S. aureus Colonization, Biofilm Production, and Phage Susceptibility in Peritoneal Dialysis Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. S. aureus Isolation and Microbiological Investigation
2.2. Biofilm Growth Using Microtiter Plate Assay
2.3. Biofilm Calculation
2.4. Bacteriophage Preparation
2.5. Bacteriophage Adaptation
2.6. Bacteriophage Lytic Activity Detection
2.7. Data Analysis
2.8. Ethical Statement
3. Results
3.1. Biofilm Growth
3.2. Bacteriophage Susceptibility and Adaptation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Whitty, R.; Bargman, J.M.; Kiss, A.; Dresser, L.; Lui, P. Residual Kidney Function and Peritoneal Dialysis—Associated Peritonitis Treatment Outcomes. Clin. J. Am. Soc. Nephrol. 2017, 12, 2016–2022. [Google Scholar] [CrossRef] [Green Version]
- Kitterer, D.; Latus, J.; Pöhlmann, C.; Alscher, M.D.; Kimmel, M. Microbiological Surveillance of Peritoneal Dialysis Associated Peritonitis: Antimicrobial Susceptibility Profiles of a Referral Center in GERMANY over 32 Years. PLoS ONE 2015, 10, e0135969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Govindarajulu, S.; Hawley, C.M.; McDonald, S.P.; Brown, F.G.; Rosman, J.B.; Wiggins, K.J.; Bannister, K.M.; Johnson, D.W. Staphylococcus Aureus Peritonitis in Australian Peritoneal Dialysis Patients: Predictors, Treatment, and Outcomes in 503 Cases. Perit. Dial. Int. 2010, 30, 311–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Tu, W.; Xu, C. Mupirocin for Preventing Exit-site Infection and Peritonitis in Patients Undergoing Peritoneal Dialysis. Nephrol. Dial. Transplant. 2010, 25, 587–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simões-Silva, L.; Ferreira, S.; Santos-Araujo, C.; Tabaio, M.; Pestana, M.; Soares-Silva, I.; Sampaio-Maia, B. Oral Colonization of Staphylococcus Species in a Peritoneal Dialysis Population: A Possible Reservoir for PD-Related Infections? Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 5789094. [Google Scholar] [CrossRef] [Green Version]
- Li, P.K.-T.; Chow, K.M. Infectious Complications in Dialysis—Epidemiology and Outcomes. Nat. Rev. Nephrol. 2012, 8, 77–88. [Google Scholar] [CrossRef]
- Kane, T.L.; Carothers, K.E.; Lee, S.W. Virulence Factor Targeting of the Bacterial Pathogen Staphylococcus Aureus for Vaccine and Therapeutics. Curr. Drug Targets. 2018, 19, 111–127. [Google Scholar] [CrossRef]
