Aminoglycosides: From Antibiotics to Building Blocks for the Synthesis and Development of Gene Delivery Vehicles †
Abstract
1. Introduction
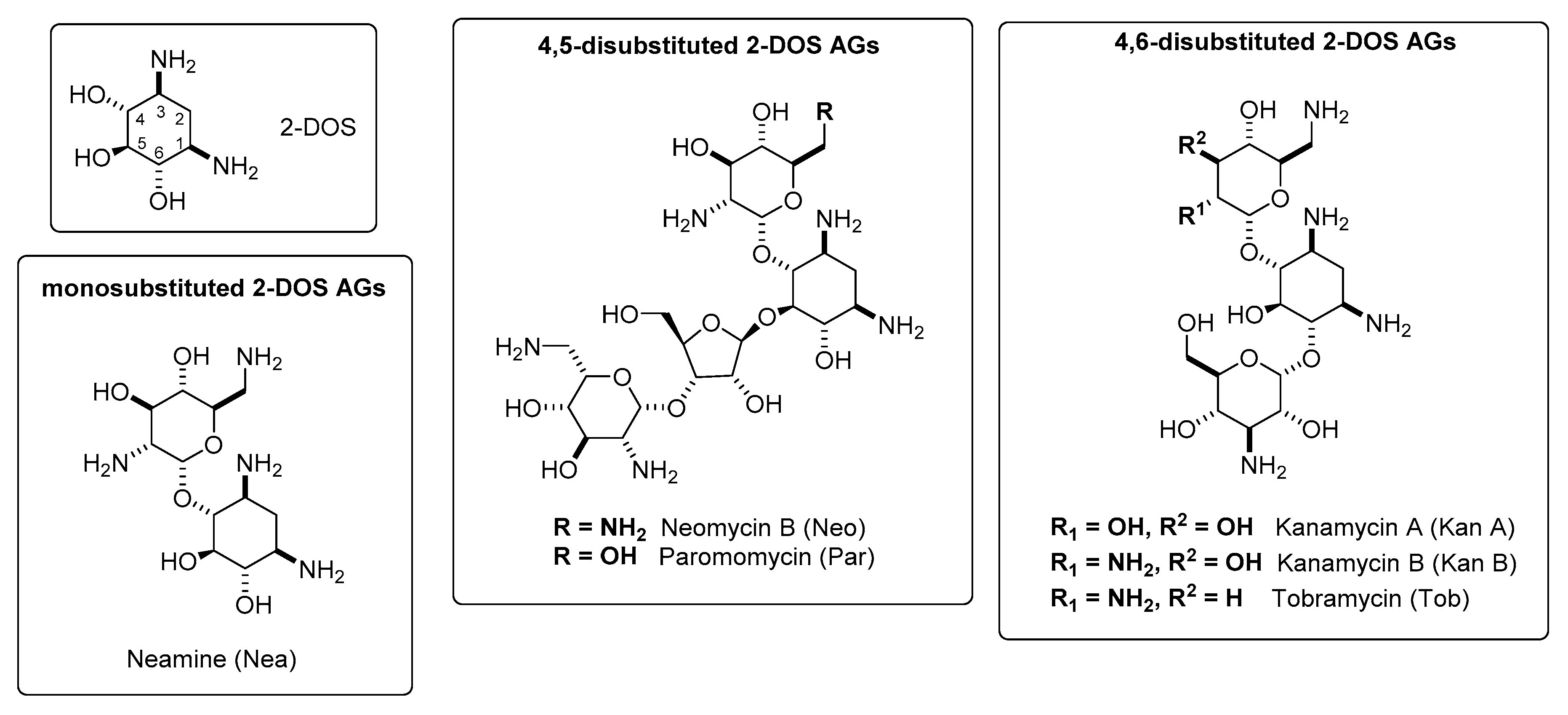
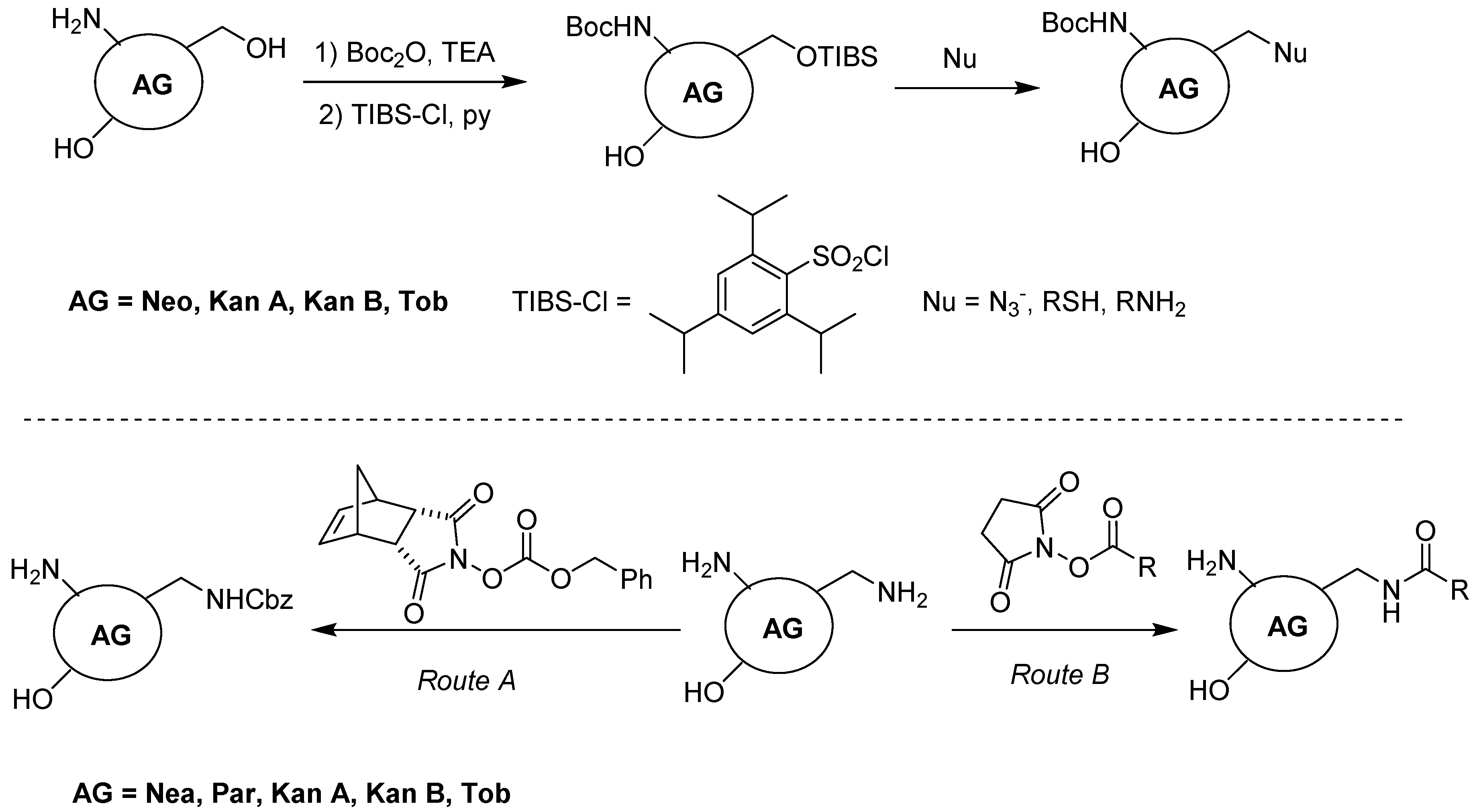
2. Classification and Chemistry of Aminoglycosides (AGs)
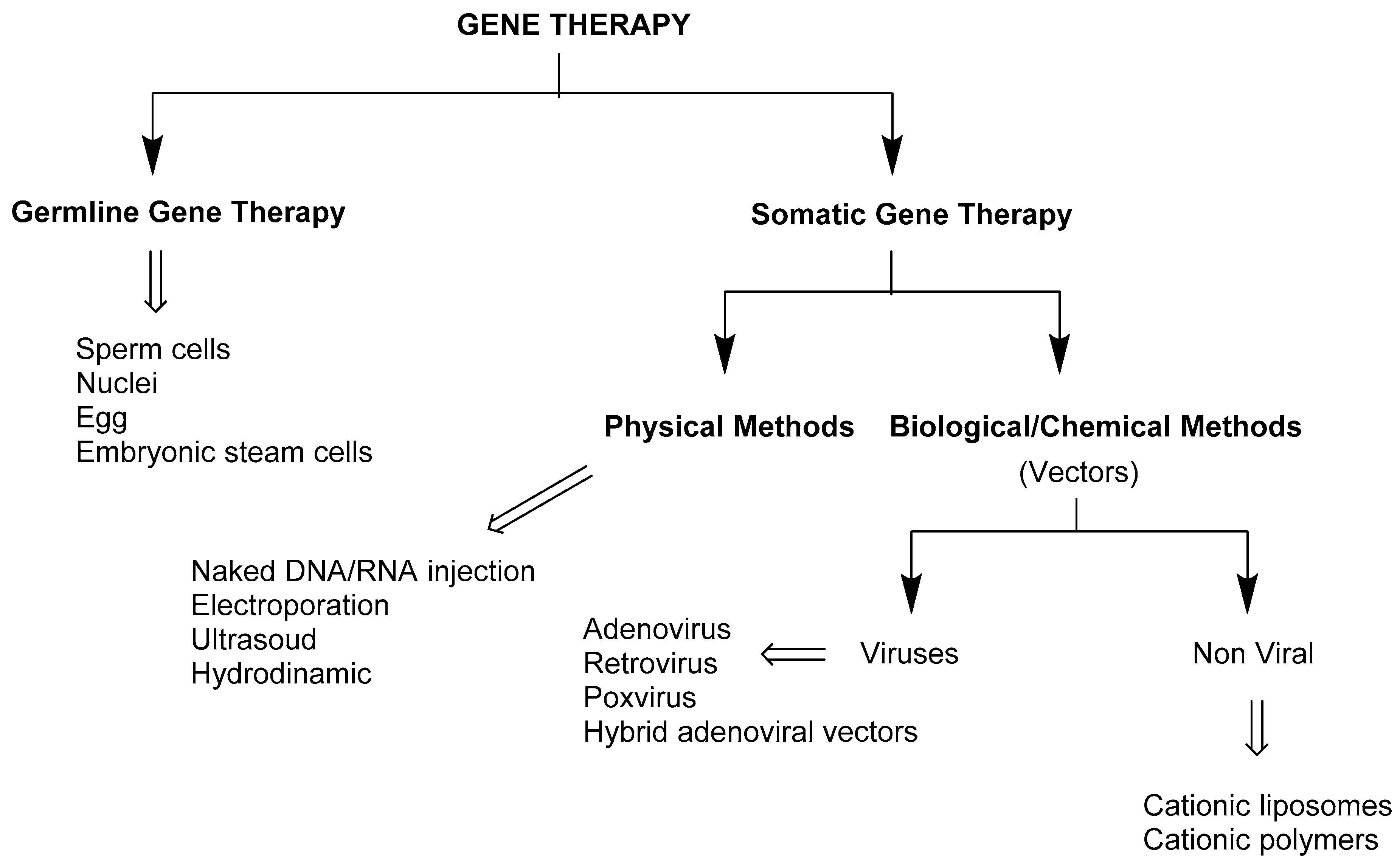
3. Gene Delivery
4. AG-Based Lipid Gene Delivery Vectors
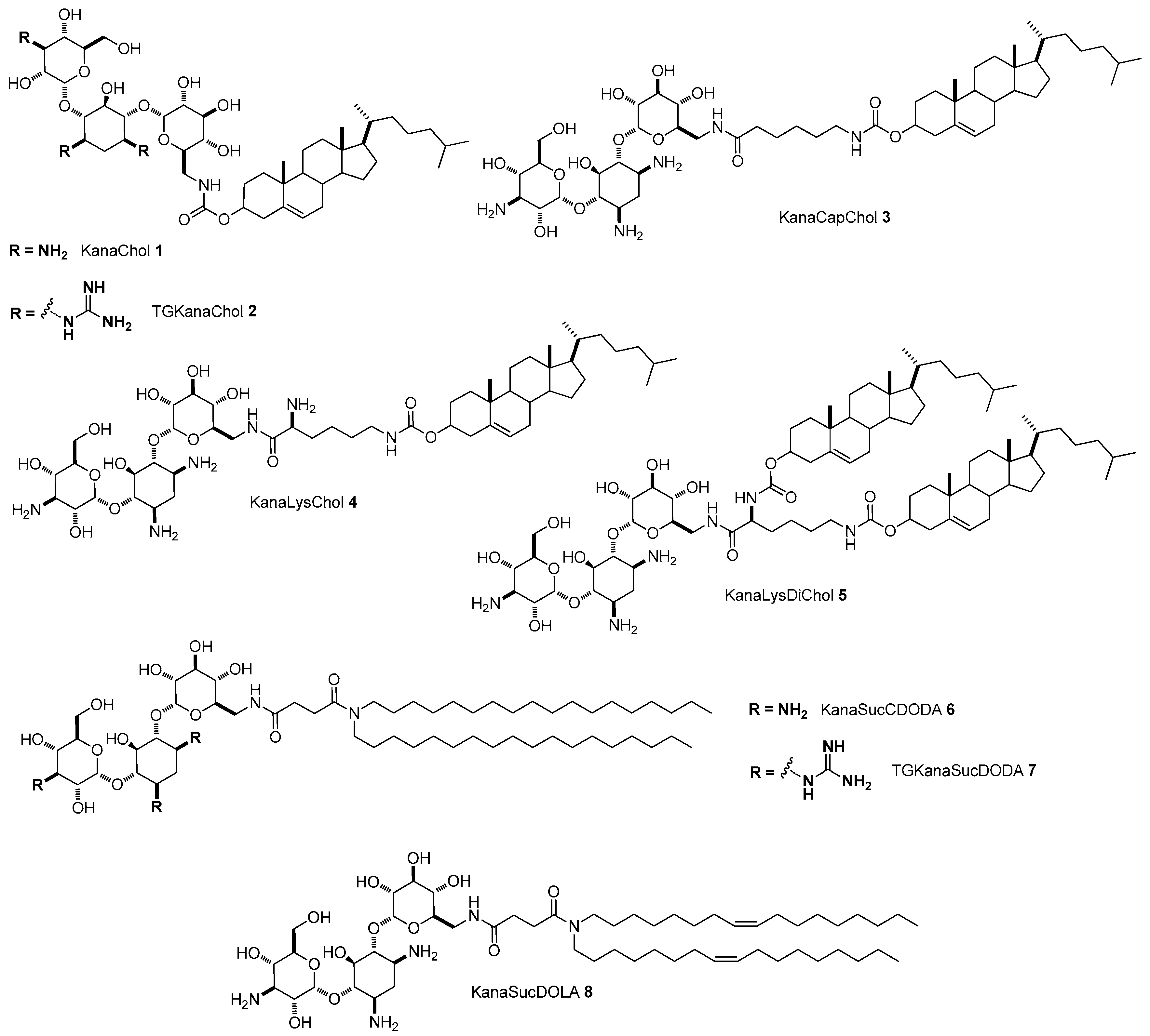
4.1. AG-Based Lipid Vectors for DNA Delivery
4.2. AG-Based Lipid Vectors for RNA Delivery
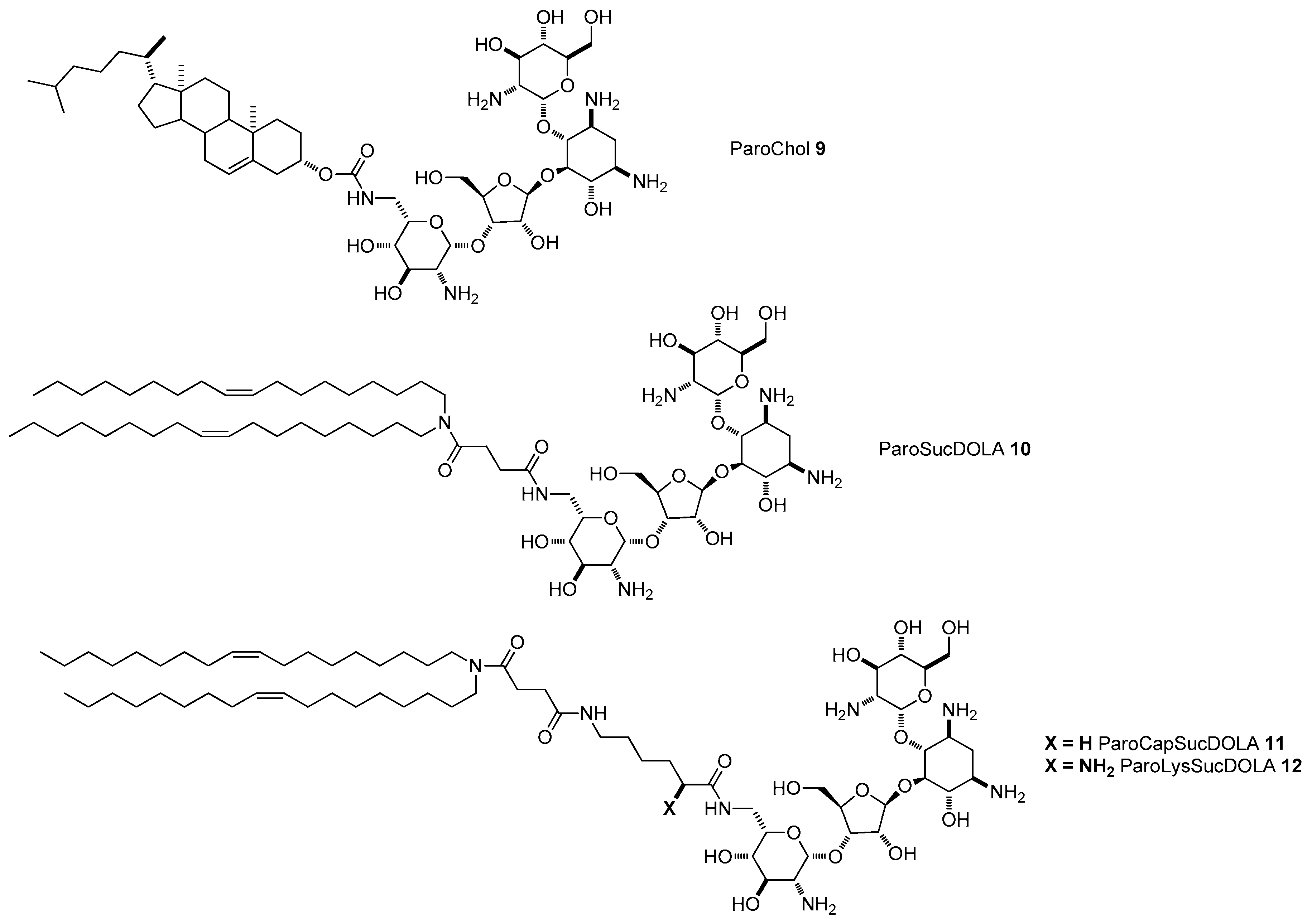
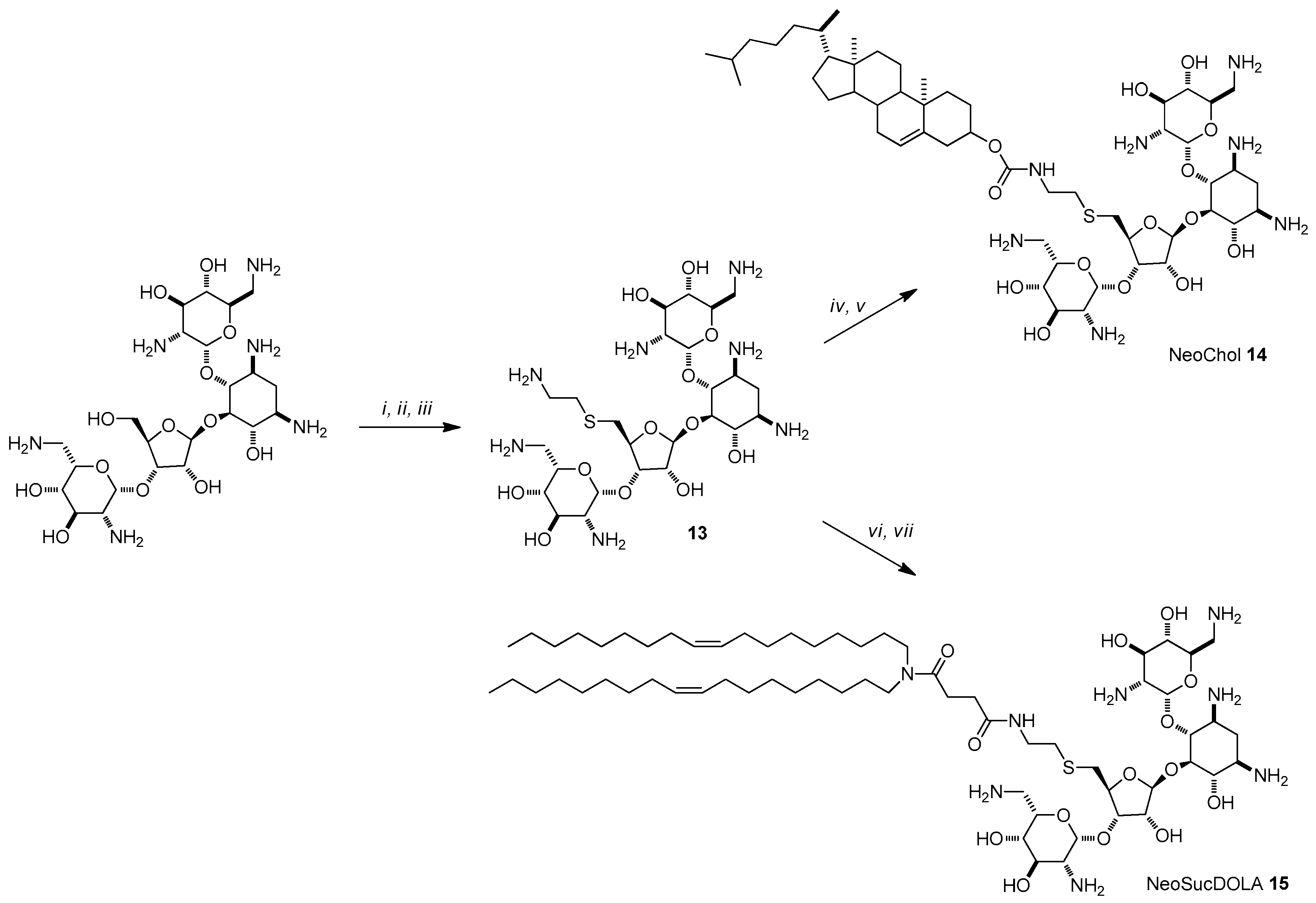
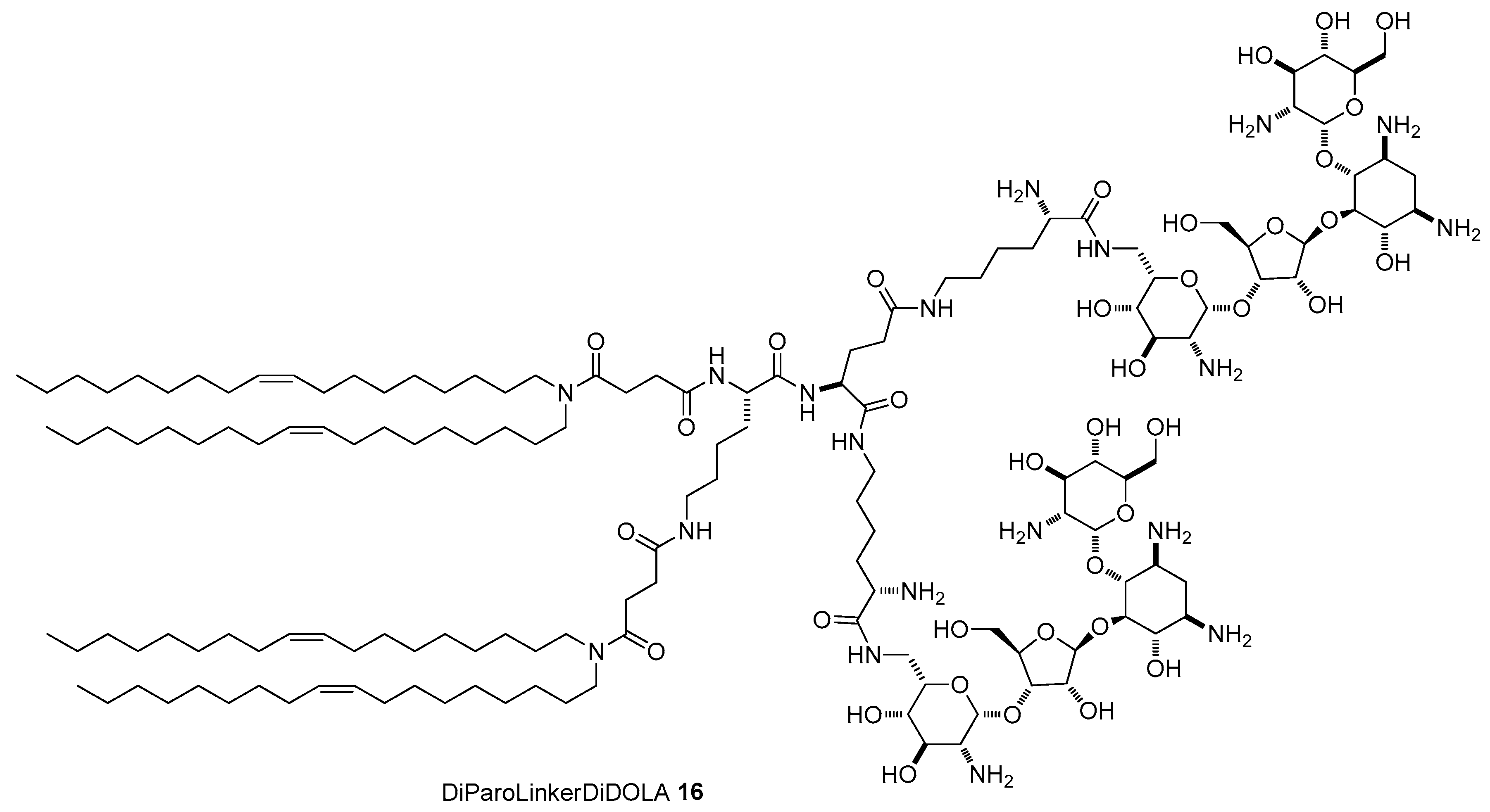
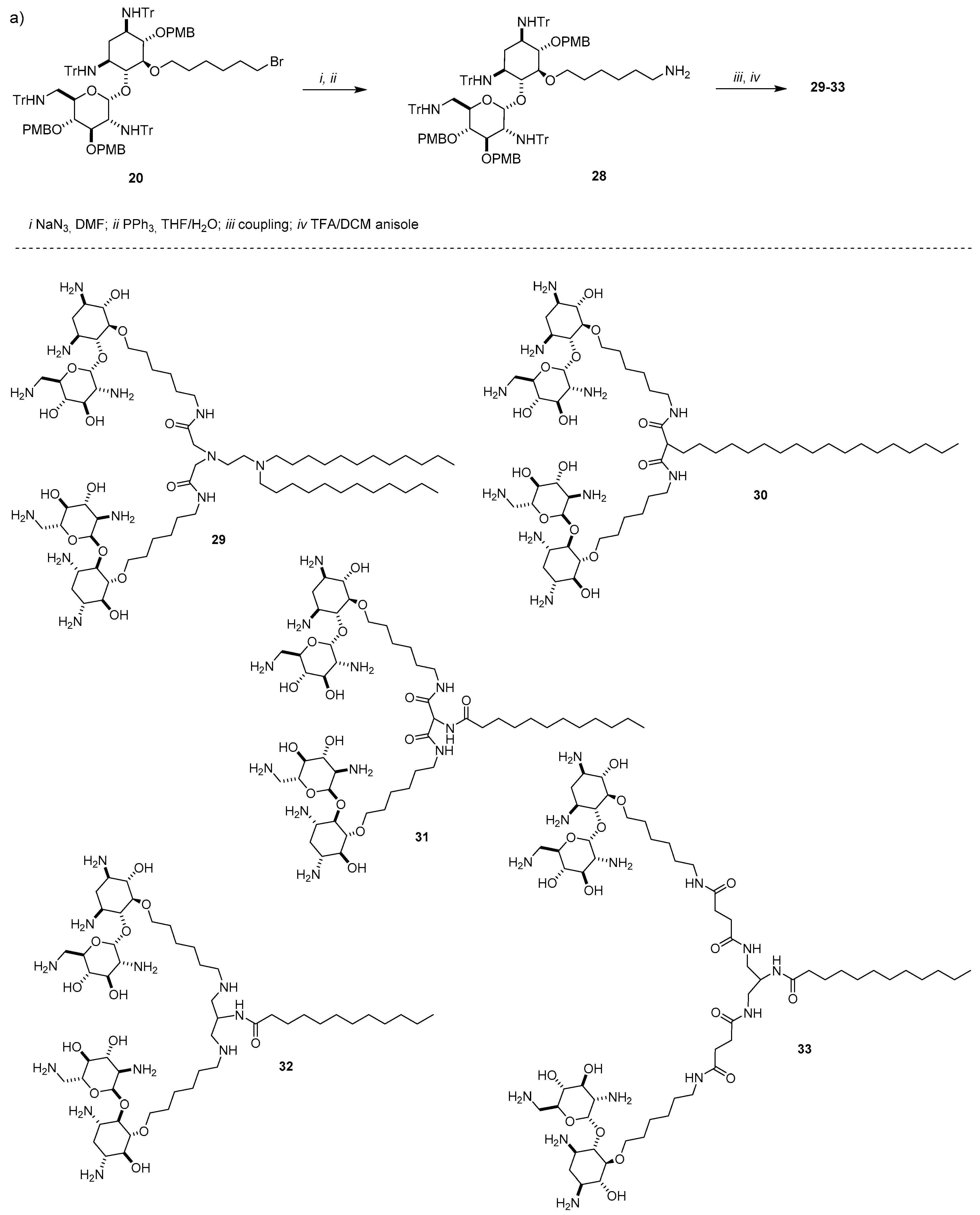
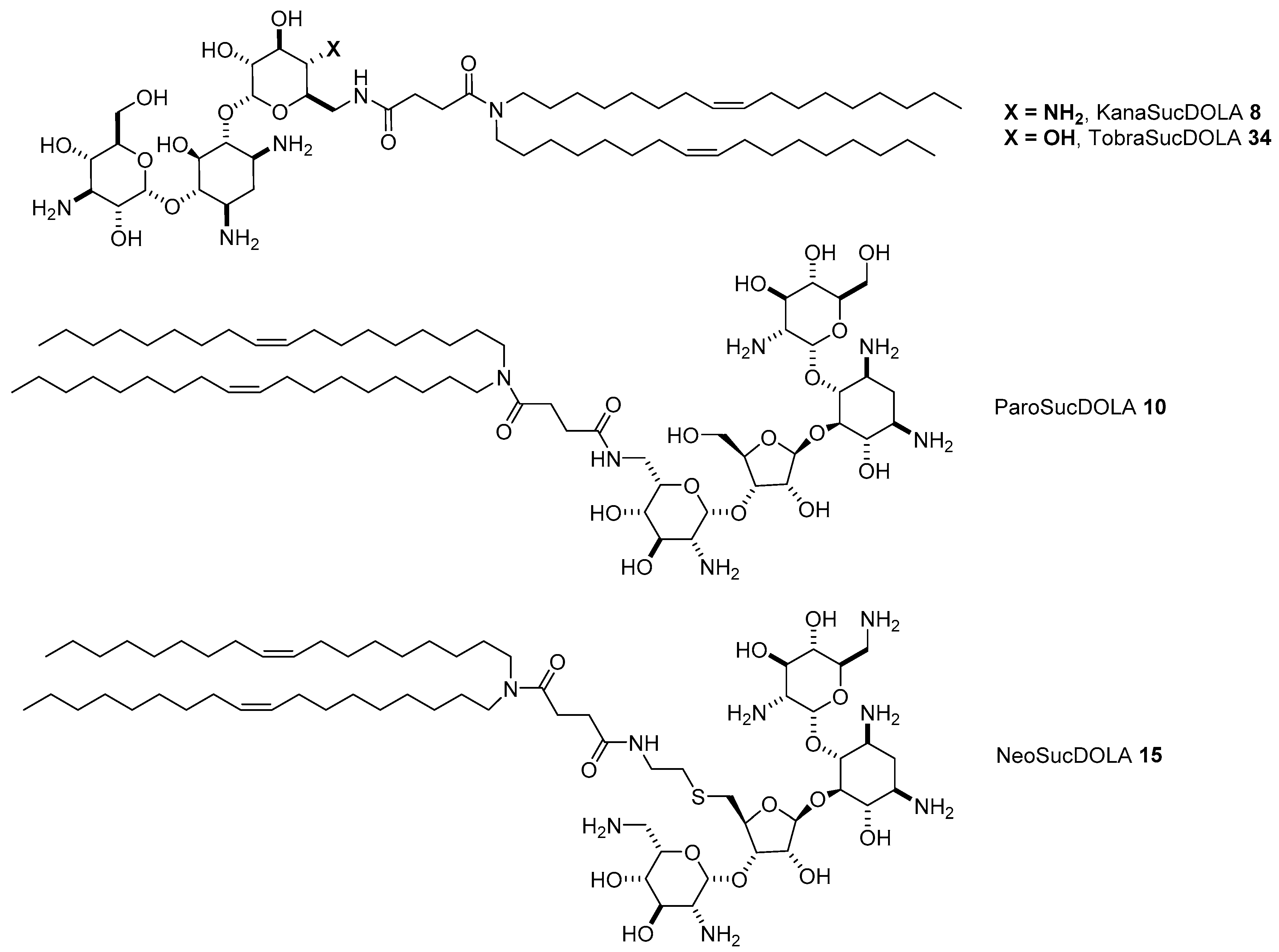
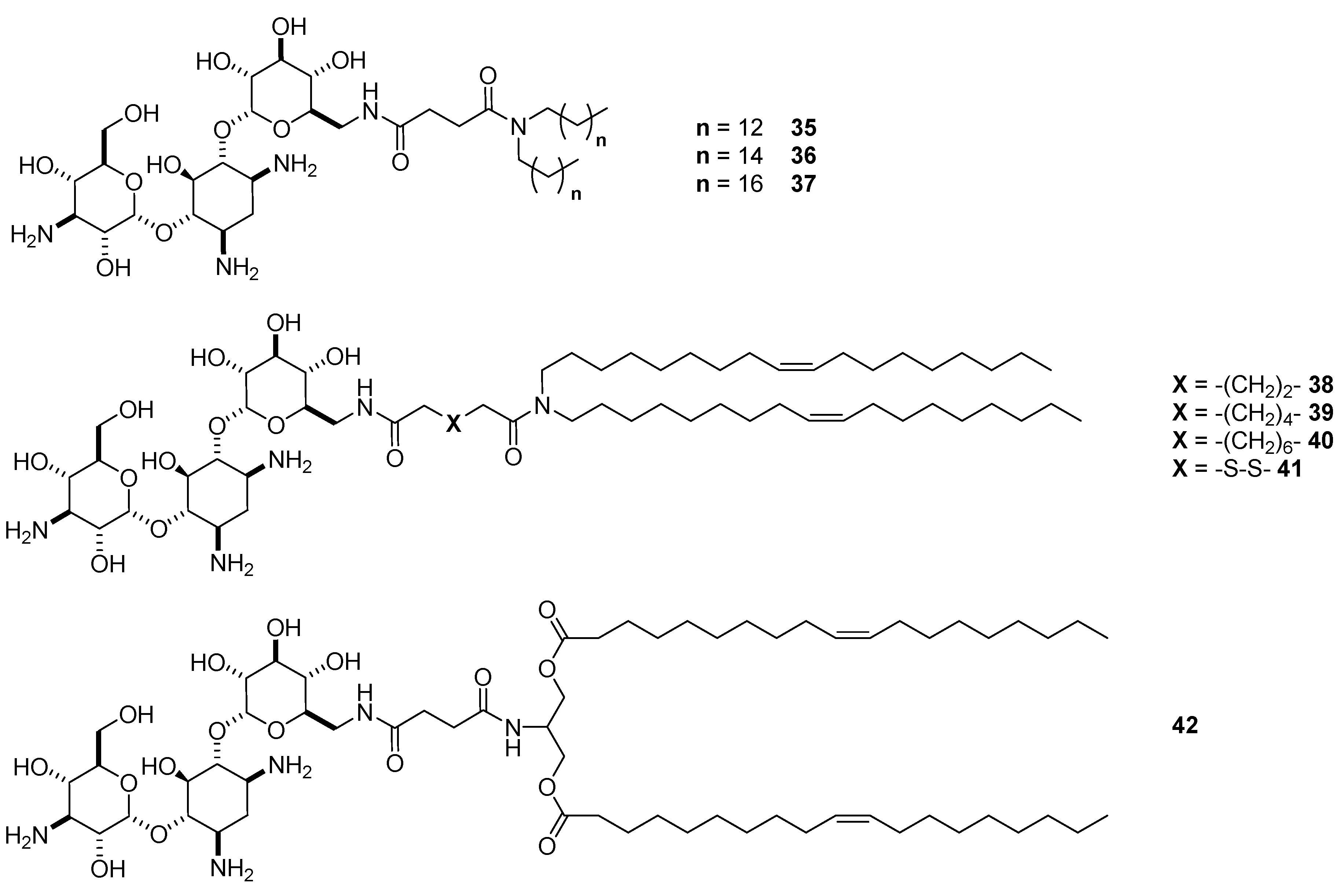
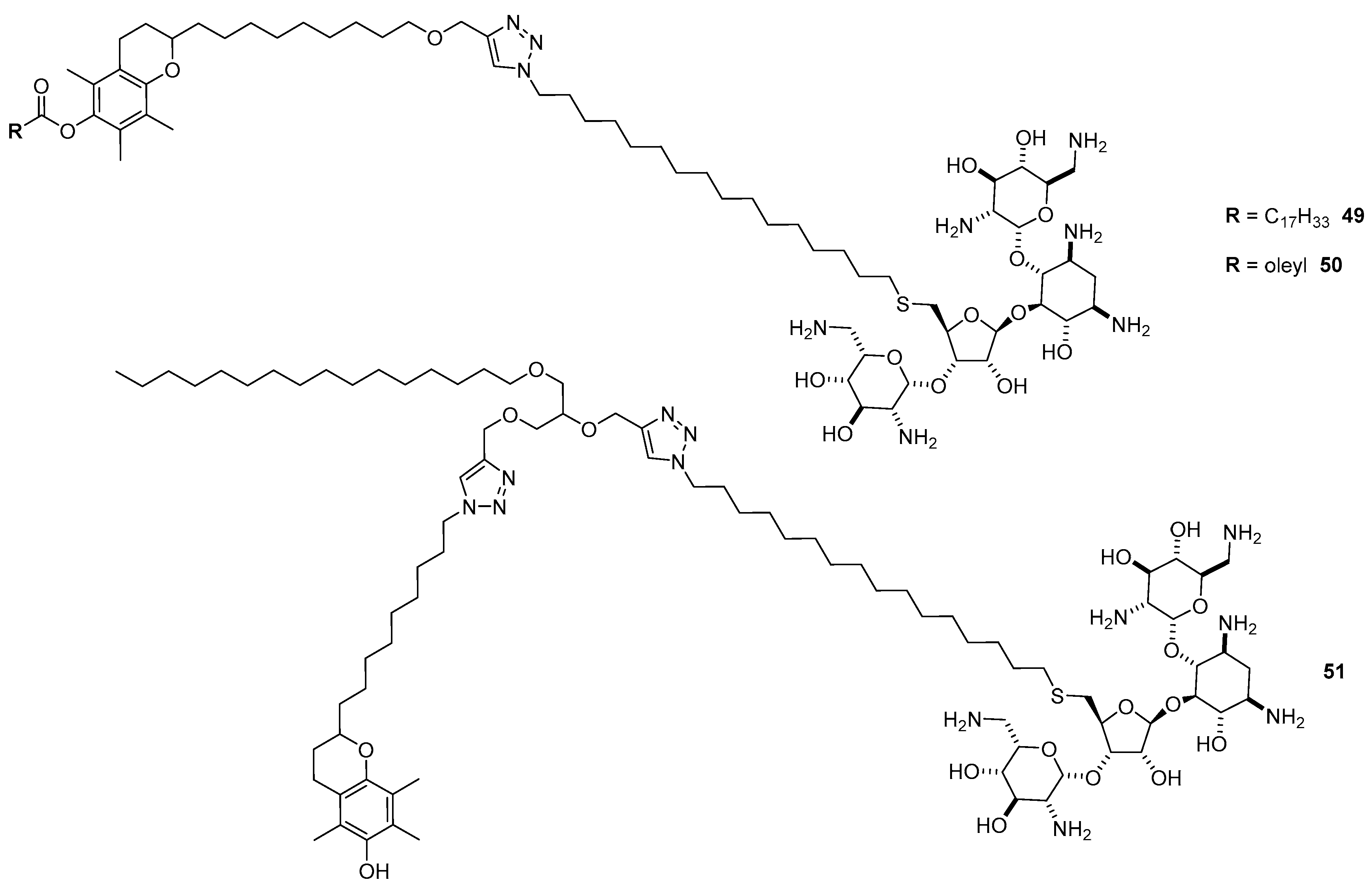
4.3. AG-Based Lipid Vectors with “Non-Conventional” Lipidic Tails
5. Polymeric AG-Based Gene Delivery Vectors
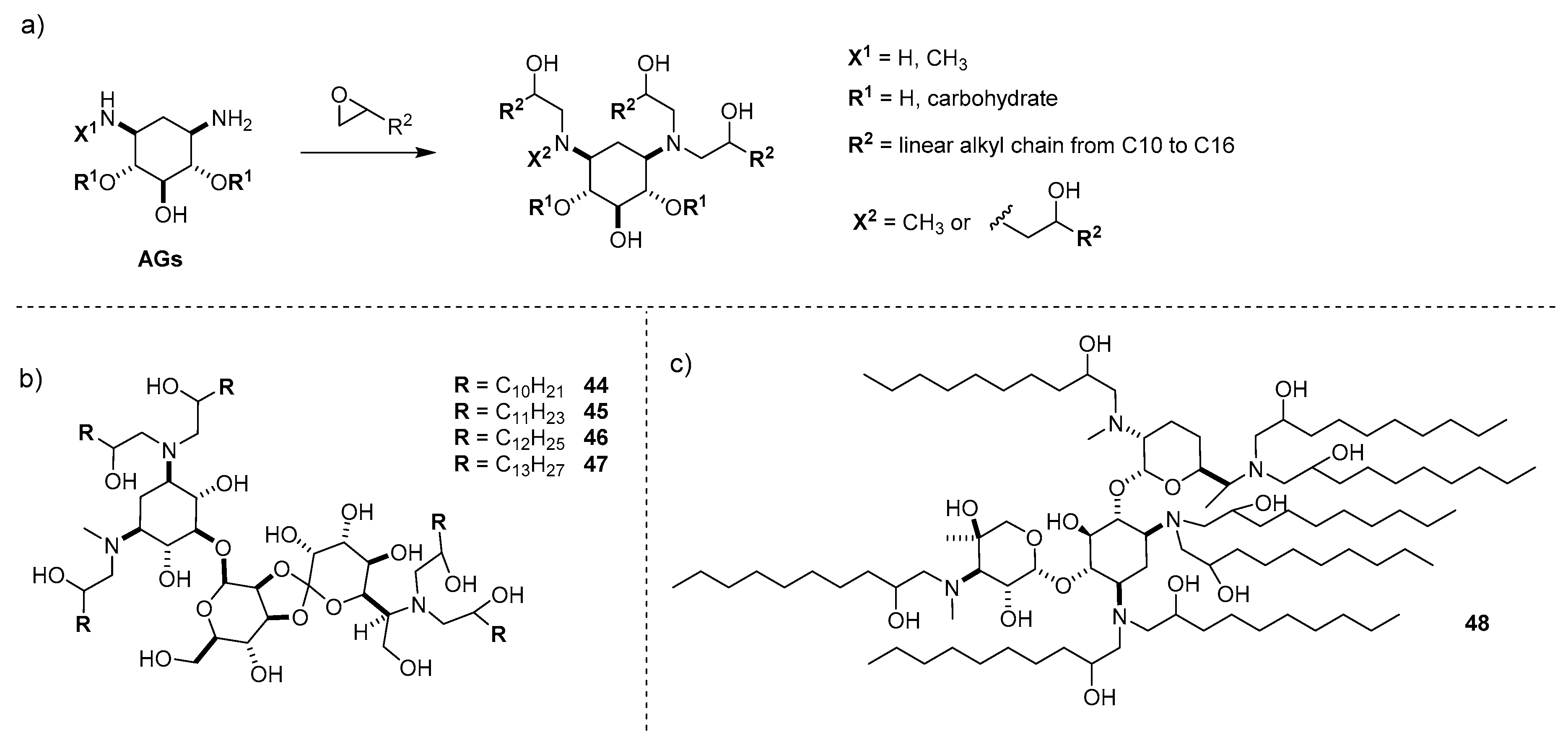
5.1. Functionalization of Polymers with AGs
5.2. Synthesis of Hyperbranched Polymers Based on AG Scaffolds
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References and Note
- Houghton, J.L.; Green, K.D.; Chen, W.; Garneau-Tsodikova, S. The future of aminoglycosides: The end or renaissance? ChemBioChem 2010, 11, 880–902. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Wright, G.D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997, 5, 234–240. [Google Scholar] [CrossRef]
- Moazed, D.; Noller, H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 1987, 327, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tor, Y. Electrostatic interactions in RNA aminoglycosides binding. J. Am. Chem. Soc. 1997, 119, 8734–8735. [Google Scholar] [CrossRef]
- Francois, B.; Russell, R.J.M.; Murray, J.B.; Aboul-ela, F.; Masquida, B.; Vicens, Q.; Westhof, E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: Role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005, 33, 5677–5690. [Google Scholar] [CrossRef] [PubMed]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of resistance to aminoglycoside antibiotics: Overview and perspectives. MedChemMed 2016, 7, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Mingeot-Leclercq, M.-P.; Tulkens, P.M. Aminoglycosides: Nephrotoxicity. Antimicrob. Agents Chemother. 1999, 43, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Selimoglu, E. Aminoglycoside-induced ototoxicity. Curr. Pharm. Des. 2007, 13, 119–126. [Google Scholar] [CrossRef]
