Broad-Spectrum Cephalosporin-Resistant and/or Fluoroquinolone-Resistant Enterobacterales Associated with Canine and Feline Urogenital Infections
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Singer, R.S. Urinary Tract Infections Attributed to Diverse ExPEC Strains in Food Animals: Evidence and Data Gaps. Front. Microbiol. 2015, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, L.; Liu, C.M.; Price, L.B. Foodborne Urinary Tract Infections: A New Paradigm for Antimicrobial-Resistant Foodborne Illness. Front. Microbiol. 2013, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Qekwana, D.N.; Phophi, L.; Naidoo, V.; Oguttu, J.W.; Odoi, A. Antimicrobial Resistance among Escherichia coli Isolates from Dogs Presented with Urinary Tract Infections at a Veterinary Teaching Hospital in South Africa. BMC Vet. Res. 2018, 14, 288. [Google Scholar] [CrossRef] [PubMed]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia coli Isolated from Different Sources: Recent Reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Antão, E.-M.; Wieler, L.H.; Ewers, C. Adhesive Threads of Extraintestinal Pathogenic Escherichia coli. Gut Pathog. 2009, 1, 22. [Google Scholar] [CrossRef]
- Johnson, J.R.; Kuskowski, M.A.; Owens, K.; Gajewski, A.; Winokur, P.L. Phylogenetic Origin and Virulence Genotype in Relation to Resistance to Fluoroquinolones and/or Extended-Spectrum Cephalosporins and Cephamycins among Escherichia coli Isolates from Animals and Humans. J. Infect. Dis. 2003, 5, 759–768. [Google Scholar] [CrossRef]
- Freitag, T.; Squires, R.A.; Schmid, J.; Elliott, J. Feline Uropathogenic Escherichia coli from Great Britain and New Zealand Have Dissimilar Virulence Factor Genotypes. Vet. Microbiol. 2005, 106, 79–86. [Google Scholar] [CrossRef]
- Smith, J.L.; Fratamico, P.M.; Gunther, N.W. Extraintestinal Pathogenic Escherichia coli. Foodborne Pathog. Dis. 2007, 4, 134–163. [Google Scholar] [CrossRef]
- Guignot, J.; Chaplais, C.; Coconnier-Polter, M.H.; Servin, A.L. The Secreted Autotransporter Toxin, Sat, Functions as a Virulence Factor in Afa/Dr Diffusely Adhering Escherichia coli by Promoting Lesions in Tight Junction of Polarized Epithelial Cells. Cell. Microbiol. 2007, 9, 204–221. [Google Scholar] [CrossRef]
- Price, L.B.; Hungate, B.A.; Koch, B.J.; Davis, G.S.; Liu, C.M. Colonizing Opportunistic Pathogens (COPs): The Beasts in All of Us. PLoS Pathog. 2017, 13, e1006369. [Google Scholar] [CrossRef]
- Liu, C.M.; Stegger, M.; Aziz, M.; Johnson, T.J.; Waits, K.; Nordstrom, L.; Gauld, L.; Weaver, B.; Rolland, D.; Statham, S.; et al. Escherichia coli ST131-H22 as a Foodborne Uropathogen. MBio 2018, 9, e00470-18. [Google Scholar] [CrossRef] [PubMed]
- Shaik, S.; Ranjan, A.; Tiwari, S.K.; Hussain, A.; Nandanwar, N.; Kumar, N.; Jadhav, S.; Semmler, T.; Baddam, R.; Islam, M.A.; et al. Comparative Genomic Analysis of Globally Dominant ST131 Clone with Other Epidemiologically Successful. MBio 2017, 8, e01596-17. [Google Scholar] [CrossRef] [PubMed]
- LeStrange, K.; Markland, S.M.; Hoover, D.G.; Sharma, M.; Kniel, K.E. An Evaluation of the Virulence and Adherence Properties of Avian Pathogenic Escherichia coli. One Heath 2017, 4, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Miller, S.; Johnston, B.; Clabots, C.; DebRoy, C. Sharing of Escherichia coli Sequence Type ST131 and Other Multidrug-Resistant and Urovirulent E. Coli Strains among Dogs and Cats within a Household. J. Clin. Microbiol. 2009, 47, 3721–3725. [Google Scholar] [CrossRef]
- Huber, H.; Zweifel, C.; Wittenbrink, M.M.; Stephan, R. ESBL-Producing Uropathogenic Escherichia coli Isolated from Dogs and Cats in Switzerland. Vet. Microbiol. 2013, 162, 992–996. [Google Scholar] [CrossRef] [PubMed]
- Madec, J.Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-Spectrum β-Lactamase/AmpC- and Carbapenemase-Producing Enterobacteriaceae in Animals: A Threat for Humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Wannemuehler, Y.; Johnson, S.J.; Stell, A.L.; Doetkott, C.; Johnson, J.R.; Kim, K.S.; Spanjaard, L.; Nolan, L.K. Comparison of Extraintestinal Pathogenic Escherichia coli Strains from Human and Avian Sources Reveals a Mixed Subset Representing Potential Zoonotic Pathogens. Appl. Environ. Microbiol. 2008, 74, 7043–7050. [Google Scholar] [CrossRef] [PubMed]
- Weese, J.S.; Blondeau, J.; Boothe, D.; Guardabassi, L.G.; Gumley, N.; Papich, M.; Jessen, L.R.; Lappin, M.; Rankin, S.; Westropp, J.L.; et al. International Society for Companion Animal Infectious Diseases (ISCAID) Guidelines for the Diagnosis and Management of Bacterial Urinary Tract Infections in Dogs and Cats. Vet. J. 2019, 247, 8–25. [Google Scholar] [CrossRef]
- Roberts, M.; White, J.; Lam, A. Prevalence of Bacteria and Changes in Trends in Antimicrobial Resistance of Escherichia coli Isolated from Positive Canine Urinary Samples from an Australian Referral Hospital over a 5-Year Period (2013–2017). Vet. Rec. Open 2019, 6, e000345. [Google Scholar] [CrossRef]