- Ricciardi, B.F.; Muthukrishnan, G.; Masters, E.; Ninomiya, M.; Lee, C.C.; Schwarz, E.M. Staphylococcus Aureus Evasion of Host Immunity in the Setting of Prosthetic Joint Infection: Biofilm and Beyond. Curr. Rev. Musculoskelet Med. 2018, 11, 389–400. [Google Scholar] [CrossRef]
- Szeto, C.-C.; Li, P.K.-T.; Johnson, D.W.; Bernardini, J.; Dong, J.; Figueiredo, A.E.; Ito, Y.; Kazancioglu, R.; Moraes, T.; Esch, S.V.; et al. ISPD Catheter-Related Infection Recommendations: 2017 Update. Perit. Dial. Int. 2017, 37, 141–154. [Google Scholar] [CrossRef]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus Aureus Biofilms: Properties, Regulation, and Roles in Human Disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [Green Version]
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus Aureus Biofilms Prevent Macrophage Phagocytosis and Attenuate Inflammation In Vivo. J. Immunol. 2011, 186, 6585–6596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, L.K.; Jensen, H.E.; Koch, J.; Bjarnsholt, T.; Eickhardt, S.; Shirtliff, M. Specific Antibodies to Staphylococcus Aureus Biofilm Are Present in Serum from Pigs with Osteomyelitis. In Vivo 2015, 29, 555–560. [Google Scholar] [PubMed]
- Singh, R.; Ray, P.; Das, A.; Sharma, M. Penetration of Antibiotics Through Staphylococcus Aureus and Staphylococcus Epidermidis Biofilms. J. Antimicrob. Chemother. 2010, 65, 1955–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barretti, P.; Moraes, T.M.C.; Camargo, C.H.; Caramori, J.C.T.; Mondelli, A.L.; Montelli, A.C.; Maria de Lourdes, R.S. Peritoneal Dialysis-Related Peritonitis Due to Staphylococcus Aureus: A Single-Center Experience over 15 Years. PLoS ONE 2012, 7, e31780. [Google Scholar] [CrossRef]
- Muenks, C.E.; Hogan, P.G.; Wang, J.W.; Eisenstein, K.A.; Burnham, C.-A.D.; Fritz, S.A. Diversity of Staphylococcus Aureus Strains Colonizing Various Niches of the Human Body. J. Infect. 2016, 72, 698–705. [Google Scholar] [CrossRef] [Green Version]
- Cespedes, C.; Saïd-Salim, B.; Miller, M.; Lo, S.-H.; Kreiswirth, B.N.; Gordon, R.J.; Vavagiakis, P.; Klein, R.S.; Lowy, F.D. The Clonality of Staphylococcus Aureus Nasal Carriage. J. Infect. Dis. 2005, 191, 444–452. [Google Scholar] [CrossRef] [Green Version]
- Le, K.Y.; Otto, M. Quorum-Sensing Regulation in Staphylococci-An Overview. Front. Microbiol. 2015, 6, 1174. [Google Scholar] [CrossRef] [Green Version]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus Aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Xu, D.; Shi, L.; Cai, R.; Li, C.; Yan, H. Association Between Agr Type, Virulence Factors, Biofilm Formation and Antibiotic Resistance of Staphylococcus Aureus Isolates from Pork Production. Front. Microbiol. 2018, 9, 1876. [Google Scholar] [CrossRef]