- Chandrika, N.T.; Garneau-Tsodikova, S. Comprehensive review of chemical strategies for the preparation of new aminoglycosides and their biological activities. Chem. Soc. Rev. 2018, 47, 1189–1249. [Google Scholar] [CrossRef]
- Bera, S.; Mondal, D.; Palit, S.; Schweizer, F. Structural modifications of the neomycin class of aminoglycosides. MedChemMed 2016, 7, 1499–1534. [Google Scholar] [CrossRef]
- Fosso, M.Y.; Li, Y.; Garneau-Tsodikova, S. New trends in the use of aminoglycosides. MedChemMed 2014, 5, 1075–1091. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, R.C. The basic science of gene therapy. Science 1993, 260, 926–932. [Google Scholar] [CrossRef] [PubMed]
- High, K.A.; Roncarolo, M.G. Gene therapy. N. Engl. J. Med. 2019, 381, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Anguela, X.M.; High, K.A. Entering the modern era of gene therapy. Annu. Rev. Med. 2019, 70, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Ibraheem, D.; Elaissari, A.; Fess, H. Gene therapy and DNA delivery systems. Int. J. Pharm. 2014, 459, 70–83. [Google Scholar] [CrossRef]
- Ebbesen, M.; Jensen, T.G. Nanomedicine: Techniques, potentials, and ethical implications. J. Biomed. Biotechnol. 2006, 5, 51516. [Google Scholar] [CrossRef]
- Coller, B.S. Ethics of human genome editing. Annu. Rev. Med. 2019, 70, 289–305. [Google Scholar] [CrossRef]
- Delhove, J.; Osenk, I.; Prichard, I.; Donnelley, M. Public acceptability of gene therapy and gene editing for human use: A systematic review. Hum. Gene Ther. 2020, 31, 20–46. [Google Scholar] [CrossRef]
- Goswami, R.; Subramanian, G.; Silayeva, L.; Newkirk, I.; Doctor, D.; Chawla, K.; Chattopadhyay, S.; Chandra, D.; Chilukuri, N.; Betapudi, V. Gene therapy leaves a vicious cycle. Front. Oncol. 2019, 9, 297. [Google Scholar] [CrossRef]
- Alsaggar, M.; Liu, D. Physical methods for gene transfer. Adv. Genet. 2015, 89, 1–124. [Google Scholar]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, A.; Jazi, M.S.; Mohammadi, S.; Nikoo, H.R.; Nazari, Z.; Hosseini, E.S.; Burtscher, I.; Mowla, S.J.; Lickert, H. Development and clinical translation of approved gene therapy products for genetic disorders. Front. Genet. 