Profile of Enterobacteria Resistant to Beta-Lactams
Abstract
1. Introduction
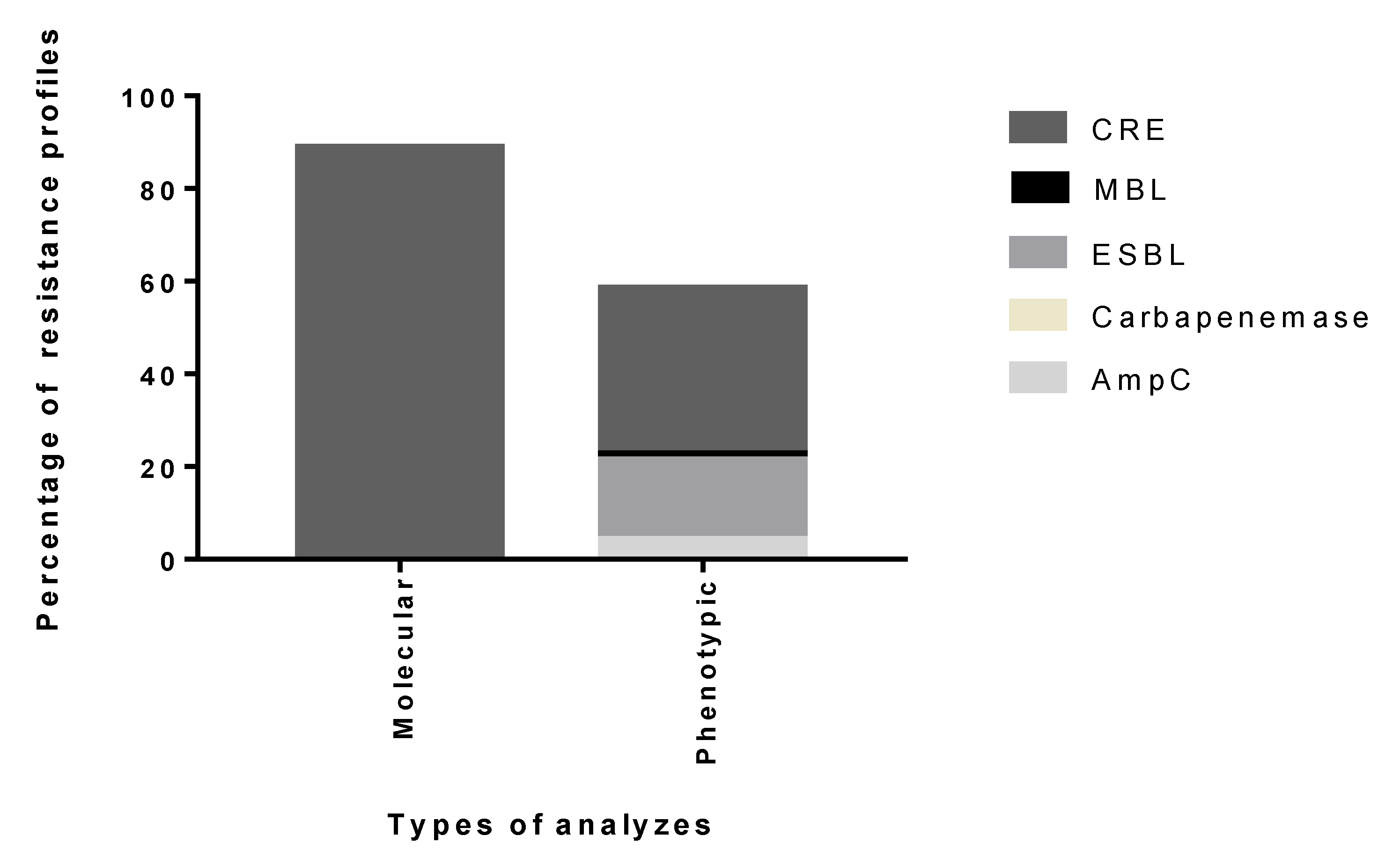
2. Results
3. Discussion
4. Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mlaga, K.D.; Lotte, R.; Montaudié, H.; Rolain, J.-M.; Ruimy, R. ‘Nissabacter archeti’ gen. nov., sp. nov., a new member of Enterobacteriaceae family, isolated from human sample at Archet 2 Hospital, Nice, France. New Microbes New Infect. 2017, 17, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Winn, W.C.; Allen, S.D.; Janda, W.M.; Koneman, S. Diagnóstico Microbiológico: Texto e Atlas colorido. Guanab. Koogan. 2008, 16, 1–1760. [Google Scholar]
- Coque, T.; Baqueiro, F.; Canton, R. Increasing prevalence of ESBL- producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 1–11. [Google Scholar]
- Lutgring, J.D.; Limbagob, B.M. The Problem of Carbapenemase-Producing-Carbapenem-Resistant Enterobacteriaceae Detection. J. Clin. Microbiol. 2016, 54, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Vega, S.; Dowzicky, M.J. Antimicrobial susceptibility among Gram-positive and Gram-negative organisms collected from the Latin American region between 2004 and 2015 as part of the Tigecycline Evaluation and Surveillance Trial. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 1–16. [Google Scholar] [CrossRef]
- Ozsurekci, Y.; Aykac, K.; Cengiz, A.B.; TanırBasaranoglu, S.; Sancak, B.; Karahan, S.; Kara, A.; Ceyhan, M. Bloodstream infections in children caused by carbapenem-resistant versus carbapenem-susceptible gram-negative microorganisms: Risk factors and outcome. Diagn. Microbiol. Infect. Dis. 2017, 87, 359–364. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Tang, L.K.; Caffrey, P.N.; Nóbrega, B.D.; Cork, C.S.; Ronksley, E.P.; Barkema, W.H.; Polachek, J.A.; Ganshorn, H.; Sharma, N.; Kellner, D.J.; et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, 316–327. [Google Scholar] [CrossRef]
- Bougnom, P.B.; Piddock, J.L. Wastewater for urban agriculture. A significant factor in dissemination of antibiotic resistance. Environ. Sci. Technol. 2017, 51, 5863–5864. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority) and ECDC (European Centre for Disease Prevention and Control). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2017. EFSA J. 2019, 17, 278. [Google Scholar] [CrossRef]
- Rossoline, G.M.; Arena, F.; Pecile, P.; Pollini, S. Update on the antibiotic resistance crisis. Curr. Opin. Pharmacol. 2014, 18, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.E.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Microbiology 2011, 18, 268–281. [Google Scholar]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. Suppl. 2004, 10, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Koraimann, G. Spread and Persistence of Virulence and Antibiotic Resistance Genes: A Ride on the F Plasmid Conjugation Module. EcoSal Plus 2018, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Chant, C.; Leung, A.; Friedrich, J.O. Optimal dosing of antibiotics in critically ill patients by using continuous/extended infusions: A systematic review and metaanalysis. Crit. Care 2013, 17, R279. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, L.; Gonzáles-López, J.J. Carbapenemases in Enterobacteriaceae: Types and molecular epidemiology. Enferm. Infecc. y Microbiol. Clin. 2014, 32, 4–9. [Google Scholar] [CrossRef]
- Bertoncheli, C.D.M.; Hörner, R. Uma revisão sobre metalo-β-lactamases. Rev. Bras. de Ciências Farm. 2008, 44, 577–599. [Google Scholar] [CrossRef]
- Lekshmi, P.N.C.J.; Sumi, B.; Viveka, S.; Jeeva, S.; Brindha, R. Antibacterial activity of nanoparticles from Allium sp. J. Microbiol. Biotechnol. Res. 2012, 2, 115–119. [Google Scholar]
- Balouiri, M.S.; Moulay, I.; Saad, K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Tuite, N.; Reddington, K.; Barry, T.; Zumla, A.; Enne, V. Rapid nucleic acid diagnostics for the detection of antimicrobial resistance in Gram-negative bacteria: Is it time for a paradigm shift. J. Antimicrob. Chemother. 2014, 69, 1729–1733. [Google Scholar] [CrossRef]
- Jabur, A.P.L.; Magalhães, L.G.; Borges, A.A.; Cardoso, A.L. Infecção urinária em gestantes atendidas em um laboratório de análises clínicas de Goiânia-GO ente 2012 e 2013. Estudos 2014, 41, 637–641. [Google Scholar]
- Agência Nacional de Vigilância Sanitária. Boletim de Segurança do Paciente e Qualidade em Serviços de Saúde nº 16 (Corrigido). 2017. Available online: https://www20.anvisa.gov.br/segurancadopaciente/index.php/publicacoes/item/boletim-seguranca-do-paciente-e-qualidade-em-servicos-de-saude-n-16-avaliacao-dos-indicadores-nacionais-das-infeccoes-relacionadas-a-assistencia-a-saude-iras-e-resistencia-microbiana-do-ano-de-2016 (accessed on 19 September 2018).