- Piechota, M.; Kot, B.; Frankowska-Maciejewska, A.; Grużewska, A.; Woźniak-Kosek, A. Biofilm Formation by Methicillin-Resistant and Methicillin-Sensitive Staphylococcus Aureus Strains from Hospitalized Patients in Poland. Biomed Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, A.; Ward, S.; Hyman, P. More Is Better: Selecting for Broad Host Range Bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasman, L.M.; Porter, L.D. Bacteriophages. StatPearls. 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK493185/ (accessed on 29 May 2020).
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological Applications of Bacteriophages: State of the Art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef] [PubMed]
- Olsen, N.M.C.; Thiran, E.; Hasler, T.; Vanzieleghem, T.; Belibasakis, G.N.; Mahillon, J.; Loessner, M.J.; Schmelcher, M. Synergistic Removal of Static and Dynamic Staphylococcus Aureus Biofilms by Combined Treatment with a Bacteriophage Endolysin and a Polysaccharide Depolymerase. Viruses 2018, 10, 438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roach, D.R.; Donovan, D.M. Antimicrobial Bacteriophage-Derived Proteins and Therapeutic Applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashel, M.; Uchiyama, J.; Ujihara, T.; Uehara, Y.; Kuramoto, S.; Sugihara, S.; Yagyu, K.-I.; Muraoka, A.; Sugai, M.; Hiramatsu, K.; et al. Efficient Elimination of Multidrug-Resistant Staphylococcus Aureus by Cloned Lysin Derived from Bacteriophage Phi MR11. J. Infect. Dis. 2007, 196, 1237–1247. [Google Scholar] [CrossRef] [Green Version]
- Chhibber, S.; Gupta, P.; Kaur, S. Bacteriophage as Effective Decolonising Agent for Elimination of MRSA from Anterior Nares of BALB/c Mice. BMC Microbiol. 2014, 14, 212. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Cirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Sergueev, K.V.; Filippov, A.A.; Farlow, J.; Su, W.; Kvachadze, L.; Balarjishvili, N.; Kutateladze, M.; Nikolich, M.P. Correlation of Host Range Expansion of Therapeutic Bacteriophage Sb-1 with Allele State at a Hypervariable Repeat Locus. Appl. Environ. Microbiol. 2019, 85, e01209–e01219. [Google Scholar] [CrossRef] [Green Version]
- Kvachadze, L.; Balarjishvili, N.; Meskhi, T.; Tevdoradze, E.; Skhirtladze, N.; Pataridze, T.; Adamia, R.; Topuria, T.; Kutter, E.; Rohde, C.; et al. Evaluation of Lytic Activity of Staphylococcal Bacteriophage Sb-1 Against Freshly Isolated Clinical Pathogens. Microb. Biotechnol. 2011, 4, 643–650. [Google Scholar] [CrossRef] [Green Version]