2019, 10, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Simoes, S.; Filipe, A.; Faneca, H.; Mano, M.; Penacho, N.; Duzgunes, N.; Pedroso de Lima, M. Cationic liposomes for gene delivery. Expert Opin. Drug Del. 2005, 2, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The headgroup evolution of cationic lipids for gene delivery. Bioconjug. Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef]
- Zhi, D.; Zhang, S.; Wang, B.; Zhao, Y.; Yang, B.; Yu, S. Transfection efficiency of cationic lipids with different hydrophobic domains in gene delivery. Bioconjug. Chem. 2010, 21, 563–577. [Google Scholar] [CrossRef]
- Zhi, D.; Bai, Y.; Yang, Y.; Cui, S.; Zhao, Y.; Chen, H.; Zhang, S. A review on cationic lipids with different linkers for gene delivery. Adv. Colloid Interface Sci. 2018, 253, 117–140. [Google Scholar] [CrossRef]
- Mintzer, M.A.; Simanek, E.E. Nonviral vectors for gene delivery. Chem. Rev. 2009, 109, 259–302. [Google Scholar] [CrossRef]
- Wightman, L.; Kircheis, R.; Rössler, V.; Carotta, S.; Ruzicka, R.; Kursa, M.; Wagner, E. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J. Gene Med. 2001, 4, 362–372. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Q.; Chang, H.; Cheng, Y. Surface-engineered dendrimers in gene delivery. Chem. Rev. 2015, 115, 5274–5300. [Google Scholar] [CrossRef]
- Tor, Y. RNA and small molecules word. Angew. Chem. Int. Ed. Engl. 1999, 38, 1579–1582. [Google Scholar] [CrossRef]
- Arya, D.P.; Cofee, R.L., Jr. DNA triple helix stabilization by aminoglycoside antibiotics. Bioorg. Med. Chem. Lett. 2000, 10, 1897–1899. [Google Scholar] [CrossRef]
- Belmont, P.; Aissaoui, A.; Hauchecorne, M.; Oudrhiri, N.; Petit, L.; Vigneron, J.-P.; Lehn, J.-M.; Lehn, P. Aminoglycoside-derived cationic lipids as efficient vectors for gene delivery in vitro and in vivo. J. Gene Med. 2002, 4, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Sainlos, M.; Belmont, P.; Vigneron, J.-P.; Lehn, P.; Lehn, J.-M. Aminoglycoside-derived cationic lipids for gene transfection: Synthesis of kanamycin A derivatives. Eur. J. Org. Chem. 2003, 2003, 2764–2774. [Google Scholar] [CrossRef]
- Luedke, N.W.; Baker, T.J.; Goodman, M.; Tor, Y. Guanidinoglycosides: A novel family of RNA ligands. J. Am. Chem. Soc. 2000, 122, 12035–12056. [Google Scholar] [CrossRef]