- Marques, C.; Gama, L.T.; Belas, A.; Bergström, K.; Beurlet, S.; Briend-Marchal, A.; Broens, E.M.; Costa, M.; Criel, D.; Damborg, P.; et al. European Multicenter Study on Antimicrobial Resistance in Bacteria Isolated from Companion Animal Urinary Tract Infections. BMC Vet. Res. 2016, 12, 213. [Google Scholar] [CrossRef]
- Obeng-Nkrumah, N.; Labi, A.K.; Blankson, H.; Awuah-Mensah, G.; Oduro-Mensah, D.; Anum, J.; Teye, J.; Kwashie, S.D.; Bako, E.; Ayeh-Kumi, P.F.; et al. Household Cockroaches Carry CTX-M-15-, OXA-48- and NDM-1-Producing Enterobacteria, and Share Beta-Lactam Resistance Determinants with Humans. BMC Microbiol. 2019, 19, 272. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Brenner, M.G.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U.; Kreienbrock, L.; et al. Whole Genome Analyses of CMY-2-Producing Escherichia coli Isolates from Humans, Animals and Food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef] [PubMed]
- Tamang, M.D.; Nam, H.M.; Jang, G.C.; Kim, S.R.; Chae, M.H.; Jung, S.C.; Byun, J.W.; Park, Y.H.; Lim, S.K. Molecular Characterization of Extended-Spectrum-β-Lactamase- Producing and Plasmid-Mediated AmpC β-Lactamase-Producing Escherichia coli Isolated from Stray Dogs in South Korea. Antimicrob. Agents Chemother. 2012, 56, 2705–2712. [Google Scholar] [CrossRef] [PubMed]
- Bortolami, A.; Zendri, F.; Maciuca, E.I.; Wattret, A.; Ellis, C.; Schmidt, V.; Pinchbeck, G.; Timofte, D. Diversity, Virulence, and Clinical Significance of Extended-Spectrum β-Lactamase- And PAmpC-Producing Escherichia coli from Companion Animals. Front. Microbiol. 2019, 10, 1260. [Google Scholar] [CrossRef]
- National Office for Animal Health UK. Critically Important Antibiotics in Veterinary Medicine: European Medicines Agency Recommendations. Available online: https://www.noah.co.uk/wp-content/uploads/2016/12/NOAH-briefing-on-CIAs-07122016.pdf (accessed on 28 May 2020).
- Smith, A.; Wayne, A.S.; Fellman, C.L.; Rosenbaum, M.H. Usage Patterns of Carbapenem Antimicrobials in Dogs and Cats at a Veterinary Tertiary Care Hospital. J. Vet. Intern. Med. 2019, 33, 1677–1685. [Google Scholar] [CrossRef]
- Dupouy, V.; Abdelli, M.; Moyano, G.; Arpaillange, N.; Bibbal, D.; Cadiergues, M.C.; Lopez-Pulin, D.; Sayah-Jeanne, S.; De Gunzburg, J.; Saint-Lu, N.; et al. Prevalence of Beta-Lactam and Quinolone/Fluoroquinolone Resistance in Enterobacteriaceae from Dogs in France and Spain—Characterization of ESBL/PAmpC Isolates, Genes, and Conjugative Plasmids. Front. Vet. Sci. 2019, 30, 279. [Google Scholar] [CrossRef]
- Singleton, D.A.; Sánchez-Vizcaíno, F.; Dawson, S.; Jones, P.H.; Noble, P.J.M.; Pinchbeck, G.L.; Williams, N.J.; Radford, A.D. Patterns of Antimicrobial Agent Prescription in a Sentinel Population of Canine and Feline Veterinary Practices in the United Kingdom. Vet. J. 2017, 224, 18–24. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 4th ed.; CLSI Supplement VET08; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Gozi, K.S.; Froes, J.R.; Ajude, L.P.; Da Silva, C.R.; Baptista, R.S.; Peiró, J.R.; Marinho, M.; Mendes, L.C.N.; Nogueira, M.C.L.; Casella, T. Dissemination of Multidrug-Resistant Commensal Escherichia coli in Feedlot Lambs in Southeastern Brazil. Front. Microbiol. 2019, 10, 1394. [Google Scholar] [CrossRef]
- Harada, K.; Nakai, Y.; Kataoka, Y. Mechanisms of Resistance to Cephalosporin and Emergence of O25b-ST131 Clone Harboring CTX-M-27 β-Lactamase in Extraintestinal Pathogenic Escherichia coli from Dogs and Cats in Japan. Microbiol. Immunol. 2012, 56, 480–485. [Google Scholar] [CrossRef]
- Maluta, R.P.; Logue, C.M.; Casas, M.R.T.; Meng, T.; Guastalli, E.A.L.; Rojas, T.C.G.; Montelli, A.C.; Sadatsune, T.; Ramos, M.D.C.; Nolan, L.K.; et al. Overlapped Sequence Types (STs) and Serogroups of Avian Pathogenic (APEC) and Human Extra-Intestinal Pathogenic (ExPEC) Escherichia coli Isolated in Brazil. PLoS ONE 2014, 9, e105016. [Google Scholar] [CrossRef] [PubMed]
- Zogg, A.L.; Simmen, S.; Zurfluh, K.; Stephan, R.; Schmitt, S.N.; Nüesch-Inderbinen, M. High Prevalence of Extended-Spectrum β-Lactamase Producing Enterobacteriaceae among Clinical Isolates from Cats and Dogs Admitted to a Veterinary Hospital in Switzerland. Front. Vet. Sci. 2018, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Gally, D.L.; Argyle, S.A. Multidrug-Resistant Escherichia coli from Canine Urinary Tract Infections Tend to Have Commensal Phylotypes, Lower Prevalence of Virulence Determinants and AmpC-Replicons. Vet. Microbiol. 2014, 169, 171–178. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nicolas-Chanoine, M.H.; Bertrand, X.; Madec, J.Y. Escherichia coli ST131, an Intriguing Clonal Group. Clin. Microbiol. Rev. 2014, 27, 543–754. [Google Scholar] [CrossRef]