- Kutateladze, M.; Adamia, R. Phage Therapy Experience at the Eliava Institute. Med. Mal. Infect. 2008, 38, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.H.; Tanji, Y. Peculiarities of Staphylococcus Aureus Phages and Their Possible Application in Phage Therapy. Appl. Microbiol. Biotechnol. 2019, 103, 4279–4289. [Google Scholar] [CrossRef] [PubMed]
- Gundogdu, A.; Bolkvadze, D.; Kilic, H. In Vitro Effectiveness of Commercial Bacteriophage Cocktails on Diverse Extended-Spectrum Beta-Lactamase Producing Escherichia Coli Strains. Front. Microbiol. 2016, 7, 1761. [Google Scholar] [CrossRef] [PubMed]
- Ajuebor, J.; Buttimer, C.; Arroyo-Moreno, S.; Chanishvili, N.; Gabriel, A.M.; O’Mahony, J.; McAuliffe, O.; Neve, H.; Franz, C.; Coffey, A. Comparison of Staphylococcus Phage K with Close Phage Relatives Commonly Employed in Phage Therapeutics. Antibiotics 2018, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- McCallin, S.; Sarker, S.A.; Sultana, S.; Oechslin, F.; Brüssow, H. Metagenome Analysis of Russian and Georgian Pyophage Cocktails and a Placebo-Controlled Safety Trial of Single Phage Versus Phage Cocktail in Healthy Staphylococcus Aureus Carriers. Environ. Microbiol. 2018, 20, 3278–3293. [Google Scholar] [CrossRef]
- Zschach, H.; Joensen, K.G.; Lindhard, B.; Lund, O.; Goderdzishvili, M.; Chkonia, I.; Jgenti, G.; Kvatadze, N.; Alavidze, Z.; Kutter, E.M.; et al. What Can We Learn from a Metagenomic Analysis of a Georgian Bacteriophage Cocktail? Viruses 2015, 7, 6570–6589. [Google Scholar] [CrossRef] [Green Version]
- Leskinen, K.; Tuomala, H.; Wicklund, A.; Horsma-Heikkinen, J.; Kuusela, P.; Skurnik, M.; Kiljunen, S. Characterization of vB_SauM-fRuSau02, a Twort-Like Bacteriophage Isolated from a Therapeutic Phage Cocktail. Viruses 2017, 9, 258. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.-P.; De Vos, D. Guidelines to Compose an Ideal Bacteriophage Cocktail. Methods Mol. Biol. 2018, 1693, 99–110. [Google Scholar] [CrossRef]
- Ljungman, S.; Jensen, J.E.; Paulsen, D.; Petersons, A.; Ots-Rosenberg, M.; Saha, H.; Struijk, D.G.; Wilkie, M.; Heimbürger, O.; Stegmayr, B.; et al. Retraining for Prevention of Peritonitis in Peritoneal Dialysis Patients: A Randomized Controlled Trial. Perit. Dial. Int. 2020, 40, 141–152. [Google Scholar] [CrossRef] [Green Version]