- Baker, T.J.; Luedke, N.W.; Tor, Y.; Goodman, M. Synthesis and anti-HIV activity of aminoglycosides. J. Org. Chem. 2000, 65, 9054–9058. [Google Scholar] [CrossRef]
- Luedke, N.W.; Carmichael, P.; Tor, Y. Cellular uptake of aminoglycosides, guanidinoglycosides, and polyarginines. J. Am. Chem. Soc. 2003, 125, 12374–12375. [Google Scholar] [CrossRef]
- Elson-Schwab, L.; Garner, O.B.; Schuksz, M.; Crawford, B.E.; Esko, J.D.; Tor, Y. Guanininylated neomycin delivers large, bioactive cargo into cells through a heparin sulfate-dependent pathway. J. Biol. Chem. 2007, 282, 13585–13591. [Google Scholar] [CrossRef]
- Wexselblatt, E.; Esko, J.D.; Tor, Y. Gneosomes: Highly lysosomotropic nanoassemblies for lysosomal delivery. ACS Nano 2015, 9, 3961–3968. [Google Scholar] [CrossRef]
- Sganappa, A.; Wexselblatt, E.; Bellucci, M.C.; Esko, J.D.; Tedeschi, G.; Tor, Y.; Volonterio, A. Dendrimeric guanidinoneomycin for cellular delivery of bio-macromolecules. ChemBioChem 2017, 18, 119–125. [Google Scholar] [CrossRef]
- Evans, H.M.; Ahmad, A.; Slack, N.L.; Safinya, C.R. Enhanced gene delivery using cholesterol and dope as third components in a ternary lipid system. Biophys. J. 2002, 82, 524. [Google Scholar]
- Sainlos, M.; Hauchecorne, M.; Oudrhiri, N.; Zertal-Zidani, S.; Aissaoui, A.; Vigneron, J.-P.; Lehn, J.-M.; Lehn, P. Kanamycin A-derived cationic lipids as vectors for gene transfection. ChemBioChem 2005, 6, 1023–1033. [Google Scholar] [CrossRef]
- Mével, M.; Sainlos, M.; Chatin, B.; Oudrhiri, N.; Hauchecorne, M.; Lambert, O.; Vigneron, J.-P.; Lehn, P.; Pitard, B.; Lehn, J.-M. Paromomycin and neomycin B derived cationic lipids: Synthesis and transfection studies. J. Control. Release 2012, 158, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, T.; Baussanne, I.; Halder, S.; Carmoy, N.; Montier, T.; Lehn, P.; Décout, J.-L. Synthesis and transfection properties of a series of lipidic neamine derivatives. Bioconjugate Chem. 2009, 20, 2032–2046. [Google Scholar] [CrossRef] [PubMed]
- Riguet, E.; Désiré, J.; Bailly, C.; Décout, J.-L. A route for preparing new neamine derivatives targeting HIV-1TAR RNA. Tetrahedron 2004, 60, 8053–8064. [Google Scholar] [CrossRef]
- Zhang, S.B.; Zhi, D.F.; Huang, L. Lipid-based vectors for siRNA delivery. J. Drug Target. 2012, 20, 724–735. [Google Scholar] [CrossRef]