- Misic, D.; Asanin, J.; Spergser, J.; Szostak, M.; Loncaric, I. OXA-72-Mediated Carbapenem Resistance in Sequence Type 1 Multidrug Colistin-Resistant Acinetobacter baumannii Associated with Urinary Tract Infection in a Dog from Serbia. Antimicrob. Agents Chemother. 2018, 62, e00219-18. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Lubbers, B.V.; Schwarz, S.; Watts, J.L. Applying Definitions for Multidrug Resistance, Extensive Drug Resistance and Pandrug Resistance to Clinically Significant Livestock and Companion Animal Bacterial Pathogens. J. Antimicrob. Chemother. 2018, 73, 1460–1463. [Google Scholar] [CrossRef]
- Khairy, R.M.; Mohamed, E.S.; Ghany, H.M.A.; Abdelrahim, S.S. Phylogenic Classification and Virulence Genes Profiles of Uropathogenic E. Coli and Diarrhegenic E. coli Strains Isolated from Community Acquired Infections. PLoS ONE 2019, 14, e0222441. [Google Scholar] [CrossRef]
- Müller, C.M.; Dobrindt, U.; Nagy, G.; Emödy, L.; Uhlin, B.E.; Hacker, J. Role of Histone-like Proteins H-NS and StpA in Expression of Virulence Determinants of Uropathogenic Escherichia coli. J. Bacteriol. 2006, 188, 5428–5438. [Google Scholar] [CrossRef]
- Mamani, R.; Flament-Simon, S.C.; García, V.; Mora, A.; Alonso, M.P.; López, C.; García-Meniño, I.; Díaz-Jiménez, D.; Blanco, J.E.; Blanco, M.; et al. Sequence Types, Clonotypes, Serotypes, and Virotypes of Extended-Spectrum β-Lactamase-Producing Escherichia coli Causing Bacteraemia in a Spanish Hospital over a 12-Year Period (2000 to 2011). Front. Microbiol. 2019, 10, 1530. [Google Scholar] [CrossRef]
- Pitout, J.D.D. Extraintestinal Pathogenic Escherichia coli: An Update on Antimicrobial Resistance, Laboratory Diagnosis and Treatment. Expert Rev. Anti-Infect. Ther. 2012, 10, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, T.; Vogler, A.J.; Thomas, A.; Sauer, S.; Hergenroether, M.; Straubinger, R.K.; Birdsell, D.; Keim, P.; Sahl, J.W.; Williamson, C.H.D.; et al. Phenotypic Characterization and Whole Genome Analysis of Extended-Spectrum Betalactamase-Producing Bacteria Isolated from Dogs in Germany. PLoS ONE 2018, 13, e0206252. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. In Antimicrobial Resistance in Bacteria from Livestock and Companion Animals; ASM Press: Washington, DC, USA, 2018; Volume 6. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae Producing Extended-Spectrum β-Lactamases (ESBLs) in the Community. J. Antimicrob. Chemother. 2005, 56, 52–59. [Google Scholar] [CrossRef]
- Loncaric, I.; Stalder, G.L.; Mehinagic, K.; Rosengarten, R.; Hoelzl, F.; Knauer, F.; Walzer, C. Comparison of ESBL—And AmpC Producing Enterobacteriaceae and Methicillin-Resistant Staphylococcus aureus (MRSA) Isolated from Migratory and Resident Population of Rooks (Corvus Frugilegus) in Austria. PLoS ONE 2013, 8, e84048. [Google Scholar] [CrossRef]
- Köck, R.; Daniels-Haardt, I.; Becker, K.; Mellmann, A.; Friedrich, A.W.; Mevius, D.; Schwarz, S.; Jurke, A. Carbapenem-Resistant Enterobacteriaceae in Wildlife, Food-Producing, and Companion Animals: A Systematic Review. Clin. Microbiol. Infect. 2018, 24, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Desvars-Larrive, A.; Ruppitsch, W.; Lepuschitz, S.; Szostak, M.P.; Spergser, J.; Feßler, A.T.; Schwarz, S.; Monecke, S.; Ehricht, R.; Walzer, C.; et al. Urban Brown Rats (Rattus Norvegicus) as Possible Source of Multidrug-Resistant Enterobacteriaceae and Meticillin-Resistant Staphylococcus Spp., Vienna, Austria, 2016 and 2017. Eurosurveillance 2019, 24, 1900149. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Saras, E.; Ponsin, C.; Dahmen, S.; Petitjean, M.; Hocquet, D.; Madec, J.Y. High Prevalence of International ESBL CTX-M-15-Producing Enterobacter cloacae ST114 Clone in Animals. J. Antimicrob. Chemother. 2016, 71, 1497–1500. [Google Scholar] [CrossRef] [PubMed]
- Borges, C.A.; Beraldo, L.G.; Maluta, R.P.; Cardozo, M.V.; Barboza, K.B.; Guastalli, E.A.L.; Kariyawasam, S.; DebRoy, C.; Ávila, F.A. Multidrug-Resistant Pathogenic Escherichia coli Isolated from Wild Birds in a Veterinary Hospital. Avian Pathol. 2017, 46, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Leung, L.M.; Chow, K.H.; Lai, E.L.; Lo, W.U.; Ng, T.K. Prevalence of Aminoglycoside Modifying Enzyme and 16S Ribosomal RNA Methylase Genes among Aminoglycoside-Resistant Escherichia coli Isolates. J. Microbiol. Immunol. Infect. 2016, 49, 123–126. [Google Scholar] [CrossRef][Green Version]
- Sun, Y.; Zeng, Z.; Chen, S.; Ma, J.; He, L.; Liu, Y.; Deng, Y.; Lei, T.; Zhao, J.; Liu, J.H. High Prevalence of BlaCTX-M Extended-Spectrum β-Lactamase Genes in Escherichia coli Isolates from Pets and Emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 2010, 16, 1475–1481. [Google Scholar] [CrossRef]
- World Health Organization. Global Priority List of Antibiotic-Resistant Batceria to Guide Research, Discovery, and Development of New Antibiotics. 2017. Available online: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 27 May 2020).