- Grothe, C.; Taminato, M.; Belasco, A.; Sesso, R.; Barbosa, D. Prophylactic Treatment of Chronic Renal Disease in Patients Undergoing Peritoneal Dialysis and Colonized by Staphylococcus Aureus: A Systematic Review and Meta-Analysis. BMC Nephrol. 2016, 17. [Google Scholar] [CrossRef] [Green Version]
- Grundmann, H.; Aanensen, D.M.; van den Wijngaard, C.C.; Spratt, B.G.; Harmsen, D.; Friedrich, A.W. Geographic Distribution of Staphylococcus Aureus Causing Invasive Infections in Europe: A Molecular-Epidemiological Analysis. PLoS Med. 2010, 7, e10000215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- North, O.I.; Sakai, K.; Yamashita, E.; Nakagawa, A.; Iwazaki, T.; Büttner, C.R.; Takeda, S.; Davidson, A.R. Phage Tail Fibre Assembly Proteins Employ a Modular Structure to Drive the Correct Folding of Diverse Fibres. Nat. Microbiol. 2019, 4, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in Vitro Evolution of Therapeutic Bacteriophages: The Appelmans Protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sybesma, W.; Zbinden, R.; Chanishvili, N.; Kutateladze, M.; Chkhotua, A.; Ujmajuridze, A.; Mehnert, U.; Kessler, T.M. Bacteriophages as Potential Treatment for Urinary Tract Infections. Front. Microbiol. 2016, 7, 465. [Google Scholar] [CrossRef]
- Harper, D.R.; Parracho, H.M.R.T.; Walker, J.; Sharp, R.; Hughes, G.; Werthén, M.; Lehman, S.; Morales, S. Bacteriophages and Biofilms. Antibiotics 2014, 3, 270–284. [Google Scholar] [CrossRef]
- Iacovelli, V.; Gaziev, G.; Topazio, L.; Bove, P.; Vespasiani, G.; Finazzi Agrò, E. Nosocomial Urinary Tract Infections: A Review. Urologia 2014, 81, 222–227. [Google Scholar] [CrossRef]
- Tozzoli, R.; Grande, L.; Michelacci, V.; Ranieri, P.; Maugliani, A.; Caprioli, A.; Morabito, S. Shiga Toxin-Converting Phages and the Emergence of New Pathogenic Escherichia Coli: A World in Motion. Front. Cell. Infect. Microbiol. 2014, 4, 80. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, S.M.; Stickler, D.J.; Mobley, H.L.T.; Shirtliff, M.E. Complicated Catheter-Associated Urinary Tract Infections Due to Escherichia Coli and Proteus Mirabilis. Clin. Microbiol. Rev. 2008, 21, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Pirnay, J.-P.; Blasdel, B.G.; Bretaudeau, L.; Buckling, A.; Chanishvili, N.; Clark, J.R.; Corte-Real, S.; Debarbieux, L.; Dublanchet, A.; De Vos, D.; et al. Quality and Safety Requirements for Sustainable Phage Therapy Products. Pharm. Res. 2015, 32, 2173–2179. [Google Scholar] [CrossRef] [Green Version]