- Meng, Z.; O’Keeffe-Ahern, J.; Lyu, J.; Pierucci, L.; Zhou, D.; Wang, W. A new developing class of gene delivery: Messenger RNA-based therapeutics. Biomater. Sci. 2017, 5, 2381–2392. [Google Scholar] [CrossRef]
- Desigaux, L.; Sainlos, M.; Lambert, O.; Chevre, R.; Letrou-Bonneval, E.; Vigneron, J.-P.; Lehn, P.; Lehn, J.-M.; Pitard, B. Self-assembled lamellar complexes of siRNA with lipidic aminoglycoside derivatives promote efficient siRNA delivery and interference. Proc. Natl. Acad. Sci. USA 2007, 104, 16534–16539. [Google Scholar] [CrossRef]
- Xu, Y.; Szoka, F.C., Jr. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry 1996, 7, 5616–5623. [Google Scholar] [CrossRef]
- Habrant, D.; Peuziat, P.; Colombani, T.; Dallet, L.; Gehin, J.; Goudeau, E.; Evrard, B.; Lambert, O.; Haudebourg, T.; Pitard, B. Design of ionizable lipids to overcome the limiting step of endosomal escape: Application in the intracellular delivery of mRNA, DNA, and siRNA. J. Med. Chem. 2016, 59, 3046–3062. [Google Scholar] [CrossRef]
- Simberg, D.; Weisman, S.; Talmon, Y.; Barenholz, Y. DOTAP (and other cationic lipids): Chemistry, biophysics, and transfection. Crit. Rev. Ther. Drug Carr. Syst. 2004, 21, 257–317. [Google Scholar] [CrossRef] [PubMed]
- Wetzer, B.; Byk, G.; Frederic, M.; Airiau, M.; Blanche, F.; Pitard, P.; Scherman, D. Reducible cationic lipids for gene transfer. Biochem. J. 2001, 356, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Pezzoli, D.; Candiani, G. Non-viral gene delivery strategies for gene therapy: A ‘‘ménage á trois’’ among nucleic acids, materials, and the biological environment. J. Nanopart. Res. 2013, 15, 1523–1550. [Google Scholar] [CrossRef]
- Ewert, K.K.; Zidovska, A.; Ahmad, A.; Bouxsein, N.F.; Evans, H.M.; McAllister, C.S.; Samuel, C.E.; Safinya, C.R. Cationic liposome–nucleic acid complexes for gene delivery and silencing: Pathways and mechanisms for plasmid DNA and siRNA. In Nucleic Acid Transfection. Topics in Current Chemistry; Bielke, W., Erbacher, C., Eds.; Springe: Berlin/Heidelberg, Germany, 2010; Volume 296. [Google Scholar]