- Johnson, J.R.; Sannes, M.R.; Croy, C.; Johnston, B.; Clabots, C.; Kuskowski, M.A.; Bender, J.; Smith, K.E.; Winokur, P.L.; Belongia, E.A. Antimicrobial Drug-Resistant Escherichia coli from Humans and Poultry Products, Minnesota and Wisconsin, 2002–2004. Emerg. Infect. Dis. 2007, 13, 838–846. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A. Molecular Epidemiology of Extraintestinal Pathogenic (Uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 2005, 295, 383–404. [Google Scholar] [CrossRef]
- Ewers, C.; Bethe, A.; Stamm, I.; Grobbel, M.; Kopp, P.A.; Guerra, B.; Stubbe, M.; Doi, Y.; Zong, Z.; Kola, A.; et al. CTX-M-15-D-ST648 Escherichia coli from Companion Animals and Horses: Another Pandemic Clone Combining Multiresistance and Extraintestinal Virulence? J. Antimicrob. Chemother. 2014, 69, 1224–1230. [Google Scholar] [CrossRef] [PubMed]
- Bouckaert, J.; Mackenzie, J.; De Paz, J.L.; Chipwaza, B.; Choudhury, D.; Zavialov, A.; Mannerstedt, K.; Anderson, J.; Piérard, D.; Wyns, L.; et al. The Affinity of the FimH Fimbrial Adhesin Is Receptor-Driven and Quasi-Independent of Escherichia coli Pathotypes. Mol. Microbiol. 2006, 61, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Harel, J.; Masson, L.; Edens, T.J.; Portt, A.; Reid-Smith, R.J.; Zhanel, G.G.; Kropinski, A.M.; Boerlin, P. Multilocus Sequence Typing and Virulence Gene Profiles Associated with Escherichia coli from Human and Animal Sources. Foodborne Pathog. Dis. 2015, 12, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; De Lorenzo, V.; Neilands, J.B. Nucleotide Sequence of the IucD Gene of the PCoIV-K30 Aerobactin Operon and Topology of Its Product Studied with PhoA and LacZ Gene Fusions. J. Bacteriol. 1988, 170, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Brett, K.N.; Ramachandran, V.; Hornitzky, M.A.; Bettelheim, K.A.; Walker, M.J.; Djordjevic, S.P. Stx1c Is the Most Common Shiga Toxin 1 Subtype among Shiga Toxin-Producing Escherichia oli Isolates from Sheep but Not among Isolates from Cattle. J. Clin. Microbiol. 2003, 41, 926–936. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Wannemuehler, Y.M.; Nolan, L.K. Evolution of the Iss Gene in Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2360–2369. [Google Scholar] [CrossRef]
- Bien, J.; Sokolova, O.; Bozko, P. Role of Uropathogenic Escherichia coli Virulence Factors in Development of Urinary Tract Infection and Kidney Damage. Int. J. Nephrol. 2012, 2012, 681473. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100S; CLSI: Wayne, PA, USA, 2018. [Google Scholar]
- Braun, S.D.; Monecke, S.; Thürmer, A.; Ruppelt, A.; Makarewicz, O.; Pletz, M.; Reißig, A.; Slickers, P.; Ehricht, R. Rapid Identification of Carbapenemase Genes in Gram-Negative Bacteria with an Oligonucleotide Microarray-Based Assay. PLoS ONE 2014, 9, e102232. [Google Scholar] [CrossRef] [PubMed]
- Gwida, M.; Awad, A.; El-Ashker, M.; Hotzel, H.; Monecke, S.; Ehricht, R.; Müller, E.; Reißig, A.; Barth, S.A.; Berens, C.; et al. Microarray-Based Detection of Resistance and Virulence Factors in Commensal Escherichia coli from Livestock and Farmers in Egypt. Vet. Microbiol. 2020, 240, 108539. [Google Scholar] [CrossRef]