- Mulzer, J.; Trampuz, A.; Potapov, E.V. Treatment of Chronic Left Ventricular Assist Device Infection with Local Application of Bacteriophages. Eur. J. Cardiothorac. Surg. 2020, 57, 1003–1004. [Google Scholar] [CrossRef]
- Zhvania, P.; Hoyle, N.S.; Nadareishvili, L.; Nizharadze, D.; Kutateladze, M. Phage Therapy in a 16-Year-Old Boy with Netherton Syndrome. Front. Med. 2017, 4, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, T.; Verbeken, G.; Vos, D.D.; Merabishvili, M.; Vaneechoutte, M.; Lavigne, R.; Jennes, S.; Zizi, M.; Pirnay, J.-P. Experimental Phage Therapy of Burn Wound Infection: Difficult First Steps. Int. J. Burns. Trauma. 2014, 4, 66–73. [Google Scholar] [PubMed]
- Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Leitner, L.; Mehnert, U.; Chkhotua, A.; Kessler, T.M.; Sybesma, W. Adapted Bacteriophages for Treating Urinary Tract Infections. Front. Microbiol. 2018, 9, 1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
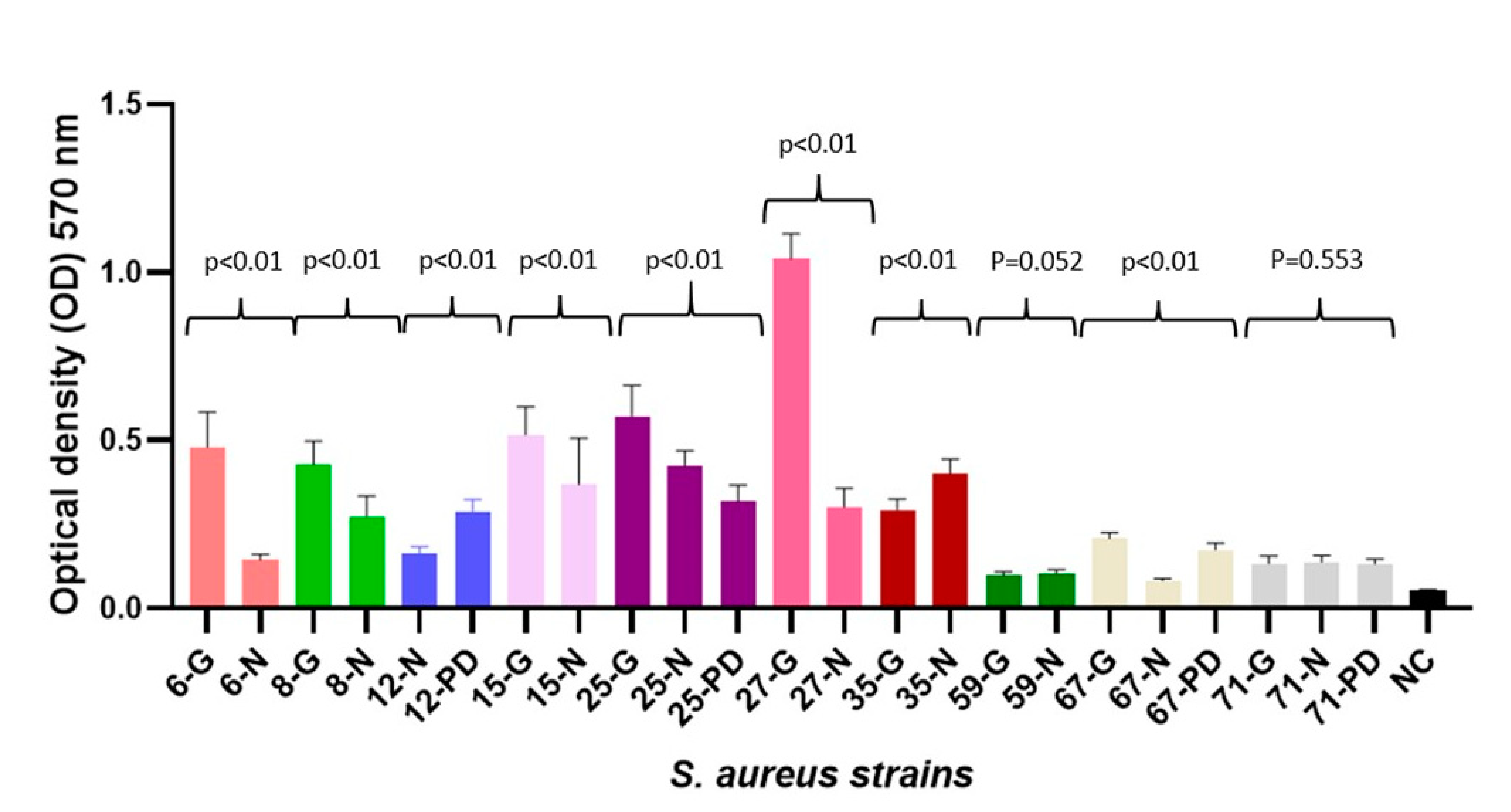
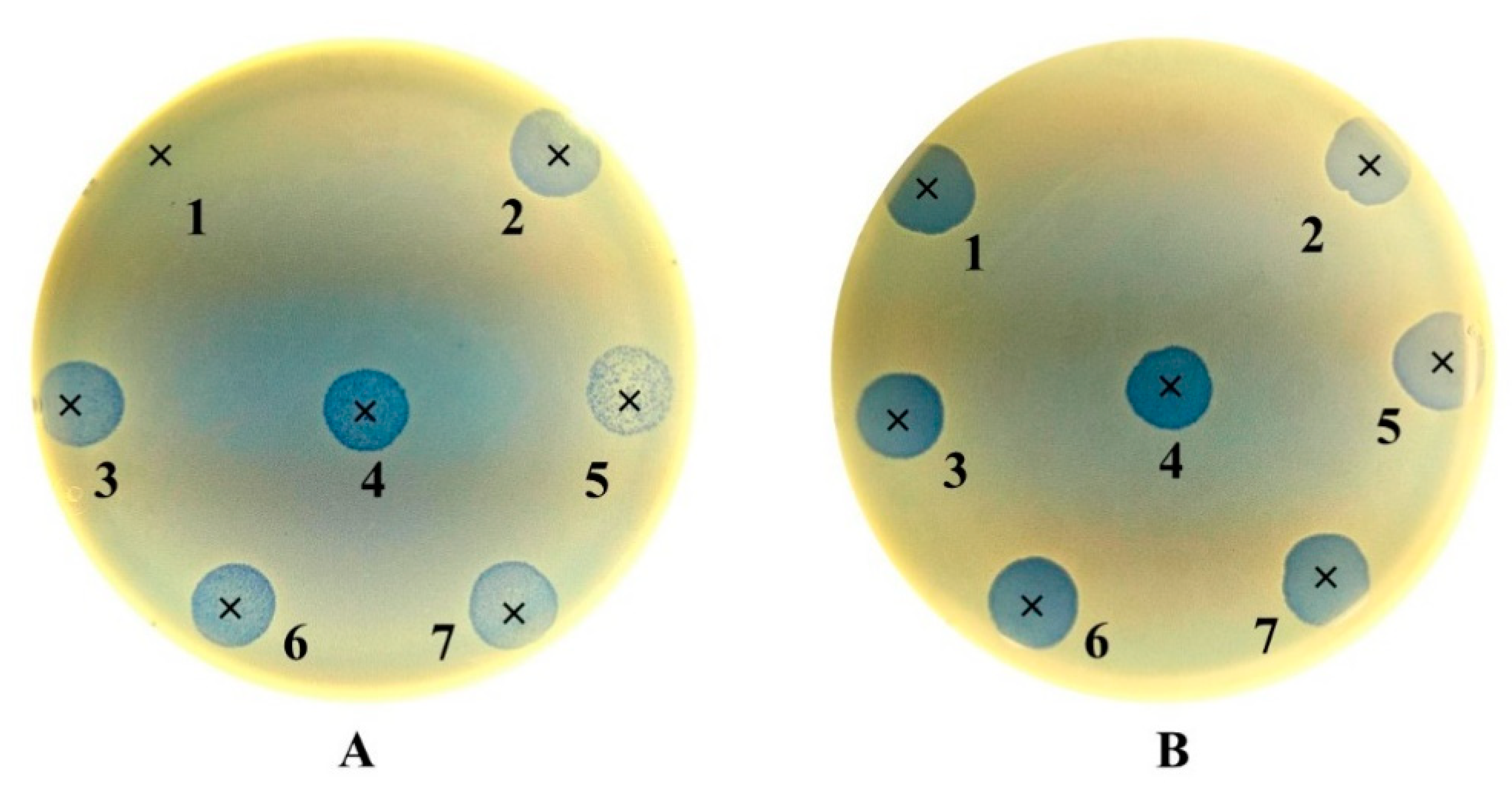
| N | Age | Sex | Cause of End-Stage Renal Disease | Carriage Site (Biofilm Production Group) | Outcome in Two Years | ||
|---|---|---|---|---|---|---|---|
| Nose | Groin | PD Catheter | |||||
| 1 | 55 | ♂ | Diabetic nephropathy | 1N strong | 1 episode of S. aureus/Pseudomonas spp. Peritonitis, transplantation | ||
| 2 | 74 | ♀ | Glomerulonephritis | 3N strong | Death | ||
| 3 | 76 | ♂ | Hypertensive nephropathy | 6N moderate | 6G strong | 3 episodes of peritonitis (2 Streptococcus spp., 1 culture-negative) | |
| 4 | 83 | ♂ | Chronic interstitial nephritis | 8N strong | 8G strong | Death | |
| 5 | 68 | ♀ | Diabetic nephropathy | 12N moderate | 12PD strong | Death | |
| 6 | 36 | ♂ | Chronic interstitial nephritis | 14N moderate | Transplantation | ||
| 7 | 64 | ♂ | Chronic interstitial nephritis | 15N strong | 15G strong | Uneventful PD | |
| 8 | 42 | ♀ | Chronic interstitial nephritis | 17N strong | Transplantation | ||
| 9 | 60 | ♂ | ADPKD | 19N moderate | Transplantation | ||
| 10 | 34 | ♂ | Glomerulonephritis | 25N strong | 25G strong | 25PD strong | Transplantation |
| 11 | 77 | ♀ | ADPKD | 26PD strong | Death | ||
| 12 | 68 | ♂ | Glomerulonephritis | 27N strong | 27G strong | Uneventful PD | |
| 13 | 69 | ♂ | Glomerulonephritis | 35N strong | 35G strong | 1 episode of culture-negative peritonitis | |
| 14 | 68 | ♂ | Diabetic nephropathy | 36N moderate | Transplantation | ||
| 15 | 69 | ♂ | Glomerulonephritis | 47G strong | Uneventful PD | ||