- Mével, M.; Haudebourg, T.; Colombani, T.; Peuziat, P.; Dallet, L.; Chatin, B.; Lambert, O.; Berchel, M.; Montier, T.; Jaffrès, P.-A.; et al. Important role of phosphoramido linkage inimidazole-based dioleyl helper lipids for liposomestability and primary cell transfection. J. Gene Med. 2016, 18, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Colombani, T.; Peuziat, P.; Dallet, L.; Haudebourg, T.; Mével, M.; Berchel, M.; Lambert, O.; Habrant, D.; Pitard, B. Self-assembling complexes between binary mixtures of lipids with different linkers and nucleic acids promote universal mRNA, DNA, and siRNA delivery. J. Control. Release 2017, 248, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Love, K.T.; Mahon, K.P.; Levins, C.G.; Whitehead, K.A.; Querbes, W.; Dorkin, J.R.; Qin, J.; Cantley, W.; Qin, L.L.; Racie, T.; et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. USA 2010, 107, 1864–1869. [Google Scholar] [CrossRef]
- Altinoglu, S.; Wang, M.; Xu, Q.B. Combinatorial library strategies for synthesis of cationic lipid-like nanoparticles and their potential medical applications. Nanomedicine 2015, 14, 643–657. [Google Scholar] [CrossRef]
- Zhang, Y.; Pelet, J.M.; Heller, D.A.; Dong, Y.; Chen, D.; Gu, Z.; Joseph, B.J.; Wallas, J.; Anderson, D.G. Lipid-modified aminoglycosides derivatives for in vivo siRNA delivery. Adv. Mater. 2013, 25, 4641–4645. [Google Scholar] [CrossRef]
- Yu, X.; Liu, S.; Cheng, Q.; Wei, T.; Lee, S.; Zhang, D.; Siegward, D.J. Lipid-modified aminoglycosides for mRNA delivery to the liver. Adv. Healthc. Mater. 2020, 9, e1901487. [Google Scholar] [CrossRef]
- Iwata, R.; Nakayama, F.; Hirochi, S.; Sato, K.; Piao, W.; Nishima, K.; Yokota, T.; Wada, T. Synthesis and properties of vitamin E analog-conjugated neomycin for delivery of RNAi drugs to liver cells. Bioorg. Med. Chem. Lett. 2015, 25, 815–819. [Google Scholar] [CrossRef]
- Yadav, S.; Mahato, M.; Pathak, R.; Jha, D.; Kumar, B.; Deka, S.R.; Gautam, H.K.; Sharma, A.K. Multifunctional self-assembled cationic peptide nanostructures efficiently carry plasmid DNA in vitro and exhibit antimicrobial activity with minimal toxicity. J. Mater. Chem. B 2014, 2, 4848–4861. [Google Scholar] [CrossRef] [PubMed]
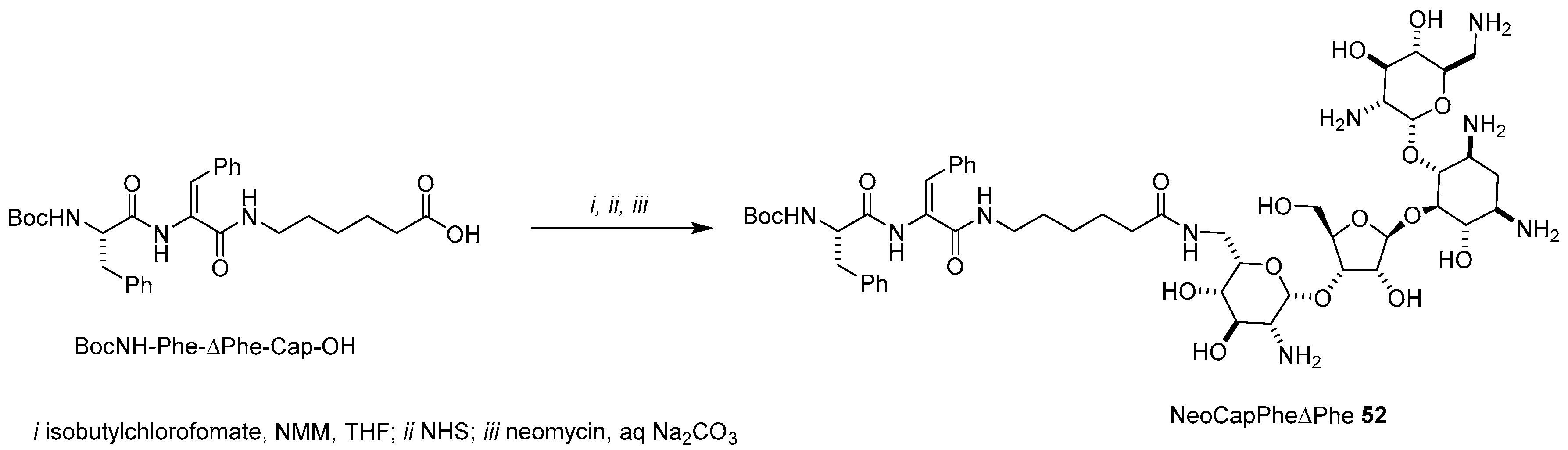
- Although the authors claimed that NeoCapPheΔPhe 47 was the only product obtained (they did not mention any HPLC purification), we think that a mixture of neomycin conjugates would have been likely recovered by coupling unprotected neomycin with NHS activated esters without any further chromatographic separation.