- Logan, L.K.; Weinstein, R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- Secchyna, F.; Gaur, R.; Sandlund, J.; Truong, C.; Tremintin, G.; Küeltz, D.; Gomez, C.; Tamburini, F.B.; Andermann, T.M.; Bhatt, A.; et al. Diverse Mechanisms of Resistance in Carbapenem-Resistant Enterobacteriaceae at a Health Care System in Silicon Valley, California. BioRxiv 2018. [Google Scholar] [CrossRef]
- Neto, M.A.R.; Rios, V.M.; Corá, L.F.; Fonseca, M.M.; Ferreira-Paim, K.; Fonseca, F.M. High rates of antimicrobial resistance of ESBL-producing Enterobacteriaceae isolated from clinical samples in Northeast of Brazil. Infect. Dis. 2018, 50, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Agência Nacional de Vigilância Sanitária. Nota Técnica Nº 01/2013:Medidas de Prevenção e Controle de Infecções por Enterobactérias Multiresistentes; Agência Nacional de Vigilância Sanitária: Brasília, Brazil, 2013. [Google Scholar]
- Wendel, A.F.; Brodner, A.H.B.; Wydra, S.; Ressina, S.; Henrich, B.; Pfeffer, K.; Toleman, M.A.; MacKenziea, C.R. Genetic Characterization and Emergence of the Metallo- β-Lactamase GIM-1 in Pseudomonas spp. and Enterobacteriaceae during a Long-Term Outbreak. Antimicrob. Agents Chemother. 2013, 57, 5162–5165. [Google Scholar] [CrossRef]
- Pfeifer, Y.; Cullik, A.; Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med Microbiol. 2010, 300, 371–379. [Google Scholar] [CrossRef]
- Verdet, C.; Benzerara, Y.; Gautier, V.; Adam, O.; Ould-Hocine, Z.; Arlet, G. Emergence of DHA-1-Producing Klebsiella spp. in the Parisian Region: Genetic Organization of the ampC and ampR Genes Originating from Morganella morganii. Antimicrob. Agents Chemother. 2006, 50, 607–617. [Google Scholar] [CrossRef]
- Kjeldsen, T.S.B.; Overgaard, M.; Nielsen, S.S.; Bortolaia, V.; Jelsbak, L.; Sommer, M.; Guardabassi, L.; Olsen, J.E. CTX-M-1 β-lactamase expression in Escherichia coli is dependent on cefotaxime concentration, growth phase and gene location. J. Antimicrob. Chemother. 2015, 70, 62–70. [Google Scholar] [CrossRef]
- Warjri, I.; Dutta, T.K.; Lalzampuia, H.; Chandra, R. Detection and characterization of extended-spectrum β-lactamases (blaCTX-M-1 and blaSHV) producing Escherichia coli, Salmonella spp. and Klebsiella pneumoniae isolated from humans in Mizoram. Vet. World 2015, 8, 599–604. [Google Scholar] [CrossRef]
- Machuca, J.; Agüerob, J.; Mirób, E.; Conejob, M.C.; Oteob, J.; Boub, G.; González-Lópezb, J.J.; Oliverb, A.; Navarrob, F.; Pascuala, A.; et al. Prevalencia en España de mecanismos de resistencia a quinolonas enenterobacterias productoras de betalactamasas de clase C adquiridasy/o carbapenemasas. Enferm. Infecc. y Microbiol. Clínica 2017, 35, 487–492. [Google Scholar] [CrossRef]
- Pietscha, M.; Eller, C.; Wendtc, C.; Holfelderc, M.; Falgenhauerd, L.; Fruthe, A.; Grössla, T.; Leistnerf, R.; Valenzag, G.; Wernera, G.; et al. Molecular characterisation of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolates from hospital and ambulatory patients in Germany. Vet. Microbiol. 2017, 200, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, P.; Wei, D.; Liu, Y.; Wan, L.; Xiang, T.; Zhang, Y. Clinical isolates of uropathogenic Escherichia coli ST131 producing NDM-7 metallo-β-lactamase in China. Int. J. Antimicrob. Agents 2016, 48, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. A resurgence of -lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int. J. Antimicrob. Agents 2015, 46, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Chen, H.; Yang, Q.; Chen, M.; Wang, H. High Prevalence of Plasmid-Mediated Quinolone Resistance Genes qnr and aac(6)-Ib-cr in Clinical Isolates of Enterobacteriaceae from Nine Teaching Hospitals in China. Antimicrob. Agents Chemother. 2008, 52, 4268–4273. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a New Metallo--Lactamase Gene, blaNDM-1, and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Sampaio, J.L.M.; Gales, A.C. Antimicrobial resistance in Enterobacteriaceae in Brazil: Focus on β-lactams and polymyxins. Braz. J. Microbiol. 2016, 47, 31–37. [Google Scholar] [CrossRef]
- Singh-Moodley, A.; Perovic, O. Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BioMed Cent. Infect. Dis. 2016, 16, 1–10. [Google Scholar] [CrossRef]
- Magagnin, C.M.; Rozales1, F.P.; Antochevis, L.; Nunes, L.S.; Martins, A.S.; Barth, A.L.; Sampaio, J.M.; Zavascki, A.P. Dissemination of blaOXA-370 gene among several Enterobacteriaceae species in Brazil. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1907–1910. [Google Scholar] [CrossRef]