| 16 | 58 | ♂ | Hypertensive nephropathy | 48N strong | Death | ||
| 17 | 65 | ♂ | Diabetic nephropathy | 58N strong | Death | ||
| 18 | 81 | ♂ | Chronic interstitial nephritis | 59N weak | 59G weak | Death | |
| 19 | 26 | ♀ | Glomerulonephritis | 67N weak | 67G moderate | 67PD moderate | 2 episodes of peritonitis (1 Streptococcus spp., 1 culture-negative) |
| 20 | 47 | ♂ | Glomerulonephritis | 70N strong | Transplantation | ||
| 21 | 49 | ♂ | Diabetic nephropathy | 71N moderate | 71G moderate | 71PD moderate | Death |
| TOTAL n (%) | 18 (53%) | 11 (32%) | 5 (15%) | ||||
| S. aureus Strain Count | |||||||
| Biofilm Production n (%) | Strong | 10 (56%) | 8 (73%) | 3 (60%) | |||
| Moderate | 6 (33%) | 2 (18%) | 2 (40%) | ||||
| Weak | 2 (11%) | 1 (9%) | |||||
| Clinical Outcomes | S. aureus Carriers | S. aureus | Total | RR | CI 95% |
|---|---|---|---|---|---|
| Noncarriers | |||||
| Number of patients | 30.0% | 70.0% | 100% (n = 70) | ||
| (n = 21) | (n = 49) | ||||
| Death | 31.1% | 16.3% | 22.9% (n = 16) | 2.33 | 1.01–5.38 |
| (n = 8) | (n = 8) | ||||
| Transplantation | 28.6% | 18.4% | 21.4% (n = 15) | 1.56 | 0.63–3.81 |
| (n = 6) | (n = 9) | ||||
| Removal of PD catheter | 0% | 20.4% | 14.3% (n = 10) | ||
| (n = 0) | (n = 10) | ||||
| Peritonitis | 19.1% | 34.7% | 30.0% (n = 21) | 0.55 | 0.21–1.44 |
| (n = 4) | (n = 17) |
| S. aureus Strain Code | Bacteriophages | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Biofilm Production Degree | Eliava BioPreparations Ltd. | Microgen Ltd. | |||||||
| Staphylococcal | Pyo | Ses | Fersisi | Enko | Intesti | Pyobacteriophag | |||
| 1.2 × 107 | 3.4 × 107 | 4.0 × 107 | 2.4 × 107 | 1.4 × 107 | 2.0 × 107 | 2.0 × 107 | |||
| Not Adapted | Adapted | ||||||||
| 1N | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 3N | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 6N | Moderate | R a | PL | PL | PL | PL | PL | PL | PL |
| 6G | Strong | R a | PL | PL | PL | PL | PL | PL | PL |
| 8N | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 8G | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 12N | Moderate | IP a | PL | PL | PL | PL | PL | PL | IP |
| 12PD | Strong | R a | PL | PL | PL | PL | PL | PL | PL |
| 14N | Moderate | PL | PL | PL | PL | PL | PL | PL | |
| 15N | Strong | R a | PL | PL | PL | PL | PL | PL | PL |
| 15G | Strong | R a | PL | PL | PL | PL | PL | PL | PL |
| 17N | Strong | R a | PL | IP | PL | PL | PL | PL | IP |
| 19N | Moderate | R a | PL | PL | PL | PL | PL | PL | IP |