- Loncaric, I.; Beiglböck, C.; Feßler, A.T.; Posautz, A.; Rosengarten, R.; Walzer, C.; Ehricht, R.; Monecke, S.; Schwarz, S.; Spergser, J.; et al. Characterization of ESBL- and AmpC-Producing and Fluoroquinolone-Resistant Enterobacteriaceae Isolated from Mouflons (Ovis Orientalis Musimon) in Austria and Germany. PLoS ONE 2016, 18, e0155786. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L.; Carrër, A.; Toleman, M.A.; Walsh, T.R. How to Detect NDM-1 Producers. J. Clin. Microbiol. 2011, 49, 718–721. [Google Scholar] [CrossRef] [PubMed]
- Everett, M.J.; Jin, Y.F.; Ricci, V.; Piddock, L.J.V. Contributions of Individual Mechanisms to Fluoroquinolone Resistance in 36 Escherichia coli Strains Isolated from Humans and Animals. Antimicrob. Agents Chemother. 1996, 40, 2380–2386. [Google Scholar] [CrossRef]
- Caroff, N.; Espaze, E.; Bérard, I.; Richet, H.; Reynaud, A. Mutations in the AmpC Promoter of Escherichia coli Isolates Resistant to Oxyiminocephalosporins without Extended Spectrum β-Lactamase Production. FEMS Microbiol. Lett. 1999, 173, 459–465. [Google Scholar] [CrossRef]
- Clermont, O.; Dhanji, H.; Upton, M.; Gibreel, T.; Fox, A.; Boyd, D.; Mulvey, M.R.; Nordmann, P.; Ruppé, E.; Sarthou, J.L.; et al. Rapid Detection of the O25b-ST131 Clone of Escherichia coli Encompassing the CTX-M-15-Producing Strains. J. Antimicrob. Chemother. 2009, 64, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Weissman, S.J.; Johnson, J.R.; Tchesnokova, V.; Billig, M.; Dykhuizen, D.; Riddell, K.; Rogers, P.; Qin, X.; Butler-Wu, S.; Cookson, B.T.; et al. High-Resolution Two-Locus Clonal Typing of Extraintestinal Pathogenic Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Vaz, C.; Monteiro, P.T.; Melo-Cristino, J.; Ramirez, M.; Carriço, J.A. PHYLOViZ: Phylogenetic inference and data visualization for sequence based typing methods. BMC BioInform. 2012, 13, 87. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and Virulence in Escherichia Coli: An Evolutionary Perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Miyoshi-Akiyama, T.; Hayakawa, K.; Ohmagari, N.; Shimojima, M.; Kirikae, T. Multilocus Sequence Typing (MLST) for Characterization of Enterobacter cloacae. PLoS ONE 2013, 8, e66358. [Google Scholar] [CrossRef]
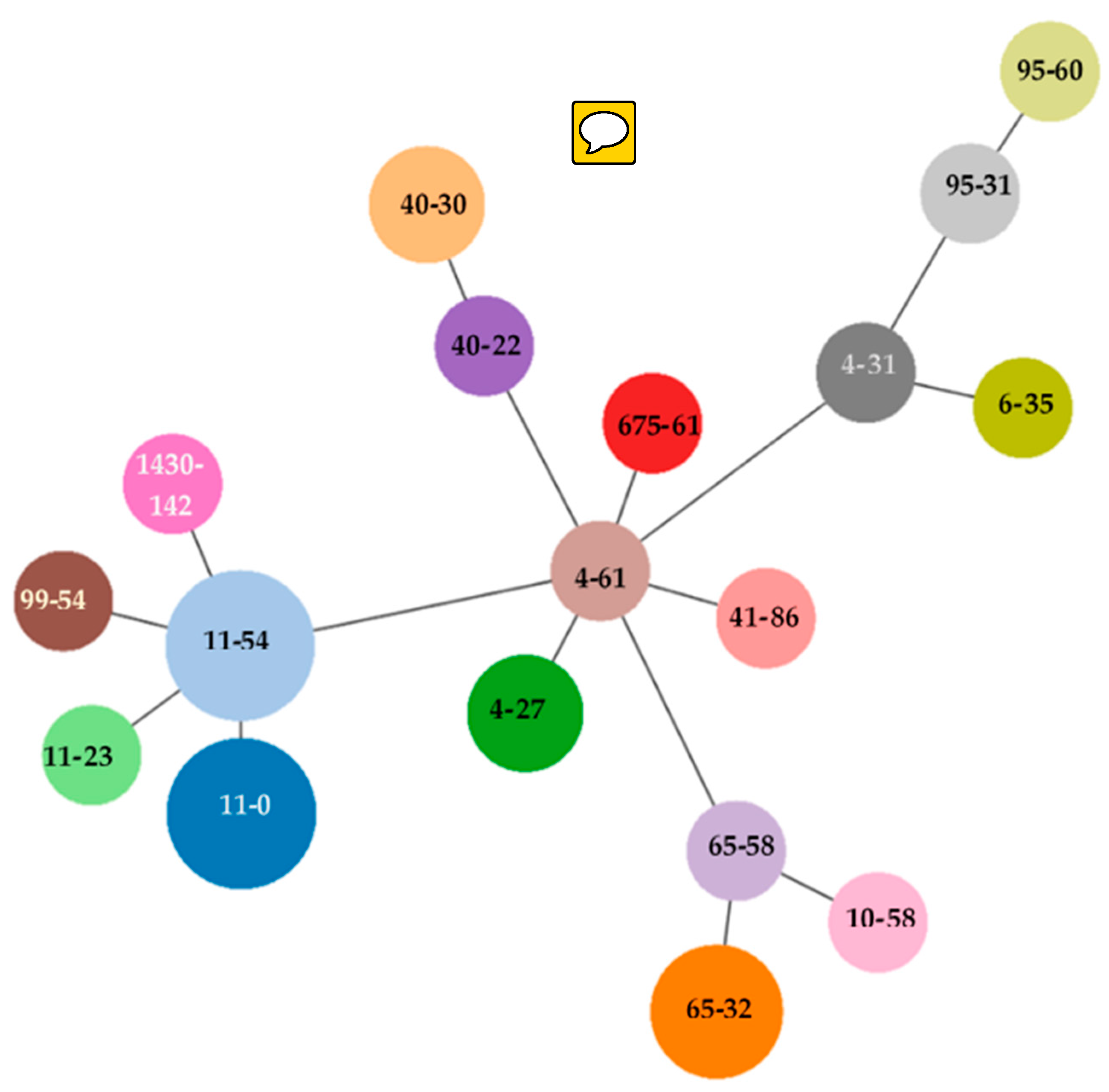
| Serogenotyping | Clonotype | ST* | Resistance Profile | Mutation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Country | Host | Source | Species | Phylogroup | O | H | fumC | fimH | Phenotype** | Genotype | ampC promoter | gyrA | parC | Previous therapy** | |
| MDR56 | SRB | dog | vaginal swab | E. coli | A | 128 | 26 | 11 | 23 | n.t. | AMP, CIP, CHL, SXT | blaTEM, catA1, tet(B), dfrA17 | Ser83Leu, Asp87Asn | Ser80Ile | AMC, ENO | |
| MDR63 | SRB | dog | urine | E. coli | C | 9 | 9 | 11 | 0 | n.t. | AMP, CAZ, CTX, CIP, TET, CHL, SXT | blaTEM, blaCMY,strA, strB, tet(B), catA1, sul1, sul2, dfrA17, dfrA19 | w.t. | Ser83Leu, Asp87Asn | Ser80Ile | not known |
| 193 | AUT | dog | bladder | E. coli | B1 | n.d. | 16 | 65 | 32 | n.t. | AMP, CAZ, CTX, CIP, TOB, TET, SXT | blaTEM, blaCMY, qnrB, aac(6), aac(6)-Ib, strB, tet(A), sul1, sul2, dfrA5 | –18, –1, +58 | Ser83Leu, Asp87Asn | Ser80Ile | not known |
| 2304 | AUT | dog | urine | E. coli | B1 | n.d. | 23 | 675 | 61 | n.t. | AMP, CAZ, CTX, CIP | blaTEM, blaCMY | –18, –1, +58 | Ser83Leu, Asp87Asn | Ser80Ile | CIP |
| 3056 | AUT | dog | urine | E. coli | unknown | 25 | 4 | 40 | 30 | n.t. | AMP, CAZ, CTX, ATM, CIP, GEN, TOB, TET | blaCTX-M-1, blaOXA-1, aac(6)-Ib, tet(A), catB3 | Ser83Leu, Asp87Asn | Ser80Ile, Glu84Val | AMC, Cephalosporin | |