- Kayama, S.; Ohge, H.; Sugai, M. Rapid discrimination of blaIMP-1, blaIMP-6, and blaIMP-34 using a multiplex PCR. J. Microbiol. Methods 2017, 135, 8–10. [Google Scholar] [CrossRef]
- Duin, D.V.; Doi, Y. The global epidemiology of carbapenemaseproducing Enterobacteriaceae. Virulence 2017, 8, 460–469. [Google Scholar] [CrossRef]
- Jeong, S.H.; Kim, H.; Kim, J.; Shin, D.H.; Kim, H.S.; Park, M.; Shin, S.; Hong, J.S.; Lee, S.S.; Song, W. Prevalence and Molecular Characteristics of Carbapenemase-Producing Enterobacteriaceae From Five Hospitals in Korea. Ann. Lab. Med. 2016, 36, 529–535. [Google Scholar] [CrossRef]
- Ingti, B.; Paul, D.; Maurya, A.P.; Bora, D.; Chanda, D.D.; Chakravarty, A.; Bhattacharjee, A. Occurrence of blaDHA-1 mediated cephalosporin resistance in Escherichia coli and their transcriptional response against cephalosporin stress: A report from India. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 1–8. [Google Scholar] [CrossRef]
- Srisrattakarn, A.; Lulitanond, A.; Wilailuckana, C.; Charoensri, N.; Wonglakorn, L.; Saenjamla, P.; Chaimanee, P.; Daduang, J.; Chanawong, A. Rapid and simple identification of carbapenemase genes, blaNDM, blaOXA-48, blaVIM, blaIMP-14 and blaKPC groups, in Gram-negative bacilli by in-house loop-mediated isothermal amplification with hydroxynaphthol blue dye. World J. Microbiol. Biotechnol. 2017, 33, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Ayala, A.T.; Acuña, H.M.B.; Calvo, M.T.A.; Morales, J.L.V.; Chacón, E.C. Emergencia de β-lactamasa AmpC plasmídica del grupo CMY-2 en Shigella sonnei y Salmonella spp. en Costa Rica, 2003–2015. Pan Am. J. Public Health 2016, 40, 70–75. [Google Scholar]
- Suwantarat, N.; Logan, L.K.; Carroll, K.C.; Bonomo, R.A.; Simner, P.J.; Rudin, S.D.; Milstone, A.M.; Tekle, T.; Ross, T.; Tamma, P.D. The Prevalence and Molecular Epidemiology of Multidrug-Resistant Enterobacteriaceae Colonization in a Pediatric Intensive Care Unit. Infect. Control Hosp. Epidemiol. 2016, 37, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.C.; Lincopan, N. Epidemiologia das betalactamases de espectro estendido no Brasil: Impacto clínico e implicações para o agronegócio. J. Bras. de Patol. e Med. Lab. 2012, 48, 91–99. [Google Scholar] [CrossRef]
- Li, X.; Plésiat, P.; Nikaidoc, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Bialvaei, A.Z.; Kafil, H.S.; Asgharzadeh, M.; Memar, M.Y.; Yousefi, M. Current methods for the identification of carbapenemases. J. Chemother. 2016, 28, 1–19. [Google Scholar] [CrossRef]
- Xia, J.; Gao, J.; Tang, W. Nosocomial infection and its molecular mechanisms of antibiotic resistance. Biosci. Trends. 2016, 10, 14–21. [Google Scholar] [CrossRef]
- Jacoby, G.A. AmpC Beta-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef]
- Rood, I.G.H.; Li, Q. Review: Molecular detection of extended spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae in a clinical setting. Diagn. Microbiol. Infect. Dis. 2017, 89, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Jones-Dias, D.; Manageiro, V.; Caniça, M. Influence of agricultural practice on mobile bla genes: IncI1-bearing CTX-M, SHV, CMY and TEM in Escherichia coli from intensive farming soils. Environ. Microbiol. 2016, 18, 260–272. [Google Scholar] [CrossRef]
- Bush, K. Overcoming β-lactam resistance in Gram-negative pathogens. Future Med. Chem. 2016, 8, 921–924. [Google Scholar] [CrossRef]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. 2011, 9, 895–896. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated Functional Classification of B-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. The ABCD’s of b-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Page, M.G.P.; Bush, K. Discovery and development of new antibacterial agents targeting Gram-negative bacteria in the era of pandrug resistance: Is the future promising? ScienceDirect 2014, 18, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Wan, L.; Yu, Y.; Xu, Q.; Deng, Q.; Cao, X.; Liu, Y. Characterization of Extended-Spectrum β-Lactamase, Carbapenemase, and Plasmid Quinolone Determinants in Klebsiella pneumoniae Isolates Carrying Distinct Types of 16S rRNA Methylase Genes, and Their Association with Mobile Genetic Elements. Microb. Drug Resist. 2015, 21, 186–193. [Google Scholar] [CrossRef]
- Rocha, D.A.C.; Campos, J.C.; Passadore, L.F.; Sampaio, S.C.F.; Nicodemo, A.C.; Sampaio, J.L.M. Frequency of Plasmid-Mediated AmpC β-Lactamases in Escherichia coli Isolates from Urine Samples in São Paulo, Brazil. Microb. Drug Resist. 2016, 22, 321–327. [Google Scholar] [CrossRef]