- Mueller, C.; Despras, G.; Lindhorst, T.K. Organizing multivalency in carbohydrate recognition. Chem. Soc. Rev. 2016, 45, 3275–3302. [Google Scholar] [CrossRef] [PubMed]
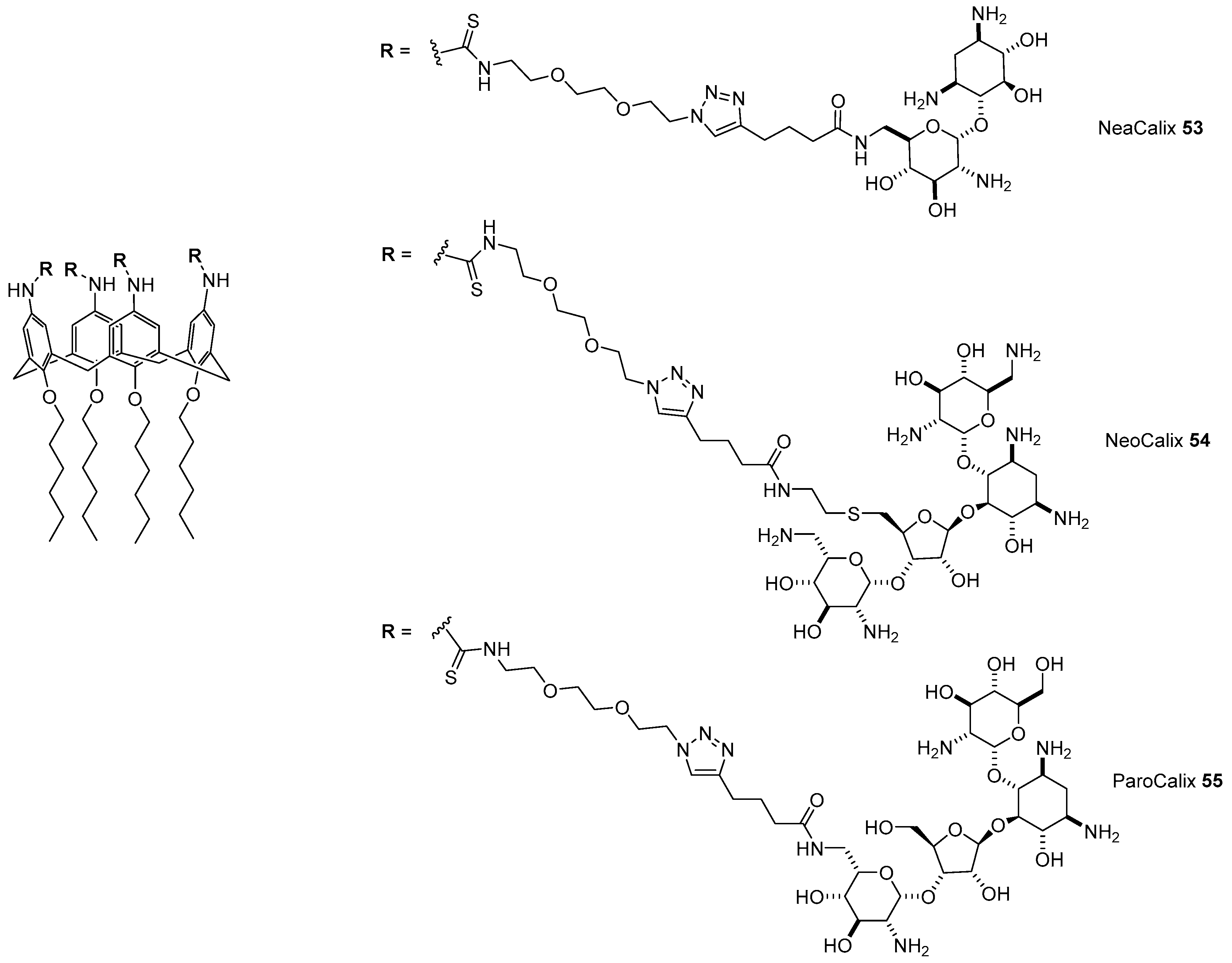
- Bagnacani, V.; Franceschi, V.; Fantuzzi, L.; Casnati, A.; Donofrio, G.; Sansone, F.; Ungaro, R. Lower Rim Guanidinocalix[4]arenes: Macrocyclic Nonviral Vectors for Cell Transfection. Bioconjug. Chem. 2012, 23, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Bagnacani, V.; Franceschi, V.; Bassi, M.; Lomazzi, M.; Donofrio, G.; Sansone, F.; Casnati, A.; Ungaro, R. Arginine clustering on calix[4]arene macrocycles for improved cell penetration and DNA delivery. Nat. Commun. 2013, 4, 1721–1728. [Google Scholar] [CrossRef] [PubMed]
- Bono, N.; Pennetta, C.; Sganappa, A.; Giupponi, E.; Sansone, F.; Volonterio, A.; Candiani, G. Design and synthesis of biologically active cationic amphiphiles built on the calix[4]arene scaffold. Int. J. Pharm. 2018, 549, 436–445. [Google Scholar] [CrossRef]
- Lächelt, U.; Wagner, E. Nucleic Acid Therapeutics Using Polyplexes: A Journey of 50 Years (and Beyond). Chem. Rev. 2015, 115, 11043–11078. [Google Scholar] [CrossRef]
- Peng, L.; Wagner, E. Polymeric Carriers for Nucleic Acid Delivery: Current Designs and Future Directions. Biomacromolecules 2019, 20, 3613–3626. [Google Scholar] [CrossRef]
- Lai, W.F.; Wong, W.T. Design of Polymeric Gene Carriers for Effective Intracellular Delivery. Trends Biotechnol. 2018, 36, 713–728. [Google Scholar] [CrossRef]
- Li, J.; Liang, H.M.; Liu, J.; Wang, Z.Y. Poly (amidoamine) (PAMAM) dendrimer mediated delivery of drug and pDNA/siRNA for cancer therapy. Int. J. Pharm. 2018, 546, 215–225. [Google Scholar] [CrossRef]
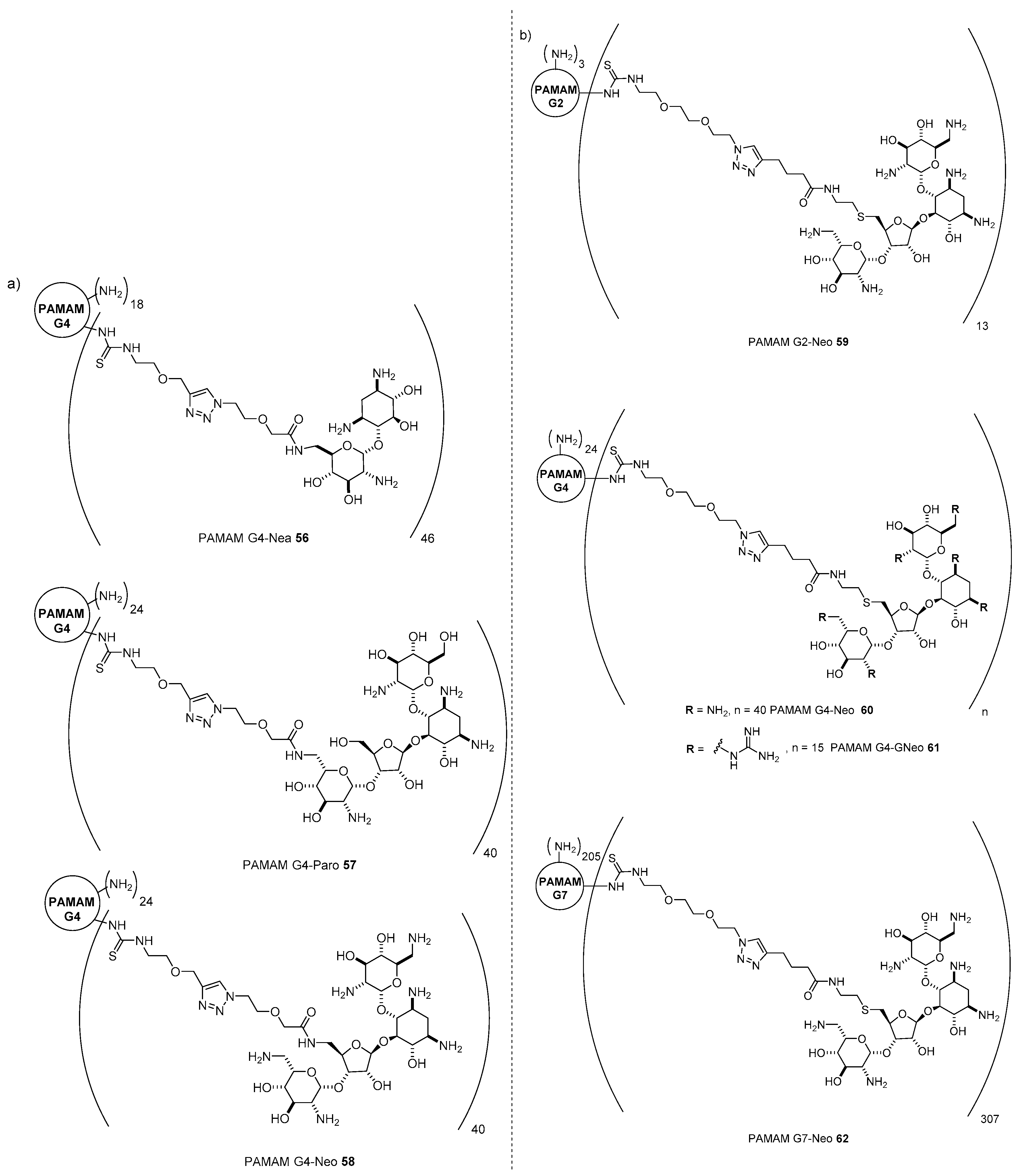
- Ghilardi, A.; Pezzoli, D.; Bellucci, M.C.; Malloggi, C.; Negri, A.; Sganappa, A.; Tedeschi, G.; Candiani, G.; Volonterio, A. Synthesis of multifunctional PAMAM-aminoglycoside conjugates with enhanced transfection efficiency. Bioconjug. Chem. 2013, 24, 1928–1936. [Google Scholar] [CrossRef]
- Bono, N.; Pennetta, C.; Belluci, M.C.; Sganappa, A.; Malloggi, C.; Tedeschi, G.; Candiani, G.; Volonterio, A. Role of generation on successful DNA delivery of PAMAM-(guanidino)neomycin conjugates. ACS Omega 2019, 4, 6796–6807. [Google Scholar] [CrossRef]