| 3168 | AUT | dog | urine | E. coli | B1 | n.d. | 16 | 65 | 32 | n.t. | AMP, CAZ, CTX, CIP, TOB, TET, SXT | blaTEM, blaCMY, qnrB, aac6Ib, tet(A), sul1, sul2, dfrA5 | –18, –1, +58 | Ser83Leu, Asp87Asn | Ser80Ile | AMC, MAR, CVN |
| 2016 | AUT | dog | urine | E. coli | B1 | n.d. | 8 | 41 | 86 | n.t. | AMP, CTX, FEP, ATM, TET | blaCTX-M-1, blaTEM, qnrS, aphA, strA, strB, tet(A), sul2 | ENO, CVN | |||
| 2058 | AUT | dog | urine | E. coli | unknown | 25 | 4 | 40 | 22 | n.t. | AMP, CTX, ATM, TET, CHL | blaCTX-M-1, blaTEM, strA, strB, tet(A), floR | none | |||
| 4125 | AUT | dog | urine | E. coli | B1 | n.d. | 16 | 65 | 58 | n.t. | AMP, CAZ, CTX, CIP, TOB, TET, SXT | blaTEM, blaCMY, qnrB, aac(6), aac(6)-Ib, strA, strB, tet(A), sul1, sul2, dfrA5 | –18, –1, +58 | Ser83Leu, Asp87Asn | Ser80Ile | CHL |
| 1286 | AUT | cat | urine | E. coli | clade II | n.d. | n.d. | 4 | 27 | n.t. | AMP, CAZ, CTX, ATM, CIP, TOB, TET, SXT | blaCMY, blaOXA-1, blaTEM, aac(6), aac(6)-Ib, aadA4, strB, catB3, tet(A), sul2, dfrA17 | +58 | Ser83Leu, Asp87Asn | Ser80Ile | none |
| 2269 | AUT | dog | ejaculate | E. coli | C | n.d. | 30 | 99 | 54 | n.t. | AMP, CIP, GEN, TOB | blaOXA-1, aac(6), aac(6)-Ib, catB3 | Ser83Leu, Asp87Asn | w.t. | none | |
| 2736 | AUT | dog | urin | E. coli | B1 | n.d. | 23 | 4 | 61 | n.t. | AMP, CIP, TET | tet(A) | Ser83Leu, Asp87Asn | Ser80Ile | ENO | |
| 3441 | AUT | dog | urin | E. coli | C | n.d. | 9 | 4 | 31 | 744 | AMP, CAZ, CTX, CIP, TET, CHL, SXT | blaCTX-M-1, blaTEM, aadA1, aadA4, aphA, strB, tet(A), tet(B), catA1, sul1, sul2, dfrA1, dfrA17 | Ser83Leu | Ser80Ile | CHL | |
| 3448 | AUT | cat | urin | E. coli | C | n.d. | 9 | 11 | 0 | 744 | AMP, CAZ, CTX, ATM, CIP, TET, CHL, SXT | blaCTX-M-1, blaTEM, aadA2, aadA4, aphA, strA, strB, tet(A), tet(B), catA1, sul1, sul2, dfrA1, dfrA17 | Ser83Leu, Asp87Asn | Ser80Ile | CHL | |
| 3534 | AUT | dog | urin | E. coli | B1 | n.d. | 9 | 11 | 54 | 744 | AMP, CAZ, CTX, CIP, GEN, TOB, TET, CHL, SXT | blaTEM, aadA1, aphA, strA, strB, tet(B), catA1, floR, sul2, dfrA1 | Ser83Leu | Ser80Ile | none | |
| 482 | AUT | dog | vagina | E. coli | C | n.d. | 9 | 1430 | 142 | n.t. | AMP, CAZ, CTX, ATM, CIP | blaCTX-M-1, blaOXA-1, blaTEM, aac(6), aac(6)-Ib, aadA4, sul2, dfrA17 | Ser83Leu, Asp87Asn | Ser80Ile | not known | |
| 4 | AUT | dog | urine | E. coli | clade II | n.d. | 6 | 10 | 58 | n.t. | AMP, CTX, ATM, CIP, GEN, TOB, TET, CHL | blaCTX-M-9, blaTEM, strB, tet(B), sul2, dfrA17 | +58 | Ser83Leu, Asp87Asn | Ser80Ile, Glu84Gly | CVN |
| 855 | AUT | dog | vaginal swab | E. coli | C | 9 | 9 | 11 | 54 | 1287 | AMP, CTX, CIP, TET, CHL, SXT | blaTEM, blaCMY,aadA1, strB, tet(B), catA1, sul2, dfrA17 | w.t. | Ser83Leu, Asp87Asn | Ser80Ile | none |
| 555 | AUT | dog | prostata cyst | E. coli | B2 | 25b | 4 | 40 | 30 | 131 | AMP, CAZ, CTX, FEP, CIP, GEN, TOB, TET, SXT | blaCTX-M-15, blaOXA-1, aac(6), aac(6)-Ib, aadA1, aadA2, aadA4, aadA5, tet(A), catB3, sul1, sul2, dfrA17 | Ser83Leu, Asp87Asn | Ser80Ile, Glu84Gly | none | |
| 260 | AUT | dog | vaginal swab | E. coli | F | n.d. | 6 | 4 | 27 | 648 | AMP, CAZ, CTX, FEP, CIP, GEN, TOB, TET, CHL, SXT | blaCTX-M-15, blaOXA-1, aac(6)-Ib, aadA1, aadA2, tet(A), catB3, sul1, sul2 | Ser83Leu, Asp87Asn | Ser80Ile | not known | |
| 655 | AUT | dog | urine | E. coli | B1 | n.d. | 7 | 4 | 31 | n.t. | AMP, CAZ, CTX, ATM, CIP, TOB, TET, SXT | blaTEM, blaCMY,aphA, strB, tet(B), sul2 | w.t. | Ser83Leu, Asp87Asn | Ser80Ile | LEX |