- Alekshun, M.N.; Levy, S.B. Molecular Mechanisms of Antibacterial Multidrug Resistance. Cell 2007, 128, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Brandt, C.; Braun, S.D.; Stein, C.; Slickers, P.; Ehricht, R.; Pletz, M.W.; Makarewicz, O. In silico serine β-lactamases analysis reveals a huge potential resistome in environmental and pathogenic species. Nature 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Olsen, I. New promising β-lactamase inhibitors for clinical use. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Papp-Wallace, K.M.; Sendi, P.; Bonomo, R.A.; Endimiani, A. Non-Phenotypic Tests to Detect and Characterize Antibiotic Resistance Mechanisms in Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2013, 77, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Rolain, J.-M. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 2014, 20, 831–838. [Google Scholar] [CrossRef]
- Poirel, L.; Wenger, A.; Bille, J.; Bernabeu, S.; Naas, T.; Nordmann, P. SME-2-Producing Serratia marcescens Isolate from Switzerland. Antimicrob. Agents Chemother. 2007, 51, 2282–2283. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile -Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Meini, M.; Llarrull, L.I.; Vila, A.J. Overcoming differences: The catalytic mechanism of metallo-β-lactamases. FEBS Lett. 2015, 589, 3419–3432. [Google Scholar] [CrossRef]
- Mlynarcik, P.; Roderova, M.; Kolar, M. Primer Evaluation for PCR and its Application for Detection of Carbapenemases in Enterobacteriaceae. Jundishapur J. Microbiol. 2016, 9, 1–6. [Google Scholar] [CrossRef]
- Pascual, A.; Pintadoc, V.; Rodríguez-Bañoa, J.; Miró, J.M. Carbapenemase-producing Enterobacteriaceae: The end of the antibiotic era? Enferm. Infecc. y Microbiol. Clínica 2014, 32, 1–3. [Google Scholar] [CrossRef]
- Bonelli, R.R.; Moreira, B.M.; Picão, R.C. Antimicrobial resistance among Enterobacteriaceae in South America: History, current dissemination status and associated socioeconomic factors. Drug Resist. Updates 2014, 17, 24–36. [Google Scholar] [CrossRef]
- Bratu, S.; Landman, D.; Haag, R.; Recco, R.; Eramo, A.; Alam, M.; Quale, J. Rapid spread of carbapenem resistant Klebsiella pneumoniae in New York City: A new threat to our antibiotic armamentarium. Arch Intern. Med. 2005, 165, 1430–1435. [Google Scholar] [CrossRef] [PubMed]
- Bush, K. Proliferation and significance of clinically relevant β-lactamases. Ann. N. Y. Acad. Sci. 2013, 1277, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Pucci, M.J. New antimicrobial agents on the horizon. Biochem. Pharmacol. 2011, 82, 1528–1539. [Google Scholar] [CrossRef]
- Clark, J.D.; Catro, J.P.F.; Compton, C.; Lee, H.; Nunery, W. Orbital cellulitis and corneal ulcer due to Cedecea: First reported case and review of the literature. Orbit 2016, 35, 140–143. [Google Scholar] [CrossRef]
- Cuzon, G.; Naas, T.; Correa, A.; Quinn, J.P.; Villegas, M.V.; Nordmann, P. Dissemination of the KPC-2 carbapenemase in non-Klebsiella pneumoniae enterobacterial isolates from Colombia. Int. J. Antimicrob. Agents 2013, 42, 59–62. [Google Scholar] [CrossRef]
- Falcone, M.; Mezzatesta, M.L.; Perilli, M.; Forcella, C.; Giordano, A. Infections with VIM-1 metallo βlactamase-producing Enterobacter cloacae and their correlation with clinical outcome. J. Clin. Microbiol. 2009, 47, 3514–3519. [Google Scholar] [CrossRef]
- Gaynes, R.; Edwards, J.R. Overview of Nosocomial Infections Caused by Gram-Negative Bacili. Clin. Infect. Dis. 2005, 41, 848–854. [Google Scholar]
- Heggendorm, L.H.; Gomes, S.W.C.; Silva, N.A.; Varges, R.G.; Póvoa, H.C.C. Epidemiological profile and antimicrobial susceptibility of microorganisms isolated from nosocomial infections. Rev. Saúde e Meio Ambiente RESMA 2016, 2, 26–47. [Google Scholar]
- Jácome, P.R.L.A.; Alves, L.R.; Jácome-Júnior, A.T.; Silva, M.J.B.; Lima, J.L.C.; Araújo, P.S.R.; Lopes, A.C.S.; Maciel, M.A.V. Detection of bla SPM-1, bla KPC, bla TEM and bla CTX-M genes in isolates of Pseudomonas aeruginosa, Acinetobacter spp. and Klebsiella spp. from cancer patients with healthcare-associated infections. J. Med. Microbiol. 2016, 65, 658–665. [Google Scholar] [CrossRef]
- Kordon, A.O.; Abdelhamed, H.; Ahmed, H.; Park, J.Y.; Karsi, A.; Pinchuk, L.M. Phagocytic and bactericidal properties of channel catfish peritoneal macrophages exposed to Edwardsiella ictaluri live attenuated vaccine and wild-type strains. Front. Microbiol. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lavagnoli, L.S.; Bassetti, B.R.; Kaiser, T.D.L.; Kutz, K.M.; Junior, C.C. Factors associated with acquisition of carbapenem-resistant Enterobacteriaceae. Rev. Lat. Am. Enfermage 2017, 25, 1–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Manageiro, V.; Ferreira, E.; Rodrigues, J.; Sampaio, D.A.; Vieira, L.; Pereira, P.; Rodrigues, P.; Palos, C.; Caniça, M. NDM-1-Producing Providencia Stuartii Isolates in a Portuguese Hospital; Réunion Interciciplinaire de Chimiothérapie Anti-Infectieuse: Paris, France, 2015; p. 35. [Google Scholar]