| 25N | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 25PD | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 25G | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 26PD | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 27N | Strong | PL | PL | IP | PL | IP | PL | PL | |
| 27G | Strong | PL | CL | PL | PL | CL | PL | PL | |
| 35N | Strong | IP a | PL | PL | PL | PL | PL | PL | PL |
| 35G | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 36N | Moderate | PL | PL | PL | PL | PL | PL | PL | |
| 47G | Strong | CL | PL | PL | PL | PL | PL | PL | |
| 48G | Strong | CL | PL | PL | PL | PL | PL | PL | |
| 58N | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 59G | Weak | CL | PL | IP | PL | PL | PL | PL | |
| 59N | Weak | CL | CL | PL | IP | PL | PL | PL | |
| 67G | Weak | R a | PL | PL | PL | PL | PL | PL | PL |
| 67N | Moderate | R a | PL | PL | PL | PL | PL | PL | PL |
| 67PD | Moderate | PL | PL | PL | PL | PL | PL | PL | |
| 70N | Strong | PL | PL | PL | PL | PL | PL | PL | |
| 71PD | Moderate | PL | PL | PL | PL | PL | PL | PL | |
| 71G | Moderate | PL | PL | PL | PL | PL | PL | PL | |
| 71N | Moderate | PL | PL | PL | PL | PL | PL | PL | |
| Phage lytic activity | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| CL (+++) | 4 (12) | 0 (0) | 2 (6) | 0 (0) | 0 (0) | 1 (3) | 0 (0) | 0 (0) | |
| PL (++) | 19 (56) | 11 (100) | 31 (91) | 32 (94) | 33 (97) | 32 (94) | 34 (100) | 31 (91) | |
| IP (+) | 2 a (6) | 0 (0) | 1 (3) | 2 (6) | 1 (3) | 1 (3) | 0 (0) | 3 (9) | |
| R (−) | 9 a (26) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Racenis, K.; Kroica, J.; Rezevska, D.; Avotins, L.; Skuditis, E.; Popova, A.; Puide, I.; Kuzema, V.; Petersons, A. S. aureus Colonization, Biofilm Production, and Phage Susceptibility in Peritoneal Dialysis Patients. Antibiotics 2020, 9, 582. https://doi.org/10.3390/antibiotics9090582
Racenis K, Kroica J, Rezevska D, Avotins L, Skuditis E, Popova A, Puide I, Kuzema V, Petersons A. S. aureus Colonization, Biofilm Production, and Phage Susceptibility in Peritoneal Dialysis Patients. Antibiotics. 2020; 9(9):582. https://doi.org/10.3390/antibiotics9090582
Chicago/Turabian StyleRacenis, Karlis, Juta Kroica, Dace Rezevska, Lauris Avotins, Edgars Skuditis, Anna Popova, Ilze Puide, Viktorija Kuzema, and Aivars Petersons. 2020. "S. aureus Colonization, Biofilm Production, and Phage Susceptibility in Peritoneal Dialysis Patients" Antibiotics 9, no. 9: 582. https://doi.org/10.3390/antibiotics9090582