| 728 | AUT | dog | vaginal swab | E. coli | B1 | n.d. | ND | 65 | 32 | 469 | AMP, CIP, TET, CHL, SXT | qnrS, tet(B), sul2, sul3, dfr12, straA | Ser83Leu, Asp87Asn | Ser80Ile | none | |
| 2189 | AUT | cat | urin | E. coli | B1 | 79 | 7 | 95 | 31 | 1463 | AMP, CIP, TET, CHL | blaTEM, tet(B), catA1 | w.t. | Ser80Ile | not known | |
| 3512 | AUT | dog | vaginal swab | E. coli | B1 | n.d. | 7 | 11 | 54 | 1642 | AMP, CIP, GEN, TOB, CHL | blaTEM, aadA4, aphA, strB, tet(B), sul1, sul2, dfrA17 | Ser83Leu, Asp87Asn | w.t. | none | |
| 2srb | SRB | dog | urin | E. coli | B1 | n.d. | 10 | 6 | 35 | n.t. | AMP, CIP, TET, SXT | blaTEM, aadA1, strA, tet(A), sul1, sul2, dfrA7, dfrA17 | -18, –1, +58 | Ser83Leu, Asp87Asn | Ser80Ile | not known |
| 1srb | SRB | dog | urin | E. coli | C | n.d. | 16 | 11 | 0 | n.t. | AMP, CIP, GEN, TET, SXT | blaTEM, blaOXA-2. aadA4, strA, strB, tet(A), sul1, sul2, dfrA17 | Ser83Leu, Asp87Asn | Ser80Ile | not known | |
| ESBL1 | SRB | cat | vaginal swab | E. coli | C | n.d. | n.d. | 11 | 0 | n.t. | AMP, CAZ, CTX, ATM, CIP, TOB, TET, SXT | blaCTX-M-1, blaOXA-1, blaTEM, aac(6), aac(6)-Ib, aadA4, strA, strB, tet(A), sul1, sul2, dfrA17 | Ser83Leu, Asp87Asn | Ser80Ile | AMC | |
| ESBL41 | SRB | dog | vaginal swab | E. coli | A | n.d. | n.d. | 95 | 60 | n.t. | AMP, CAZ, CTX, CIP, GEN, TOB, TET, CHL, SXT | blaCTX-M-1, blaTEM, aadA1, aadA2, aphA, strA, strB, tet(A), sul1, sul2, dfrA14 | Ser83Leu, Asp87Asn | Ser80Ile | AMC | |
| Resistance Profile | Mutation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Country | Host | Source | Species | ST* | Phenotype** | Genotype | gyrA | parC | Previous therapy** |
| 3srb | SRB | dog | vaginal swab | Serratia marcescens | n.t. | AMP, CTX, TET, CHL, SXT | blaTEM, aadA4, aphA, strB, tet(B), catA1 | not known | ||
| 1919 | AUT | dog | urine | Klebsiella pneumoniae | n.t. | AMP, CTX, CIP, GEN, TOB, TET | blaCTX-M-1, blaOXA-1, qnrB, aac(6), aac(6)-Ib, catB3 | Ser83Ile | codon 79Asp silent mutation (GAT>GAC) | not known |
| 3938 | AUT | dog | urin | Klebsiella pneumoniae | n.t. | AMP, CAZ, CTX, CIP, GEN, TOB, TET, CHL, SXT | blaOXA-1, blaSHV, aac(6), aac(6)-Ib, qrnB, qnrS, aphA, catB3, sul1 | w.t. | Ser80Ile | not known |
| 3824 | AUT | dog | prostata swab | Klebsiella pneumoniae | n.t. | AMP, CAZ, CTX, FEP, CIP, TOB, SXT, NIT | blaOXA-1, blaSHV, aac(6), aac(6)-Ib, aphA, qrnB, catB3, sul1, dfrA12 | Asp87Asn | Ser80Ile, Glu84Gly | AMC, MAR |
| 1049 | AUT | cat | urin | Raoultella ornithinolytica | n.t. | AMP, CAZ, CTX, CIP, GEN, TOB, TET, CHL, SXT | blaTEM, aadA1, aphA, tet(B), sul1, catA1, sul2 | w.t. | w.t. | not known |
| 210 | AUT | dog | urine | Proteus mirabilis | n.t. | AMP, CTX, FEP, CHL, SXT | blaTEM, aadA1, aphA, strA, strB, catA1, sul2, dfrA1 | none | ||
| 3362 | AUT | cat | bladder wall | Citrobacter portucalensis | n.t. | AMP, CAZ, CTX, ATM, FEP, CIP, SXT | blaCTX-M-1, blaCMY, qrnB, strA, strB, sul2, dfrA1 | Ser83Leu | w.t. | MAR |
| ESBL40 | SRB | Dog | vaginal swab | Enterobacter cloacae | 114 | AMP, CAZ, CTX, MEM, ATM, FEP, CIP, TOB, TET, CHL, SXT, FOF | blaNDM-1, blaCTX-M-15, blaOXA-1, blaTEM, qnrB, aadA1, aac(6), aac(6)-Ib, aadA2, aphA, strA, strB, tet(A), sul2, dfrA12, dfrA14 | Ser83Leu | w.t. | AMC, ENO |
| Isolate | Country | Host | Source | Species | Virulence Factors |
|---|---|---|---|---|---|