- Pontes, D.S.; Araujo, R.S.A.; Dantas, N.; Scott, L.; Scott, M.T.; Moura, R.O.; Mendonça-Junios, F.J.B. Genetic Mechanisms of Antibiotic Resistance and the Role of Antibiotic Adjuvants. Curr. Top. Med. Chem. 2018, 18, 42–74. [Google Scholar] [CrossRef]
- Assawatheptawee, K.; Tansawai, U.; Kiddee, A.; Thongngen, P.; Punyadi, P.; Romgaew, T.; Kongthai, P.; Sumpradit, T.; Niumsup, P.R. Occurrence of Extended-Spectrum and AmpC-Type β-Lactamase Genes in Escherichia coli Isolated from Water Environments in Northern Thailand. Microbes Environ. 2017, 32, 293–296. [Google Scholar] [CrossRef]
- Dixon, N.; Fowler, R.C.; Yoshizumi, A.; Horiyama, T.; Ishii, Y.; Harrison, L.; Geyer, C.N.; Moland, E.S.; Thomson, K.; Hanson, N.D. IMP-27: A Unique Metallo-β-Lactamase Identified in Geographically Distinct Isolates of Proteus mirabilis. Antimicrob. Agents Chemother. 2016, 60, 6418–6421. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.A.; Wang, W.; Sun, C.; Gu, L.; Liu, Z.; Yu, T.; Ahmad, Y.; Jiang, Z.; Hou, J. Molecular Characterization of Pathogenic Salmonella Spp From Raw Beef In Karachi, Pakistan. Antibiotics 2020, 73, 1–16. [Google Scholar]
- Song, W.; Lee, K.M.; Kim, H.S.; Kim, J.S.; Kim, J.; Jeons, S.H.; Roh, K.H. Clonal spread of both oxyimino-cephalosporin- and cefoxitin-resistant Klebsiella pneumoniae isolates co-producing SHV-2a and DHA-1 -lactamase at a burns intensive care unit. Int. J. Antimicrob. Agents 2006, 28, 520–524. [Google Scholar] [CrossRef]
- Sugumar, M.; Kumar, K.M.; Manoharan, A.; Anbarasu, A.; Ramaiah, S. Detection of OXA-1 b-Lactamase Gene of Klebsiella pneumoniae from Blood Stream Infections (BSI) by Conventional PCR and In-Silico Analysis to Understand the Mechanism of OXA Mediated Resistance. PLoS ONE 2014, 9, 1–8. [Google Scholar] [CrossRef]
- Cubero, M.; Calatayud, L.; Tubau, F.; Ayats, J.; Peña, C.; Martín, R.; Liñares, J.; Domínguez, M.A.; Ardanuy, C. Clonal spread of Klebsiella pneumoniae producing OXA-1 betalactamase in a Spanish hospital. Int. Microbiol. 2013, 16, 227–233. [Google Scholar]
- Mahalleh, R.G.D.; Dahmardeh, J.; Rad, N.S. The frequency of bla Verona imipenemase and bla imipenemase genes in clinical isolates of Pseudomonas aeruginosa in therapeutic centers of Zahedan. Immunopathol. Persa 2018, 4, e31. [Google Scholar] [CrossRef]
- Shi, W.F.; Li, K.; Ji, Y.; Jiang, Q.B.; Wang, Y.Y.; Shi, M.; Mi, Z.H. Carbapenem and cefoxitin resistance of Klebsiella pneumoniae strains associated with porin OmpK36 loss and DHA-1 beta-lactamase production. Braz. J. Microbiol. 2013, 44, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, A.A.; Abdelaziz, N.A.; Amin, M.A.; Aziz, R.K. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci. Rep. 2019, 9, 4224. [Google Scholar] [CrossRef] [PubMed]
- Hajjej, Z.; Gharsallah, H.; Naija, H.; Boutiba, I.; Labbene, I.; Ferjani, M. Idcases. Successful treatment of a Carbapenem-resistant Klebsiella pneumoniae carrying bla(OXA-48), bla(VIM-2), bla(CMY-2) and bla(SHV-) with high dose combination of imipenem and amikacin. Idcases 2016, 4, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Pournajaf, A.; Rajabnia, R.; Razavi, S.; Solgi, S.; Ardebili, A.; Yaghoubi, S.; Khodabandeh, M.; Yahyapour, Y.; Emadi, B.; Irajian, G. Molecular characterization of carbapenem-resistant Acinetobacter baumannii isolated from pediatric burns patients in an Iranian hospital. Trop. J. Pharm. Res. 2018, 17, 135–141. [Google Scholar] [CrossRef]
- Gajul, S.V.; Mohite, S.T.; Datkhile, K.D.; Kakade, S.V.; Mangalagi, S.S.; Wavare, S.M. Prevalence of Extended Spectrum Beta Lactamase Genotypes in Klebsiella pneumoniae from Respiratory Tract Infections at Tertiary Care Hospital. J. Krishna Inst. Med. Sci. Univ. 2019, 8, 66–75. [Google Scholar]
- Chikwendu, C.I.; Ibe, S.N.; Okpokwasili, G.C. Detection of bla(SHV) and bla(TEM) beta-lactamase genes in multi-resistant Pseudomonas isolates from environmental sources. Afr. J. Microbiol. Res. 2011, 5, 2067–2074. [Google Scholar] [CrossRef]
- Dhanji, H.; Patel, R.; Wall, R.; Doumith, M.; Patel, B.; Hope, R.; Livermore, D.M. Woodford N. Variation in the genetic environments of bla(CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J. Antimicrob. Chemother. 2011, 66, 1005–1012. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhu, X.H.; Hou, H.Y.; Lu, Y.F.; Yu, J.; Mao, L.; Mao, L.Y.; Sun, Z.Y. Characteristics of diarrheagenic Escherichia coli among children under 5 years of age with acute diarrhea: A hospital based study. BMC Infect. Dis. 2018, 18, 63. [Google Scholar] [CrossRef]
