streptococcus uberis. Antibiotics. 2020; 9(9):621. https://doi.org/10.3390/antibiotics9090621
Chicago/Turabian StyleVander Elst, Niels, Sara B. Linden, Rob Lavigne, Evelyne Meyer, Yves Briers, and Daniel C. Nelson. 2020. "Characterization of the Bacteriophage-Derived Endolysins PlySs2 and PlySs9 with In Vitro Lytic Activity against Bovine Mastitis Streptococcus uberis" Antibiotics 9, no. 9: 621. https://doi.org/10.3390/antibiotics9090621
APA StyleVander Elst, N., Linden, S. B., Lavigne, R., Meyer, E., Briers, Y., & Nelson, D. C. (2020). Characterization of the Bacteriophage-Derived Endolysins PlySs2 and PlySs9 with In Vitro Lytic Activity against Bovine Mastitis Streptococcus uberis. Antibiotics, 9(9), 621. https://doi.org/10.3390/antibiotics9090621