| MDR56 | SRB | dog | vaginal swab | E. coli | tsh, mchF, hemL, intl1, iroN, iss, fimH, iucD |
| MDR63 | SRB | dog | urin | E. coli | senB, hemL, intl1, iss, iucD |
| 193 | AUT | dog | bladder | E. coli | mchF, hemL, intl1, iroN, iss |
| 2304 | AUT | dog | urine | E. coli | lpfA, mchF, hemL, iroN, iss, fimH |
| 3056 | AUT | dog | urine | E. coli | iha, prfB, cnf1, sat, iss. fimH, iucD, papC |
| 3168 | AUT | dog | urine | E. coli | hemL, iss, fimH, iucD |
| 2016 | AUT | dog | urine | E. coli | lpfA, cma, hemL, iroN, iss, fimH |
| 2058 | AUT | dog | urine | E. coli | prfB, mchF, iroN, iss, fimH, iucD, papC |
| 4125 | AUT | dog | urine | E. coli | lpfA, mchF, hemL, intl1, iroN, iss, fimH, iucD |
| 1286 | AUT | cat | urine | E. coli | prfB, hemL, iss, fimH, papC |
| 2269 | AUT | dog | ejaculate | E. coli | hemL, fimH, iucD |
| 2736 | AUT | dog | urin | E. coli | lpfA, hemL, fimH |
| 3441 | AUT | dog | urin | E. coli | tsh, cba, cma, mchF, hemL, intl1, iron, iss, fimH, iucD |
| 3448 | AUT | cat | urin | E. coli | tsh, cba, cma, mchF, hemL, intl1, iron, iss, fimH, iucD |
| 3534 | AUT | dog | urin | E. coli | prfB, mcmA, hemL, intl2, ireA, fimH, iucD |
| 482 | AUT | dog | vagina | E. coli | hemL, fimH |
| 4 | AUT | dog | urine | E. coli | hemL, iss, fimH |
| 855 | AUT | dog | vaginal swab | E. coli | hemL, intl1, iroN, iss, iucD |
| 555 | AUT | dog | prostata cyst | E. coli | iha, nfaE, prfB, sat, senB, intl1, iss, fimh, iucD, papC |
| 260 | AUT | dog | vaginal swab | E. coli | hemL, iss, fimH, iucD |
| 655 | AUT | dog | urine | E. coli | lpfA, hemL, iss, fimH, iucD |
| 728 | AUT | dog | vaginal swab | E. coli | |
| 2189 | AUT | cat | urin | E. coli | mchF, hemL, iroN, iss |
| 3512 | AUT | dog | vaginal swab | E. coli | lpfA, hemL, intl1, iss, fimH, iucD |
| 2srb | SRB | dog | urin | E. coli | lpfA, cma, hemL, intl1, iroN, iss, fimH, iucD |
| 1srb | SRB | dog | urin | E. coli | iha, prfB, astA, sat, senB, hemL, fimH, papC |
| ESBL1 | SRB | cat | vaginal swab | E. coli | iucD |
| ESBL41 | SRB | dog | vaginal swab | E. coli | fimH, iucD |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loncaric, I.; Misic, D.; Szostak, M.P.; Künzel, F.; Schäfer-Somi, S.; Spergser, J. Broad-Spectrum Cephalosporin-Resistant and/or Fluoroquinolone-Resistant Enterobacterales Associated with Canine and Feline Urogenital Infections. Antibiotics 2020, 9, 387. https://doi.org/10.3390/antibiotics9070387
Loncaric I, Misic D, Szostak MP, Künzel F, Schäfer-Somi S, Spergser J. Broad-Spectrum Cephalosporin-Resistant and/or Fluoroquinolone-Resistant Enterobacterales Associated with Canine and Feline Urogenital Infections. Antibiotics. 2020; 9(7):387. https://doi.org/10.3390/antibiotics9070387
Chicago/Turabian StyleLoncaric, Igor, Dusan Misic, Michael P. Szostak, Frank Künzel, Sabine Schäfer-Somi, and Joachim Spergser. 2020. "Broad-Spectrum Cephalosporin-Resistant and/or Fluoroquinolone-Resistant Enterobacterales Associated with Canine and Feline Urogenital Infections" Antibiotics 9, no. 7: 387. https://doi.org/10.3390/antibiotics9070387
APA StyleLoncaric, I., Misic, D., Szostak, M. P., Künzel, F., Schäfer-Somi, S., & Spergser, J. (2020). Broad-Spectrum Cephalosporin-Resistant and/or Fluoroquinolone-Resistant Enterobacterales Associated with Canine and Feline Urogenital Infections. Antibiotics, 9(7), 387. https://doi.org/10.3390/antibiotics9070387