- Chen, T.L.; Chang, W.C.; Kuo, S.C.; Lee, Y.T.; Chen, C.P.; Siu, L.K.; Cho, W.L.; Fung, C.P. Contribution of a Plasmid-Borne bla(OXA-58) Gene with Its Hybrid Promoter Provided by IS1006 and an ISAba3-Like Element to beta-Lactam Resistance in Acinetobacter Genomic Species 13TU. Antimicrob. Agents Chemother. 2010, 54, 3107–3112. [Google Scholar] [CrossRef]
- Asgin, N.; Otlu, B.; Cakmakliogullari, E.K.; Celik, B. High prevalence of TEM, VIM, and OXA-2 beta-lactamases and clonal diversity among Acinetobacter baumannii isolates in Turkey. J. Infect. Dev. Ctries. 2019, 13, 794–801. [Google Scholar] [CrossRef]
- Li, J.L.; Ji, X.L.; Deng, X.H.; Zhou, Y.F.; Ni, X.Q.; Liu, X.K. Detection of the SHV genotype polymorphism of the extended-spectrum beta-lactamase-producing Gram-negative bacterium. Biomed. Rep. 2015, 3, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Mathys, D.A.; Mathys, B.A.; Mollenkopf, D.F.; Daniels, J.B.; Wittum, T.E. Enterobacteriaceae Harboring AmpC (bla(CMY)) and ESBL (bla(CTX-M)) in Migratory and Nonmigratory Wild Songbird Populations on Ohio Dairies. Vector-Borne Zoonotic Dis. 2017, 17, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Khari, F.I.M.; Karunakaran, R.; Rosli, R.; Tay, S.T. Genotypic and Phenotypic Detection of AmpC beta-lactamases in Enterobacter spp. Isolated from a Teaching Hospital in Malaysia. PLoS ONE 2016, 11, e0150643. [Google Scholar]
- Pruthvishree, B.S.; Kumar, O.R.V.; Sivakumar, M.; Tamta, S.; Sunitha, R.; Sinha, D.K.; Singh, B.R. Molecular characterization of extensively drug resistant (XDR), extended spectrum beta-lactamases (ESBL) and New Delhi Metallo beta-lactamase-1 (blaNDM1) producing Escherichia coli isolated from a male dog-a case report. Vet. Arhiv 2018, 88, 139–148. [Google Scholar] [CrossRef]
- Kim, Y.T.; Kim, T.U.; Baik, H.S. Characterization of extended spectrum beta-lactamase genotype TEM, SHV, and CTX-M producing Klebsiella pineumoniae isolated from clinical specimens in Korea. J. Microbiol. Biotechnol. 2006, 16, 889–895. [Google Scholar]
- Ma, L.; Siu, L.K.; Lin, J.C.; Wu, T.L.; Fung, C.P.; Wang, J.T.; Lu, P.L.; Chuang, Y.C. Updated molecular epidemiology of carbapenem-non-susceptible Escherichia coli in Taiwan: First identification of KPC-2 or NDM-1-producing E-coli in Taiwan. BMC Infect. Dis. 2013, 13, 599. [Google Scholar] [CrossRef]
- Yang, D.K.; Liang, H.J.; Gao, H.L.; Wang, X.W.; Wang, Y. Analysis of drug-resistant gene detection of blaOXA-like genes from Acinetobacter baumannii. Genet. Mol. Res. 2015, 14, 18999–19004. [Google Scholar] [CrossRef]
- Azzab, M.M.; El-Sokkary, R.H.; Tawfeek, M.M.; Gebriel, M.G. Multidrug-resistant bacteria among patients with ventilator-associated pneumonia in an emergency intensive care unit, Egypt. East. Mediterr. Health J. 2016, 22, 894–903. [Google Scholar] [CrossRef]
- Balkan, I.I.; Aygün, G.; Aydın, S.; Mutcalı, S.I.; Kara, Z.; Kuşkucu, M.; Öztürk, R. Blood stream infections due to OXA-48-like carbapenemase-producing Enterobacteriaceae: Treatment and survival. Int. J. Infect. Dis. 2014, 26, 51–56. [Google Scholar] [CrossRef]
- Kis, Z.; Toth, A.; Janvari, L.; Damjanova, I. Countrywide dissemination of a DHA-1-type plasmid-mediated AmpC beta-lactamase-producing Klebsiella pneumoniae ST11 international high-risk clone in Hungary, 2009–2013. J. Med. Microbiol. 2016, 65, 1020–1027. [Google Scholar] [CrossRef]
- Gugliandolo, A.; Caio, C.; Mezzatesta, M.L.; Rifici, C.; Bramanti, P.; Stefani, S.; Mazzon, E. Successful ceftazidime-avibactam treatment of MDR-KPC-positive Klebsiella pneumoniae infection in a patient with traumatic brain injury A case report. Medicine 2017, 31, e7664. [Google Scholar] [CrossRef] [PubMed]
- Farzana, R.; Shamsuzzaman, S.M.; Mamun, K.Z. Isolation and molecular characterization of New Delhi metallo-beta-lactamase-1 producing superbug in Bangladesh. J. Infect. Dev. Ctries. 2013, 7, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Souli, M.; Galani, I.; Antoniadou, A.; Papadomichelakis, E.; Poulakou, G. An outbreak of infection due to β-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: Molecular characterization, epidemiology, and outcomes. Clin. Infect. Dis. 2010, 50, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Zhou, D.; Wang, Q.; Feng, J.; Feng, W.; Luo, W.; Zhang, D.; Liu, Y.; Qiul, X.; Yin, Z.; et al. The first report of detecting the blaSIM-2gene and determining the complete sequence of the SIM-encoding plasmid. Clin. Microbiol. Infect. 