- Christensen, E.I.; Birn, H.; Storm, T.; Weyer, K.; Nielsen, R. Endocytic receptors in the renal proximal tubule. Physiology 2012, 27, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Christensen, E.I.; Birn, H. Megalin and cubilin in proximal tubule protein reabsorption: From experimental models to human disease. Kidney Int. 2016, 89, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.M.; Noolvi, M.N.; Sharma, P.; Jaiswal, V.; Bansal, S.; Lohan, S.; Kumar, S.S.; Abbot, V.; Dhiman, S.; Bhardwaj, V. Quantitative structure-activity relationship (QSAR) studies as strategic approach in drug discovery. Med. Chem. Res. 2014, 23, 4991–5007. [Google Scholar] [CrossRef]
- Potta, T.; Zhen, Z.; Grandhi, T.S.P.; Christensen, M.D.; Ramos, J.; Breneman, C.M.; Rege, K. Discovery of antibiotics-derived polymers for gene delivery using combinatorial synthesis and cheminformatics modeling. Biomaterials 2014, 35, 1977–1988. [Google Scholar] [CrossRef]
- Miryala, B.; Zhen, Z.; Potta, T.; Breneman, C.M.; Rege, K. Parallel synthesis and quantitative structure-activity relathionship (QSAR) modelling of aminoglycoside-derived lipopolymers for transgene ecpression. ACS Biomater. Sci. Eng. 2015, 1, 656–668. [Google Scholar] [CrossRef]
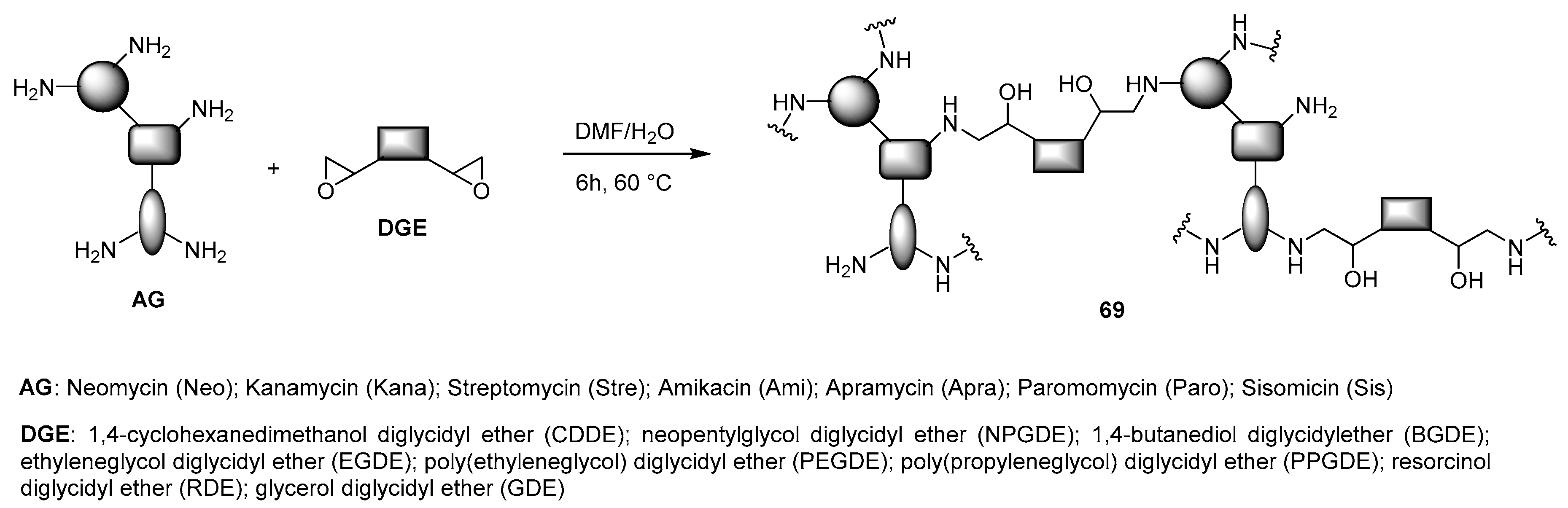
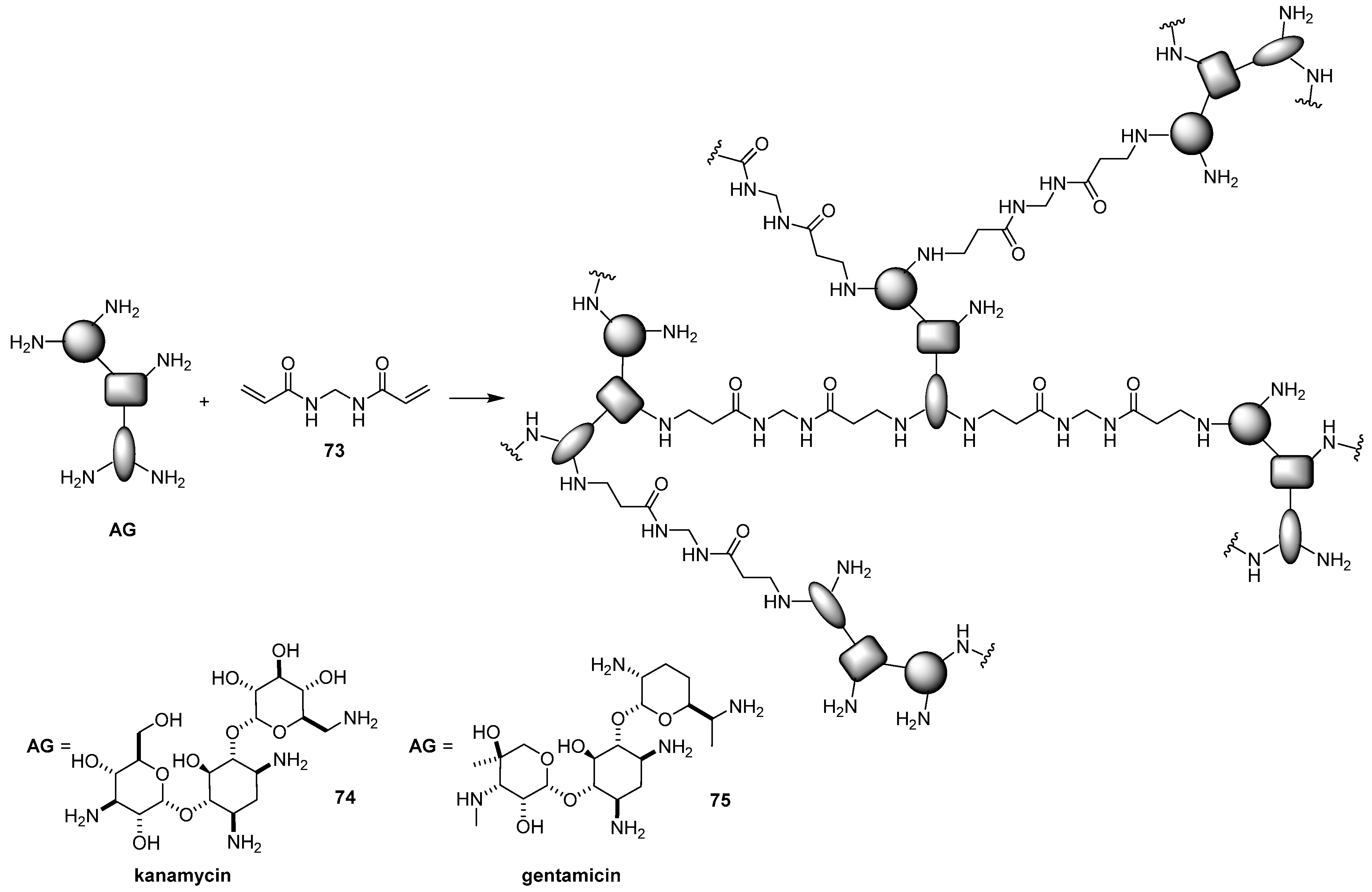
- Chen, M.; Wu, J.; Zhou, L.; Jin, C.; Tu, C.; Zhu, B.; Wu, F.; Zhu, Q.; Zhu, X.; Yan, D. Hyperbranched glycoconjugated polymers from natural small molecule kanamycin as safe and efficient gene vector. Polym. Sci. 2011, 2, 2674–2682. [Google Scholar] [CrossRef]
- Chen, M.; Hu, M.; Wang, D.; Wang, G.; Zhu, X.; Yan, D.; Sun, J. Multifunctional hyperbranched glycoconjugated polymers based on natural aminoglycosides. Bioconjug. Chem. 2012, 23, 1189–1199. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellucci, M.C.; Volonterio, A. Aminoglycosides: From Antibiotics to Building Blocks for the Synthesis and Development of Gene Delivery Vehicles. Antibiotics 2020, 9, 504. https://doi.org/10.3390/antibiotics9080504
Bellucci MC, Volonterio A. Aminoglycosides: From Antibiotics to Building Blocks for the Synthesis and Development of Gene Delivery Vehicles. Antibiotics. 2020; 9(8):504. https://doi.org/10.3390/antibiotics9080504
Chicago/Turabian StyleBellucci, Maria Cristina, and Alessandro Volonterio. 2020. "Aminoglycosides: From Antibiotics to Building Blocks for the Synthesis and Development of Gene Delivery Vehicles" Antibiotics 9, no. 8: 504. https://doi.org/10.3390/antibiotics9080504
APA StyleBellucci, M. C., & Volonterio, A. (2020). Aminoglycosides: From Antibiotics to Building Blocks for the Synthesis and Development of Gene Delivery Vehicles. Antibiotics, 9(8), 504. https://doi.org/10.3390/antibiotics9080504