2016, 22, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Toombs-Ruane, L.J.; Benschop, J.; Priest, P.; Murdoch, D.R.; French, N.P. Multidrug resistant Enterobacteriaceae in New Zealand: A current perspective. N. Z. Veter. J. 2017, 65, 62–70. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; Xu, Z.; Ying, C.; Yu, W.; Xiao, Y. Identification of Raoultella terrigena as a Rare Causative Agent of Subungual Abscess Based on 16S rRNA and Housekeeping Gene Sequencing. Can. J. Infect. Dis. Med. Microbiol. 2016, 1, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Marques, J.B.; Bonez, P.C.; Agertt, V.A.; Flores, V.C.; Dalmolin, T.V.; Rossi, G.G.; Forno, N.L.F.D.; Bianchini, B.V.; Mizdal, C.R.; Siqueira, F.S.; et al. Molecular characterization of Enterobacteriaceae resistant to carbapenem antimicrobials. Rev. Bras. de Patol. Médica Lab. 2015, 51, 162–165. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 1–9. [Google Scholar] [CrossRef]
- Stefaniak, L.A.; Duarte, E.L.; Nishiyama, S.A.B.; Nakano, V. Resistência bacteriana: A importância das beta-lactamases. Rev. UNINGÁ 2005, 4, 123–137. [Google Scholar]
- Álvarez, L.M.A.; García, J.M.G.; Hernández, M.D.P.; González, S.M.; Gutiérrez, J.J.P. Utility of Phenotypic and Genotypic Testing in the Study of Mycobacterium tuberculosis Resistance to First-Line Anti-Tuberculosis drugs. Arch. Broncopneumol. 2017, 53, 192–198. [Google Scholar] [CrossRef]
- Filho, D.B.F.; Rocha, E.C.; Júnior, J.A.S.; Paranhos, R.; Neves, J.A.B.; Silva, M.B. Desvendando os Mistérios do Coeficiente de Correlação de Pearson: O retorno. Leviathan-Cad. Pesqui. Políica 2014, 8, 66–96. [Google Scholar]
- Rodrigues, R.L.; Nascimento, H.F.; Menezes, G.L.; Lopes, A.R.; Nevoa, J.C.; Soares, W.C.S.; Santiago, S.B.; Barbosa, M.S. Contribuição ao estudo comparativo do diagnóstico laboratorial clássico e molecular de Helicobacter pylori: Uma abordagem investigativa. Rev. Acadêmica do Inst. de Ciências da Saúde 2016, 2, 18–25. [Google Scholar]
- Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. Extended broad-spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Brazilian Committee on Antimicrobial Suscetibility Testing. Orientações do EUCAST para Detecção de Mecanismos de Resistência e Resistências Específicas de Importância Clinica e/ou Epidemiológica. 2015. EUCAST. Versão 1.0. Available online: http://brcast.org.br/documentos (accessed on 16 February 2018).
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing-Informational Supplement M100-S22; Versão 27; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
| Antibiotics | Percentage of Antimicrobial Resistance (%) | |||||
|---|---|---|---|---|---|---|
| Manual Resuscitators | Human Cornea | Human Tonsilas | Veterinary Hospital | Animal Bladder | Animal Uterus | |
| Ampicillin | 42.85 | 65.21 | 0.2 | 90.47 | 100 | 100 |
| Aztreonam | 0 | 26.08 | 0 | 85.71 | 0 | 0 |
| Amoxicillin-clavalunate | 0 | 39.13 | 0 | 90.47 | 100 | 100 |
| Ceftazidime | 100 | 30.43 | 0 | 85.71 | 100 | 0 |
| Cefoxitin | 42.85 | 39.13 | 0.2 | 76.19 | 0 | 0 |
| Cefazolin | 0 | 52.17 | 100 | 80.95 | 100 | 0 |
| Cefepime | 100 | 30.43 | 0 | 95.23 | 0 | 0 |
| Ceftriaxone | 0 | 26.08 | 0 | 0 | 0 | 0 |
| Cefuroxime | 100 | 26.08 | 0 | 76.19 | 0 | 0 |
| Imipenem | 100 | 30.43 | 0.2 | 28.57 | 100 | 0 |
| Piperacillin-tazobactam | 100 | 17.39 | 0 | 67.66 | 100 | 0 |
| Antimicrobials | Molecular Detection Rate (%) | Phenotypic Detection Rate (%) | Descriptive Statistics | ||
|---|---|---|---|---|---|
| Standard Deviation | Default Error | Variance | |||
| Ampicillin | 83.33 | 74.28 | 6.39931637 | 4.525 | 40.95125 |
| Aztreonam | 94.44 | 34.28 | 42.53954396 | 30.08 | 1809.6128 |
| Amoxicilina + Clavalunate | 88.88 | 62.85 | 18.40599 | 13.015 | 338.7805 |
| Ceftazidime | 94.44 | 51.42 | 30.41973 | 21.51 | 925.3602 |
| Cefoxitine | 94.44 | 41.42 | 37.4908 | 26.51 | 1405.56 |
| Cefazoline | 38.88 | 54.28 | 10.88944 | 7.7 | 118.58 |
| Cefepime | 88.88 | 44.28 | 31.53696 | 22.3 | 994.58 |
| Ceftriaxone | 88.88 | 41.42 | 33.55929 | 23.73 | 1126.226 |
| Cefuroxime | 72.22 | 8.57 | 45.00735 | 31.825 | 2025.661 |
| Imipenem | 88.88 | 35.71 | 37.59687 | 26.585 | 1413.524 |
| Piperacillin + Tazobactam | 94.44 | 41.42 | 37.4908 | 26.51 | 1405.56 |
| Antibiotics | blaOXA | blaIMP | blaNDM | blaSME | blaDHA | blaCMY | blaTEM | blaKPC | blaSPM | blaCTX-M | blaVIM | blaSIM | blaGIM | blaSHV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin | + | - | - | + | - | + | + | + | + | + | - | + | - | + |
| Aztreonam | + | + | + | - | + | + | + | + | + | + | + | + | - | + |
| Amoxicillin + Clavanulate | + | + | + | - | - | + | + | + | - | + | + | - | - | + |
| Ceftazidime | + | + | + | - | + | + | + | + | + | + | + | + | - | + |
| Cefoxitin | + | - | + | - | + | + | + | + | + | + | - | - | - | + |
| Cefazolin | + | + | + | - | + | + | + | + | - | + | - | - | - | + |
| Cefepime | + | + | + | - | + | + | + | + | + | + | + | + | - | + |
| Ceftriaxone | + | + | + | - | + | + | + | + | - | + | - | - | - | + |
| Cefuroxime | + | - | + | - | - | - | + | + | + | + | + | - | - | + |
| Imipenem | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Piperacillin + Tazobactam | + | + | + | - | - | + | + | + | + | + | + | + | - | + |
| Bacterial Genus | Number of Samples |
|---|---|
| Cedecea neteri | 2 |
| Citrobacter freundii | 3 |
| Edwardsiella ictaluri | 1 |
| Enterobacter aerogenes | 12 |
| Enterobacter agglomerans | 6 |
| Erwinia persicina | 1 |
| Escherichia blattae | 1 |
| Escherichia coli | 12 |
| Hafnia alvei | 3 |
| Klebsiella spp. | 7 |
| Morganella morganii | 2 |
| Proteus mirabilis | 4 |
| Providencia rustigiani | 1 |
| Providencia spp. | 1 |
| Raoultella terrigena | 1 |
| Salmonella paratyphia | 1 |
| Salmonella spp. | 2 |
| Salmonella typhi | 2 |
| Serratia marcecens | 6 |
| Yersinia ruckeri | 1 |
| Yersinia spp. | 1 |
| Genes | Gene Sequence from 5′ to 3′ | Temperature of Ringing | Quantity of Bases | Access at the GenBank | Amplified Fragment Size |
|---|---|---|---|---|---|
| blaOXA | Sense: GGCAGCGGGTTCCCTTGTC | 49.7 | 19 | FN396876.1 | 171pb |
| Reverso: CGATAATGGGCTGCAGCGG | 49.7 | 19 | |||
| blaIMP | Sense: CCAGCGTACGGCCCACAGA | 49.6 | 19 | NG035455.1 | 138pb |
| Reverso: GGTGATGGCTGTTGCGGCA | 50.3 | 19 | |||
| blaNDM | Sense: CGGCCGCGTGCTGGTG | 49.8 | 16 | JN711113.1 | 182pb |
| Reverso: GGCATAAGTCGCAATCCCCG | 50.2 | 20 | |||
| blaSME | Sense: GGCGGCTGCTGTTTTAGAGAGG | 50.9 | 25 | KJ188748.1 | 184pb |
| Reverso: TGCAGCAGAAGCCATATCACCTAAT | 50.3 | 22 | |||
| blaDHA | Sense: GCGGGCGAATTGCTGCAT | 49.8 | 18 | NG041043.1 | 183pb |
| Reverso: TGGGTGCCGGGGTAGCG | 50.1 | 17 | |||
| blaCMY | Sense: GGATTAGGCTGGGAGATGCTGAA | 50.1 | 23 | NG041279.1 | 158pb |
| Reverso: CCAGTGGAGCCCGTTTTATGC | 49.6 | 21 | |||
| blaTEM | Sense: TCCGTGTCGCCCTTATTCCC | 49.6 | 20 | KJ923009 | 165pb |
| Reverso: CCTTGAGAGTTTTCGCCCCG | 49.6 | 20 | |||
| blaSHV | Sense: GGCAGCGGGTTCCCTTGTC | 49.7 | 19 | FN396876.1 | 171pb |
| Reverso: CGATAATGGGCTGCAGCGG | 49.7 | 19 | |||
| blaVIM | Sense: GTTATGCCGCACCCACCCC | 50.3 | 19 | NG036099.1 | 194 pb |
| Reverso: ACCAAACACCATCGGCAATCTG | 49.7 | 22 | |||
| blaSPM | Sense: CGAAAATGCTTGATGGGACCG | 50.3 | 21 | DQ145284.1 | 147pb |
| Reverso: CACCCGTGCCGTCCAAATG | 49.7 | 19 | |||
| blaCTX | Sense: CTGAGCTTAGCGCGGCCG | 50.1 | 18 | FJ815279.1 | 189pb |
| Reverso: AATGGCGGTGTTTAACGTCGG | 50.0 | 21 | |||
| blaGIM | Sense: CGGTGGTAACGGCGCAGTG | 50.2 | 19 | JX566711.1 | 149pb |
| Reverso: TGCCCTGCTGCGTAACATCG | 50.2 | 20 | |||
| blaKPC | Sense: GGCGGCTCCATCGGTGTG | 49.5 | 18 | AF297554.1 | 155pb |
| Reverso: GTGTCCAGCAAGCCGGCCT | 50.4 | 19 | |||
| blaSIM | Sense: GCACCACCGGCAAGCGC | 50.8 | 17 | EF125010.1 | 156pb |
| Reverso: TGTCCTGGCTGGCGAACGA | 50.0 | 19 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, A.L.; dos Santos, A.P.; Ito, C.R.M.; Queiroz, P.H.P.d.; de Almeida, J.A.; de Carvalho Júnior, M.A.B.; de Oliveira, C.Z.; Avelino, M.A.G.; Wastowski, I.J.; Gomes, G.P.L.A.; et al. Profile of Enterobacteria Resistant to Beta-Lactams. Antibiotics 2020, 9, 410. https://doi.org/10.3390/antibiotics9070410
Santos AL, dos Santos AP, Ito CRM, Queiroz PHPd, de Almeida JA, de Carvalho Júnior MAB, de Oliveira CZ, Avelino MAG, Wastowski IJ, Gomes GPLA, et al. Profile of Enterobacteria Resistant to Beta-Lactams. Antibiotics. 2020; 9(7):410. https://doi.org/10.3390/antibiotics9070410
Chicago/Turabian StyleSantos, Andressa Liberal, Adailton Pereira dos Santos, Célia Regina Malveste Ito, Pedro Henrique Pereira de Queiroz, Juliana Afonso de Almeida, Marcos Antonio Batista de Carvalho Júnior, Camila Zanatta de Oliveira, Melissa Ameloti G. Avelino, Isabela Jubé Wastowski, Giselle Pinheiro Lima Aires Gomes, and et al. 2020. "Profile of Enterobacteria Resistant to Beta-Lactams" Antibiotics 9, no. 7: 410. https://doi.org/10.3390/antibiotics9070410
APA StyleSantos, A. L., dos Santos, A. P., Ito, C. R. M., Queiroz, P. H. P. d., de Almeida, J. A., de Carvalho Júnior, M. A. B., de Oliveira, C. Z., Avelino, M. A. G., Wastowski, I. J., Gomes, G. P. L. A., Souza, A. C. S. e., Vasconcelos, L. S. N. d. O. L., Santos, M. d. O., da Silva, C. A., & Carneiro, L. C. (2020). Profile of Enterobacteria Resistant to Beta-Lactams. Antibiotics, 9(7), 410. https://doi.org/10.3390/antibiotics9070410





